Abstract
The success of a rhizobial inoculant in the soil depends to a large extent on its capacity to compete against indigenous strains. M403, a Sinorhizobium meliloti strain with enhanced competitiveness for nodule occupancy, was recently constructed by introducing a plasmid containing an extra copy of a modified putA (proline dehydrogenase) gene. This strain and M401, a control strain carrying the same plasmid without the modified gene, were used as soil inoculants for alfalfa in a contained field release experiment at León, Spain. In this study, we determined the effects of these two strains on the indigenous microbial community. 16S rRNA genes were obtained from the rhizosphere of alfalfa inoculated with strain M403 or strain M401 or from noninoculated plants by amplification of DNA from soil with bacterial group-specific primers. These genes were analyzed and compared by restriction fragment length polymorphism and temperature gradient gel electrophoresis. The results allowed us to differentiate between alterations in the microbial community apparently caused by inoculation and by the rhizosphere effect and seasonal fluctuations induced by the alfalfa plants and by the environment. Only moderate inoculation-dependent effects could be detected, while the alfalfa plants appeared to have a much stronger influence on the microbial community.
Inoculation of legume crops with nitrogen-fixing soil bacteria, collectively known as rhizobia, has been used widely to improve legume productivity in the field. However, the introduced inoculants often fail to become established in soils with indigenous rhizobial populations (8, 45, 46, 48). In such soils, the better-adapted indigenous populations tend to outcompete the inoculants for nodule formation and saprophytic survival. Thus, for an inoculant to be effective, the strains used must not only fix nitrogen efficiently but also be highly competitive. Genetic engineering can be used to improve the competitiveness of rhizobial inoculants. Indeed, highly competitive strains of Rhizobium etli have been produced by introducing trifolitoxin-encoding genes into this bacterium (33). However, before the manipulation of genes conferring increased competitiveness can be considered for construction of an ideal rhizobial inoculant, the ecological risks associated with their use should be evaluated by determining their effects on the indigenous microbial community.
Proline dehydrogenase converts proline to glutamate and has been implicated in the nodulation competitiveness and colonization ability of Sinorhizobium meliloti (18, 19). Therefore, to improve the competitiveness of an S. meliloti strain, we constructed a new strain, M403, by introducing multicopy plasmids containing a highly expressed proline dehydrogenase gene (putA) (49). We investigated the effects of this genetic modification on nodulation competitiveness in the field by inoculating pots buried in an agricultural field at Riego de la Vega, León, Spain, with this strain and a control strain, M401, which harbors the same multicopy plasmids without the modified putA gene. We found that the strain containing the modified putA gene occupied a higher proportion of nodules than the control strain after 1 month (49). Data concerning the persistence of M403 in soil and the plasmid stability in this strain have shown that the modified putA gene tends to disappear with time. This suggests that the probability of gene transfer to natural populations is low. However, a rhizobial strain with a putative metabolic advantage, such as M403, may displace unrelated indigenous proline-dependent microbial populations from the rhizosphere.
In most field release studies to date, nonrhizobial genetically modified microorganisms were shown to cause only slight, transient perturbations in the microbial communities indigenous to the rhizospheres of various crops (3, 6, 28, 29). Similarly, lysimeter experiments (field experiments with polyvinyl chloride cylinders containing soil columns) with rhizobial luciferase gene-tagged S. meliloti strain L33 and a RecA− derivative, L1, showed that these inoculants did not affect the indigenous microbial community (39). However, Schwieger and Tebbe (40) reported that release of L33 into the field reduced the number of types of γ-proteobacteria and resulted in an increase in the number of members of the α-proteobacteria in the rhizosphere of alfalfa.
In most cases, culture-based methods, such as plating and most-probable-number techniques, have been used to determine the effects of genetically modified microorganisms in the soil following their release into the environment. However, these methods are time-consuming and indirect, and the results often reflect information for only a small proportion of the total microbial community (38). This has led to an increase in the use of culture-independent techniques based on amplification of conserved genes, such as small-subunit rRNA genes, from bacterial DNA or RNA isolated directly from the soil (11). Several techniques have been developed to distinguish between amplicons with slightly different sequences by size, single-strand conformation, or specific melting behavior. These techniques include restriction fragment length polymorphism (RFLP) (32), terminal RFLP (22), single-strand conformation polymorphism (5, 38), and temperature gradient gel electrophoresis (TGGE)-denaturing gradient gel electrophoresis (26).
The aim of this work was to investigate the effects of M401 and the modified putA gene in M403 on the soil rhizosphere bacterial communities. To detect alterations in the microbial communities, we used a cultivation-independent approach by amplifying 16S ribosomal DNA (rDNA) genes directly from soil DNA during the course of the experiment. To increase the breadth and resolution of the data, primers with various degrees of specificity were used together with RFLP and TGGE.
MATERIALS AND METHODS
Bacterial strains, field site, and inoculation.
A description of the bacterial strains, field site, and inoculation technique has been presented elsewhere (49). Briefly, a field site at Riego de la Vega in northern Spain was partitioned into nine plots, each containing nine partially buried pots filled with displaced soil. On 25 May 1999 three plots were sown with alfalfa (Medicago sativa L. cv. Aragón) seeds coated with the inoculant M401 (5 × 105 bacteria/seed), three plots were sown with alfalfa seeds coated with M403 (1.3 × 105 bacteria/seed), and the remaining three plots were sown with seeds coated with sterile peat, as controls. Both types of inoculant were derived from S. meliloti strain M4, which is naturally resistant to streptomycin and is tagged with the gusA gene in the nonessential region of the chromosome, between the recA and alaS genes (41). M401 harbors the multicopy plasmid pBBRHG1, which confers mercury resistance (49). M403 harbors a derivative of this plasmid, pBBRHG3, which contains the putA gene of S. meliloti preceded by the constitutive promoter of the kanamycin resistance gene. Despite the constitutive nature of this promoter, proline nonetheless induces an increase in expression of this putA gene (49). Authorization for the field experiments was obtained from the Spanish Ministry of the Environment (resolutions B/ES/98/50 and B/ES/98/52).
Sampling.
Rhizosphere soil samples (soil adhering to the roots) were obtained from one pot from each of the nine plots 60, 98, and 159 days after inoculation, on 24 July 1999, 31 August 1999, and 31 October 1999, respectively. This resulted in three replicate samples for each of the three treatments. Samples of bulk soil (soil not associated with plant roots) were also taken from each plot. Microbial DNA was extracted from the soil as described below (32). To obtain rhizosphere soil, alfalfa roots were washed in 50 ml of sterile phosphate-buffered saline (140 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM NaH2PO4) for 20 min. The resulting soil suspension was then centrifuged for 5 min at 3,000 × g at 4°C. DNA was extracted from the pellet, which corresponded to the rhizosphere soil.
DNA extraction.
Total DNA was extracted from the soil by the protocol described by Porteous et al. (32). Briefly, 500 mg of soil was homogenized in sodium dodecyl sulfate lysis buffer containing guanidine isothiocyanate by ultrasonication. The bacteria were lysed by incubation at 68°C, and the DNA was precipitated with potassium acetate, polyethylene glycol 8000, and a glycogen carrier. The resulting crude DNA extract was further purified by dissolution in hexadecyltrimethylammonium bromide buffer, followed by sonication, incubation at 68°C, and extraction with chloroform. The DNA was precipitated with isopropanol, collected by centrifugation, dissolved in 2.5 M ammonium acetate, and precipitated again with ethanol. Finally, the DNA was dissolved in 1× Tris-acetate-EDTA (TAE) buffer (37) so that it could be further purified and concentrated with a Microcon-100 microconcentrator (Amicon, Beverly, Mass.). Typically, about 10 ng of high-quality soil DNA μl−1 was isolated by this protocol.
Primers and PCR conditions.
All PCRs were performed with a Robocycler 40 apparatus (Stratagene). For RFLP analysis, PCR was used to amplify 16S rRNA genes from total DNA extracted from soil with universal primers F536 and R1406 or primers F67 and R685 specific for β- and γ-proteobacteria (Table 1). On the other hand, for TGGE analysis 16S rRNA genes from total DNA extracted from soil were amplified by using the nested PCR approach. In the first PCR, specific primers (Table 1), such as primers F203α and L to amplify the 16S rRNA genes of α-proteobacteria, primers F948β and L to amplify the 16S rRNA genes of β-proteobacteria, primers F243 and R1378 to amplify the 16S rRNA genes of bacteria with high G+C contents (HGC primers), and primers F984 and R1378 to amplify the 16S rRNA genes of most eubacteria, were used. The second PCR was performed with universal primer R1378 in combination with primer F984GC (Table 1), which consisted of primer F984 attached to a GC clamp and was designed to prevent complete melting of the amplification product during separation in the denaturing gradient (27).
TABLE 1.
Primers used in PCR experiments
| Primer | Positiona | Sequence 5′→3′ | Annealing temp (°C) | Refer- ence(s) |
|---|---|---|---|---|
| F536 | 519-536 | CAGCAGCCGCGGTAATAC | 62 | 32 |
| R1406 | 1406-1389 | ACGGGCGGTGTGTACAAG | 62 | 32 |
| F67 | 50-67 | AACACATGCAAGTCGAAC | 55 | 32 |
| R685 | 685-666 | TCTACGCATTTCAC(CT)GCTAC | 55 | 32 |
| F984 | 968-984 | AACGCGAAGAACCTTAC | 53 | 15, 17 |
| F984GC | 968-984 | CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGAACGCGAAGAACCTTAC | 53 | 15 |
| R1378 | 1401-1378 | CGGTGTGTACAAGGCCCGGGAACG | 53, 63 | 15, 17 |
| F243 | 226-243 | GGATGAGCCCGCGGCCTA | 63 | 15 |
| F203α | 175-203 | CCGCATACGCCCTACGGGGGAAAGATTTAT | 56 | 13 |
| F948β | 931-948 | CGCACAAGCGGTGGATGA | 64 | 13 |
| L | 1514-1494 | CTACGG(AG)TACCTTGTTACGAC | 56, 64 | K. Smalla |
Escherichia coli numbering (4).
The PCR mixtures used for RFLP analysis (final volume, 30 μl) contained 50 to 100 ng of template DNA, each deoxyribonucleoside triphosphate (dNTP) at a concentration of 0.25 mM, 10× reaction buffer, 3.75 mM MgCl2, 2 U of AmpliTaq DNA polymerase (Stoffel fragment; Perkin-Elmer), 2.5 μg of T4 gene 32 protein (Boehringer), and each primer at a concentration of 0.25 μM. The PCR program for universal primers F536 and R1406 consisted of heating the mixture to 94°C for 5 min, followed by 25 cycles of 1 min of denaturation at 95°C, 1 min of annealing at 62°C, and 2 min of extension at 72°C. The reaction was terminated with a final extension step consisting of 72°C for 5 min. The PCR program used for β- and γ-proteobacterial primers (F67 and R685) consisted of heating the mixture at 94°C for 5 min, followed by 20 cycles of 1 min of denaturation at 95°C, 1 min of annealing at 55°C, and 2 min of extension at 72°C and then a final extension step of 72°C for 5 min.
The first amplification mixture for the nested PCR for TGGE analysis (final volume, 25 μl) contained 1 μl of total DNA as the template, 2.5 μl of 10× reaction buffer, MgCl2 at a concentration of 2.5 mM (for β-proteobacterial and HGC primers) or 3.75 mM (for α-proteobacterial and universal primers), each dNTP at a concentration of 0.2 mM, 2.5 μg (for β-proteobacterial primers) or 5 μg (for α-proteobacterial, HGC, and universal primers) of bovine serum albumin to prevent inhibition of the reaction by impurities (36), 1 to 2.5 U of AmpliTaq DNA polymerase, and each primer at a concentration of 0.2 μM. The mixture was heated at 94°C for 5 min. It was then subjected to 28 cycles of 1 min of denaturation at 94°C, 1 min of annealing at the specific annealing temperature of each primer set (Table 1), and 2 min of extension at 72°C. The reaction ended with a final extension step of 72°C for 10 min.
TABLE 2.
Bands isolated from TGGE gels
| Band | Length (bp) | Closest database reference (GenBank accession no.) | % Simi- larity |
|---|---|---|---|
| 1 | 393 | Pseudomonas mendocina ATCC 25411 (AF094734) | 100 |
| Pseudomonas aeruginosa ATCC 33350 (AF094720) | 100 | ||
| 2 | 394 | Pseudomonas corrugata clone B5 (AF060534) | 100 |
| Pseudomonas brassicacearum CFBP 11706 (AF100321) | 100 | ||
| Pseudomonas sp. strain 3-1 (AF126101) | 100 | ||
| 3 | 394 | Rahnella aquatilis ATCC 33989 (X79939) | 100 |
| 4 | 400 | Streptomyces pseudovenezuelae ISP 5212(AJ399481) | 99.5 |
| 5a | 413 | Kluyvera georgiana ATCC 51603 (AF047186) | 93.8b |
| 6 | 389 | Unidentified activated sludge bacterial clone T83 (Z93969) | 98.4c |
| 7 | 389 | β-Proteobacterium B7 (AF035053) | 93.3d |
| 8 | 389 | Burkholderia graminis AUS35 (U96941) | 97.2 |
| 9 | 394 | Enterobacter amnigenus isolate Q3-25-6 (AF060977) | 99.7 |
Possible heteroduplex molecule.
Value for 241 bp.
Value for 383 bp.
Value for 363 bp.
The second amplification of the nested PCR analysis was performed by using 1 μl of the products of the first reaction as the template. The reaction mixture (final volume, 25 μl) contained 2.5 μl of 10× reaction buffer, 3.75 mM MgCl2, each dNTP at a concentration of 0.2 mM, 4% acetamide, 2 U of AmpliTaq DNA polymerase, 0.2 μM GC clamp primer F984GC, and 0.2 μM primer R1378. The PCR was performed by using an annealing temperature of 53°C and the same cycles that were used for the first amplification.
RFLP.
The RLFP analysis was performed as described by Porteous et al. (32). Briefly, DNA amplified with universal primers or primers specific for β-and γ-proteobacteria was cleaned by precipitation with an equal volume of a mixture containing 20% polyethylene glycol 8000 and 2.5 M NaCl. After incubation at 37°C for 15 min, the DNA was collected by centrifugation at 12,000 × g for 5 min. The precipitate was then washed with 200 μl of 80% ethanol (−10°C) and dried. The DNA was digested with the restriction enzyme HaeIII. The digestion procedure consisted of resuspending the DNA pellet in a 15-μl mixture containing 0.3 μl (3 U) of HaeIII, 1.5 μl of buffer, and 13.2 μl of water. The reaction mixture was incubated at 37°C for 1 h, and samples were electrophoresed at 200 V on an 8% polyacrylamide gel (8 ml of a 40% acrylamide-bisacrylamide solution [29:1], 4 ml of 10× TBE buffer [37], 500 μl of a 10% ammonium persulfate solution, 40 μl of N,N,N′,N′-tetramethylethylenediamine, and enough water to bring the final volume to 40 ml).
TGGE.
We used the Maxi TGGE (Biometra) system according to the manufacturer's instructions. The polyacrylamide gels used consisted of 0.17% (vol/vol) N,N,N′,N′-tetramethylethylenediamine, 0.03% ammonium persulfate, 5% acrylamide-bisacrylamide (29:1), 1× TAE buffer (37), 2% glycerol, 8 M urea, and 20% formamide deionized with AG501-X8 mixed-bed resin (Bio-Rad). Aliquots of the final PCR products (4 to 5 μl) for each of the nine samples at each time point were loaded onto the gel and electrophoresed horizontally in 1× TAE buffer for 16 h at a constant voltage of 130 V and with a temperature gradient of 45 to 60°C. Gels were stained as described by Heuer et al. (15). Selected bands were excised from SYBR Green I-stained gels, and the corresponding DNA was recovered by crushing the gel fragment containing a band in 100 μl of water and incubating the mixture overnight at 4°C. The mixture was centrifuged (13,000 × g for 15 min), and the supernatant was concentrated in a SpeedVac to 3 to 4 μl. We used 1 μl of the concentrate for reamplification with universal primers under the appropriate PCR conditions and inserted the amplified DNA product into the pGEM-T Easy vector (Promega) by following the manufacturer's instructions. The resulting clones were sequenced with primers specific for the pGEM-T vector.
RESULTS
Inoculation-independent variations in the microbial community.
Variations occurring within the soil microbial communities in the absence of the inoculant strains were analyzed by studying the reproducible patterns generated by RFLP analysis (Fig. 1) or TGGE (Fig. 2) performed with noninoculated samples throughout the experiment. Two types of inoculant-independent changes could be distinguished. The first and most obvious changes could be observed in both RFLP and TGGE gels regardless of the primer sets used and involved differences between bulk soil samples taken before inoculation and rhizosphere samples taken after 60 days. These changes consisted of new bands that appeared and other bands that disappeared, suggesting that a selection process took place.
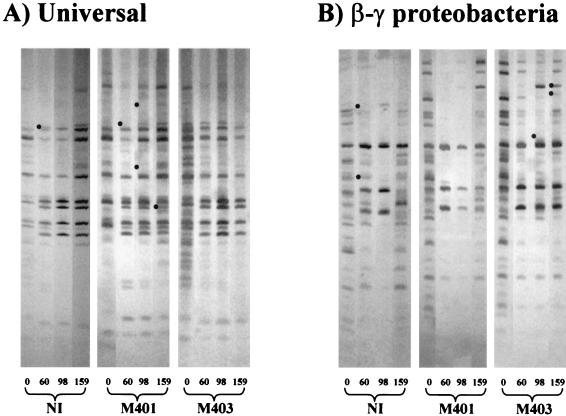
FIG. 1.
RFLP gels obtained with DNA from rhizosphere soil amplified with universal primers (A) or primers specific for β- and γ-proteobacteria (B). Each gel shows the pattern obtained at each time point for a representative sample from the three replicate plots subjected to each inoculation treatment. The dots indicate bands present or absent in some of the replicate samples. These nonreproducible bands were not considered when we determined inoculation-dependent and -independent changes. The numbers below the gels indicate the numbers of days after inoculation. NI, noninoculated; M401, inoculated with M401; M403, inoculated with M403.
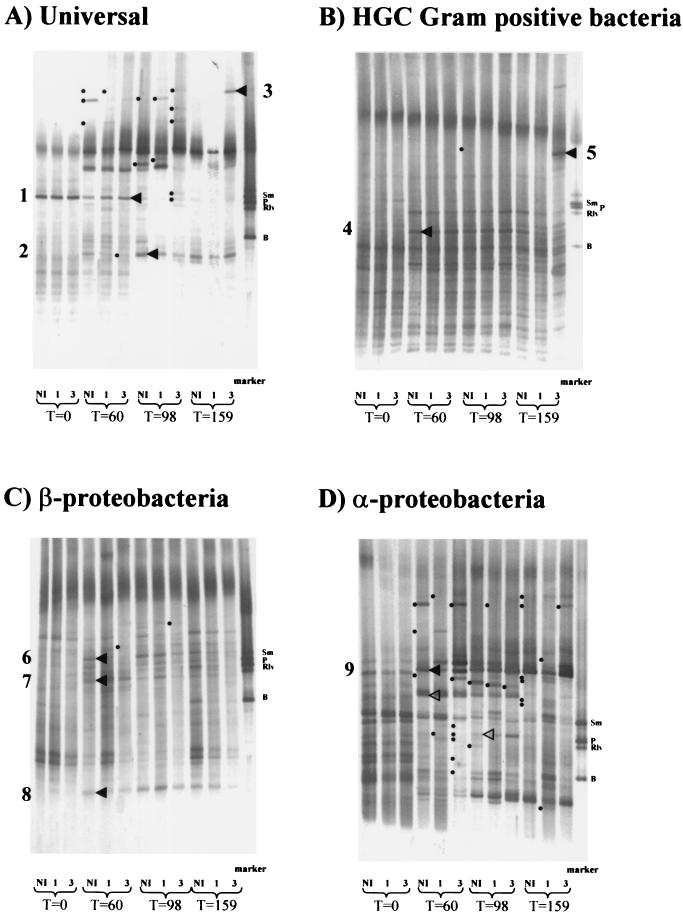
FIG. 2.
TGGE gels obtained with DNA from rhizosphere soil amplified with universal primers (A) or primers specific for gram-positive bacteria with high G+C DNA contents (HGC primers) (B), β-proteobacteria (C), or α-proteobacteria (D). Each gel shows the pattern obtained at each time point for a representative sample from the three replicate plots subjected to each inoculation treatment. The dots indicate bands present or absent in some of the replicate samples. These nonreproducible bands were not considered when we determined inoculation-dependent and -independent changes. The arrowheads indicate bands that have been sequenced. The open arrowheads in panel D indicate examples of temporally affected populations. T=0, T=60, T=98, and T=159, 0, 60, 98, and 159 days after inoculation; NI, noninoculated; 1, inoculated with M401; 3, inoculated with M403. The marker consisted of amplified pure cultures of S. meliloti GR4 (Sm), Pseudomonas sp. strain M2L32 (P), Rhizobium leguminosarum bv. viciae GRB11 (Rlv), and Bacillus sp. strain 1R18.2 (B).
The second type of alterations involved seasonal fluctuations within rhizosphere bacterial populations from 60 days onward, but the changes were evident only when certain primer set-fingerprint combinations were used. For instance, the RFLP patterns obtained from the rhizosphere of noninoculated plants with universal primers did not change significantly from 60 days onward, but the patterns obtained with β- and γ-proteobacterial primers at 159 days clearly differed from those obtained with the same primers at 60 and 98 days (Fig. 1). Similarly, the TGGE pattern obtained with universal primers showed that there was a seasonal shift in a prominent band (Fig. 2A, band 1), which was present in bulk soil and at 60 days but was much less prominent at 98 days. On the other hand, in the same gel a band which was less prominent at 60 days was much more prominent at 98 days (Fig. 2A, band 2) and remained prominent until the end of the experiment. Seasonal fluctuations could also be observed in α-proteobacterial populations when nonreproducible bands were ignored. For example, a band which was present since 60 days apparently disappeared at 159 days (Fig. 2D). Meanwhile, another population amplified with the same primers was more prominent at 98 days (Fig. 2D) but was much less prominent at 159 days. On the other hand, seasonal fluctuations could not clearly be distinguished in samples from 60 days onward by using the TGGE patterns obtained with the HGC and β-proteobacterium-specific primers (Fig. 2B and C, respectively).
Changes in the microbial community caused by environmental release of strains M401 and M403.
Regardless of the primers or fingerprint technique used, all patterns obtained with bulk soil before inoculation were very similar, indicating that the soil populations were homogeneous in all plots before the experiment commenced. The amounts of M401 and M403 released were 3 orders of magnitude greater than the size of the indigenous S. meliloti population (49). However, bands corresponding to S. meliloti could not clearly be defined in RFLP (data not shown) or TGGE (Fig. 2) gels. It has been suggested that only populations that account for more than 0.1% of the total microbial population can be detected by TGGE (16, 24); therefore, it appears that in our experiment, even in inoculated plots, S. meliloti was not a dominant member of the microbial community.
To determine if inoculation with increased amounts of S. meliloti caused variations in the bacterial communities, we compared the results obtained for M401-inoculated plots with those obtained for noninoculated plots. The RFLP profiles produced with universal primers did not differ (Fig. 1A). However, the RFLP patterns obtained with β- and γ-proteobacterial primers showed that the seasonal change observed in noninoculated plots at 159 days did not occur in plots inoculated with M401. This change also did not occur in M403-inoculated plots (Fig. 1B). On the other hand, the results obtained with TGGE gels did not reveal any differences between M401-inoculated plots and noninoculated plots (Fig. 2), even when β-proteobacterium-specific primers were used. Therefore, the results suggest that inoculation with S. meliloti permitted certain γ-proteobacterial populations to be maintained longer in the rhizosphere of alfalfa and that other populations remained unaffected.
Finally, to determine the effect of the modified putA gene on indigenous microbial communities, we compared the patterns obtained at each sampling time for plots inoculated with M403 to the patterns obtained for either noninoculated or M401-inoculated plots. Neither the patterns obtained by RFLP analysis with universal or β- and γ-proteobacterial primers nor the patterns obtained by TGGE analysis with β-proteobacterial or α-proteobacterial primers revealed differences between M403-inoculated plots and the noninoculated or M401-inoculated plots. Nevertheless, after 159 days, an M403-specific band could be discerned in the TGGE gel produced with the universal primers (Fig. 2A, band 3), and another band could be discerned in the TGGE gel produced with the HGC primers (Fig. 2B, band 5). Neither of these bands was found in the M401-inoculated or noninoculated rhizosphere.
Analysis of the sequences of DNA isolated from TGGE bands.
Some of the bands from TGGE gels, but not the bands from RFLP gels, could be excised and sequenced to obtain phylogenetic information. As a result, we analyzed five bands from the rhizosphere of alfalfa which appeared at 60 days but were not present in bulk soil samples, two bands representing populations showing seasonal dominance in the rhizosphere of alfalfa, and the two bands which appeared only in M403-inoculated samples.
Populations represented by bands 4, 6, 7, 8, and 9 appeared to be enriched in the rhizosphere of alfalfa. The DNA of band 4 displayed sequence similarity to DNA from Streptomyces pseudovenezuelae, the DNA of band 6 displayed sequence similarity to DNA from the unidentified sludge bacterium clone T83, and the DNA of band 7 displayed sequence similarity to DNA from the β-proteobacterial strain B7 (Table 2). Curiously, the DNA sequences corresponding to bands 6 and 7 were also very similar to each other (data not shown). The sequence obtained from band 8 displayed similarity to Burkholderia graminis DNA, while the sequence obtained from band 9 displayed sequence similarity to Enterobacter amnigenus DNA (Table 2).
The populations represented by bands 1 and 2 appeared to be seasonally affected as the plant developed. The DNA sequence of band 1, a band that became less prominent with time, displayed equally high levels of similarity to sequences from Pseudomonas mendocina and Pseudomonas aeruginosa (Table 2). On the other hand, the DNA sequence of band 2, a band which appeared as the plant developed, displayed equal levels of similarity to sequences from the soil bacteria Pseudomonas sp. strain 3-1, Pseudomonas brassicacearum, and Pseudomonas corrugata (Table 2).
The populations represented by bands 3 and 5 (Fig. 2A and B) appeared to be specifically correlated to inoculation with M403, which harbors the modified putA gene. The sequence of the DNA corresponding to band 3 was very similar to a sequence from Rahnella aquatilis (Table 2). In contrast, the DNA from band 5 appeared to be a heteroduplex molecule (12, 51) consisting of a sequence not listed in databases fused to a sequence very similar to a sequence from Kluyvera georgiana (Table 2).
DISCUSSION
In this work, we studied the effect of contained environmental release of the genetically modified S. meliloti strains M401 and M403 on the indigenous microbial community in the rhizosphere of alfalfa. M403, which harbors a plasmid containing an inducible putA gene with high basal levels of expression, was previously shown to transiently increase nodule occupancy under field conditions compared to the nodule occupancy observed with M401, which lacks the modified gene (49). The indigenous microbial populations, which could have been affected, were evaluated during the experiment by comparing RFLP and TGGE molecular fingerprints of 16S rDNA genes directly amplified from soil DNA with primers with various degrees of specificity. On the whole, both fingerprint techniques produced highly reproducible results, although there were some exceptions for TGGE performed with α-proteobacterium-specific primers. However, since at each time point the same DNA was used with other primer sets, which gave highly reproducible patterns, it is unlikely that any lack of reproducibility was caused by problems associated with the techniques used. Instead, the variations were probably due to plot- or pot-specific factors to which α-proteobacterial populations were especially sensitive. Therefore, nonreproducible bands were not taken into account, which allowed us to detect inoculation-dependent and -independent changes in the structure of the microbial communities.
Inoculation-independent effects.
Inoculation-independent factors which could affect microbial community structure include soil type, crop species, environmental factors, and anthropogenic effects, such as those caused by farming practices. However, in our experiment these factors were similar for all treatments. The quality and quantity of root exudates produced by alfalfa depend on its age and stage of development (14). As root exudates are an important source of nutrition for many rhizosphere microorganisms, changes in their composition may affect the patterns and activities of rhizobacterial populations. In fact, alfalfa has been shown to increase the concentrations of amino acids and sugars, CO2 respiration, and enzyme activities in the soil (2). If possible microenvironmental variations are ignored, the inoculation-independent changes observed in our experiment could, therefore, have been due to a large degree to the growing alfalfa plants.
Fluctuations in the rhizosphere microbial populations associated with different plants have been studied by several authors (9, 10, 13, 21, 30, 40, 43). These authors observed that some plants enrich certain populations from the surrounding soil in what is known as the rhizosphere effect. For instance, development of the maize plant specifically affected populations of Burkholderia cepacia (7), Paenibacillus azotofixans (42), and Pseudomonas (31). In addition to maize, a generalized rhizosphere effect has been detected for strawberry, potato, and oilseed rape (13, 43). Additionally, seasonal fluctuations were often observed in the rhizosphere microbial communities established with these plants. Curiously, neither the rhizosphere effect nor seasonal fluctuations were observed in chrysanthemum or barley rhizosphere communities (9, 10, 30). Therefore, different plants appear to have different effects on the indigenous microbial populations.
Recently, the bulk soil and rhizosphere microbial communities associated with alfalfa also have been studied (52). The authors, who used TGGE with universal primers, detected minor seasonal shifts in the rhizosphere but did not detect systematic community shifts in bulk soil. This suggests that alfalfa also can have a rhizosphere effect. In our experiment we could discern differences between the bulk soil community before inoculation and the rhizosphere community 60 days later. If we assume that the bulk soil microbial community remained unchanged, our data likewise suggest that alfalfa enriches certain populations in the rhizosphere.
In this work, detection of seasonal fluctuations in alfalfa rhizosphere populations depended on the type of primer set and fingerprint technique used. For instance, seasonal changes were observed in RFLP patterns only when primers specific for β- and/or γ-proteobacteria were used. Seasonal changes were also detected by TGGE performed with universal and α-proteobacterial primers but not with β-proteobacterial or HGC primers. This suggests that only the RFLP patterns of the γ-proteobacterial populations had been affected. Our results are similar to those obtained for the maize rhizosphere (13), although with the latter crop greater variation was observed in the β-proteobacterial populations. The differences in microbial communities associated with alfalfa and maize, together with the differences observed between other plants (23, 40, 43, 52), suggest again that rhizosphere bacterial community structure depends on the plant species.
The DNA sequences of bands that appeared as a result of a possible rhizosphere effect exhibited similarities with the sequences of bacteria which could be expected to be found in soil. For instance, Streptomyces is typically found in soil, while B. graminis and E. amnigenus have been isolated from the rhizospheres of crops such as maize, wheat (50), and potato (17). Other organisms, such as the activated sludge clone T83 and the β-proteobacterium B7, were found in activated sludge (44) and drinking water (20) and could have been introduced via irrigation. The two seasonally affected bands analyzed both showed high sequence similarity to Pseudomonas species, which have also been found in soil and aqueous environments. Here, the population represented by band 1 could be a major component of the bulk soil microbial community, as it was found with nonspecific universal primers. On the other hand, the population corresponding to band 2, which became more dominant after 98 days, was a population of Pseudomonas species, which have been found specifically in the rhizospheres of common crops (1, 31, 17).
Effect of inoculation.
In this study, only two effects could be attributed to inoculation of M401 and M403. On the one hand, the seasonal changes observed in β- and/or γ-proteobacterial populations by RFLP analysis in noninoculated plots were not seen in plots inoculated with either strain. A comparison of this observation with the data obtained with other primers, including those specific for β-proteobacteria, suggests that the persistence of certain γ-proteobacterial populations in the rhizosphere of alfalfa was extended by inoculation with M401 or M403 or, more likely, by the increased quantity of S. meliloti introduced into the soil upon inoculation. On the other hand, TGGE data show that inoculation with M403, but not inoculation with M401, resulted in the appearance of two extra components in the microbial populations detected with the universal and HGC primers. One of these two components appeared to form part of a heteroduplex molecule. This indicates that the presence of the modified putA gene has only a modest effect on indigenous microbial populations, although the physiological significance of the two affected populations remains unknown. Previously, van Dillewijn et al. (49) reported that the concentrations of M401 and M403 decreased from the 105 cells of each strain per g of seeds originally released to below the detection levels (<102 CFU g of soil−1) in bulk soil within 98 days, while in the rhizosphere the concentration of M401 decreased to 104 CFU g of soil−1 and the concentration of M403 decreased to 102 CFU g of soil−1 at 159 days. Therefore, the apparently modest effect of the inoculants on the indigenous microbial communities could be due to their low levels of persistence.
Our results are comparable to those obtained after the release of other rhizobial strains. L33 is an S. meliloti strain tagged with the luciferase gene (41). When this strain was released in the field, single-strand conformation polymorphism profiles of a 16S rRNA gene region and amplified ribosomal DNA restriction analysis of isolated bacteria suggested that it caused a reduction in the number of types of γ-proteobacteria and an increase in the number of members of α-proteobacteria in the rhizosphere of alfalfa (40). Similar to field release of M403, field release of an R. etli strain containing genes encoding trifolitoxin resulted in increased nodulation competitiveness under agricultural conditions (34). However, trifolitoxin kills many α-proteobacteria (47). As a result, ribosomal intergenic spacer analysis indicated that the inoculant caused a dramatic reduction in the diversity of indigenous α-proteobacteria but did not affect other microbial populations (35). In this study, TGGE analysis showed that α-proteobacterial populations, although apparently sensitive to plot- or pot-specific factors, do not appear to be affected by inoculation with M401 or M403. On the other hand, inoculation with greater quantities of S. meliloti could have affected the persistence of certain γ-proteobacterial populations in the rhizosphere of alfalfa, although the lack of effective γ-proteobacterial primers did not allow us to study this phenomenon in more detail. Moreover, the presence of the modified putA gene in M403 appeared to affect only two populations, which were detected with primers which had not been used in the other studies.
Concluding remarks.
Previously, M403 containing a modified putA gene was shown to be more competitive than the control strain M401 with the local alfalfa-nodulating bacteria for nodule formation. Here, we present results which permit an in-depth view of the interactions (i) between alfalfa and the indigenous microbial community, (ii) between S. meliloti inoculation and the indigenous microbial community, and (iii) M403 inoculation or increased competitiveness bestowed by the modified putA gene and the indigenous microbial community. The data suggest that the alfalfa plant has a greater influence on the microbial populations in the rhizosphere than the inoculants. Inoculation with S. meliloti (M401 and M403) in soils with small indigenous S. meliloti populations appeared to affect some populations, but the effect of the modified putA gene in M403 seemed to be limited to only two populations. Therefore, in this case increased competitiveness only moderately affected microbial communities.
When the results presented in this paper are interpreted, limitations of the techniques should be taken into account. DNA isolation from soil (32), PCR amplification (51), and RFLP and TGGE analyses of 16S rDNA genes (16, 25) each have biases which could affect the outcome of the patterns presented. However, our results indicate that using primers with different bacterial group specificities and different fingerprint techniques effectively increased the resolution. In this way, RFLP provided better resolution for demonstrating the effect of general S. meliloti inoculation (inoculation of both M401 and M403), while TGGE proved to be better for identifying specific M403-dependent changes. Moreover, TGGE permitted phylogenetic information to be obtained from isolated bands (26). Nevertheless, it is possible that underrepresented but ecologically important groups could have been affected by inoculation but were not detected by the techniques used in this work or that the effects were overshadowed by the effects of the host plant. Thus, although the ecological risks associated with the presence of M403 or control strain M401 appear to be low, we still could debate whether the few effects of the engineered strains on the rhizosphere population were significant. Nevertheless, our results suggest that at least in principle the higher nodulation competitiveness and small ecological effects associated with the modified putA gene could be combined with traits such as increased nitrogen fixation to obtain safe and highly efficient inoculants.
Acknowledgments
This work was supported by Comisión Asesora de Investigación Científica y Técnica grant BIO96-0397 and by European Union grant BIO4-CT98-0483.
We thank María Isabel López-Díaz, Santiago Martínez-Doral, and Encarna Velázquez for valuable assistance with the field experiments. We also thank Kornelia Smalla for assistance with TGGE and for providing primers and Frederique Ampe for advice regarding DNA isolation from gels and subsequent amplification procedures.
REFERENCES
- 1.Achouak, W., L. Sutra, T. Heulin, J. M. Meyer, N. Fromin, S. Degraeve, R. Christen, and L. Gardan. 2000. Pseudomonas brassicacearum sp. nov. and Pseudomonas thivervalensis sp. nov., two root-associated bacteria isolated from Brassica napus and Arabidopsis thaliana. Int. J. Syst. E vol. Microbiol. 50:9-18. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann, G., and H. Kinzel. 1992. Physiological and ecological aspects of the interactions between plant roots and rhizosphere soil. Soil Biol. Biochem. 24:543-552. [Google Scholar]
- 3.Bolton, H., Jr., J. K. Fredrickson, J. M. Thomas, S. W. Li, D. J. Workman, S. A. Bentjen, and J. L. Smith. 1991. Field calibration of soil-core microcosms: ecosystem structural and functional comparisons. Microb. Ecol. 21:175-189. [DOI] [PubMed] [Google Scholar]
- 4.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 5.Chen, X., T. Baumstark, G. Steger, and D. Riesner. 1995. High resolution SSCP by optimization of the temperature by transverse TGGE. Nucleic Acids Res. 23:4524-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Leij, F. A. A. M., E. J. Sutton, J. M. Whipps, J. S. Fenlon, and J. M. Lynch. 1995. Impact of field release of genetically modified Pseudomonas fluorescens on indigenous microbial populations of wheat. Appl. Environ. Microbiol. 61:3443-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Cello, F., A. Bevivino, L. Chiarini, R. Fani, D. Paffetti, S. Tabachioni, and C. Dalmastri. 1997. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 63:4485-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowling, D. N., and W. J. Broughton. 1986. Competition for nodulation of legumes. Annu. Rev. Microbiol. 40:131-157. [DOI] [PubMed] [Google Scholar]
- 9.Duineveld, B. M., A. S. Rosado, J. D. Van Elsas, and J. A. van Veen. 1998. Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl. Environ. Microbiol. 64:4950-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duineveld, B. M., G. A. Kowalchuk, A. Keijzer, J. D. van Elsas, and J. A. van Veen. 2001. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felske, A., B. Engelen, U. Nübel, and H. Backhaus. 1996. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl. Environ. Microbiol. 62:4162-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferris, M. J., and D. M. Ward. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes, N. C. M., H. Heuer, J. Schönfeld, R. Costa, L. Mendonça-Hagler, and K. Smalla. 2001. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232:167-180. [Google Scholar]
- 14.Hamlen, R., F. Lukezic, and J. Bloom. 1972. Influence of age and stage of development on the neutral carbohydrate components in root exudates from alfalfa plants grown in a gnotobiotic environment. Can. J. Plant Sci. 52:633-642. [Google Scholar]
- 15.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heuer, H., and K. Smalla. 1997. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities, p. 353-373. In J. D. Van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker Inc., New York, N.Y.
- 17.Heuer, H., K. Hartung, G. Wieland, I. Kramer, and K. Smalla. 1999. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl. Environ. Microbiol. 65:1045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiménez-Zurdo, J. I., P. van Dillewijn, M. J. Soto, M. R. de Felipe, J. Olivares, and N. Toro. 1995. Characterization of a Rhizobium meliloti proline dehydrogenase mutant altered in nodulation efficiency and competitiveness on alfalfa roots. Mol. Plant-Microbe Interact. 8:492-498. [DOI] [PubMed] [Google Scholar]
- 19.Jiménez-Zurdo, J. I., F. M. García-Rodríguez, and N. Toro. 1997. The Rhizobium meliloti putA gene: its role in the establishment of the symbiotic interaction with alfalfa. Mol. Microbiol. 23:85-93. [DOI] [PubMed] [Google Scholar]
- 20.Kalmbach, S., W. Manz, J. Wecke, and U. Szewzyk. 1999. Aquabacterium gen. nov., with description of Aquabacterium citratiphilum sp. nov., Aquabacterium parvum sp. nov. and Aquabacterium commune sp. nov., three in situ dominant bacterial species from the Berlin drinking water system. Int. J. Syst. Bacteriol. 49:769-777. [DOI] [PubMed] [Google Scholar]
- 21.Lottmann, J., H. Heuer, J. de Vries, A. Mahn, K. Düring, W. Wackernagel, K. Smalla, and G. Berg. 2000. Establishment of introduced antagonistic bacteria in the rhizosphere of transgenic potatoes and their effect on the bacterial community. FEMS Microbiol. Ecol. 33:41-49. [DOI] [PubMed] [Google Scholar]
- 22.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and nontransgenic potato plants. FEMS Microbiol. Ecol. 32:241-247. [DOI] [PubMed] [Google Scholar]
- 23.Miethling, R., G. Wieland, H. Backhaus, and C. C. Tebbe. 2000. Variation of microbial rhizosphere communities in response to crop species, soil origin, and inoculation with Sinorhizobium meliloti L33. Microb. Ecol. 40:43-56. [DOI] [PubMed] [Google Scholar]
- 24.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 26.Muyzer, G. 1999. DGGE/TGGE, a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 27.Myers, R. M., S. G. Fischer, L. S. Lerman, and T. Maniatis. 1985. Nearly all single base substitutions in DNA fragments joined to a GC clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res. 13:3131-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nacamulli, C., A. Bevivino, C. Dalmastri, S. Tabacchioni, and L. Chiarini. 1997. Perturbation of maize microflora following seed bacterization with Burkholderia cepacia MCI 7. FEMS Microbiol. Ecol. 23:183-193. [Google Scholar]
- 29.Natsch, A., C. Keel, N. Hebecker, E. Laasik, and G. Défago. 1997. Influence of biocontrol strain Pseudomonas fluorescens CHA0 and its antibiotic-overproducing derivative on the diversity of resident root-colonizing pseudomonads. FEMS Microbiol. Ecol. 23:341-352. [Google Scholar]
- 30.Normander, B., and J. I. Prosser. 2000. Bacterial origin and community composition in the barley phytosphere as a function of habitat and presowing conditions. Appl. Environ. Microbiol. 66:4372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picard, C., F. Di Cello, M. Ventura, R. Fani, and A. Gluckert. 2000. Frequency and biodiversity of 2,4-diacetylphloroglucinol-producing bacteria isolated from the maize rhizosphere at different stages of plant growth. Appl. Environ. Microbiol. 66:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porteous, L. A., R. J. Seidler, and L. S. Watrud. 1997. An improved method for purifying DNA from soil for polymerase chain reaction amplification and molecular ecology applications. Mol. Ecol. 6:787-791. [Google Scholar]
- 33.Robleto, E. A., A. J. Scupham, and E. W. Triplett. 1997. Trifolitoxin production in Rhizobium etli strain CE3 increases competitiveness for rhizosphere colonization and root nodulation of Phaseolus vulgaris in soil. Mol. Plant-Microbe Interact. 10:228-233. [Google Scholar]
- 34.Robleto, E. A., K. Kmiecik, E. S. Oplinger, J. Nienhaus, and E. W. Triplett. 1998. Trifolitoxin production increases nodulation competitiveness of Rhizobium etli CE3 under agricultural conditions. Appl. Environ. Microbiol. 64:2630-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robleto, E. A., J. Borneman, and E. W. Triplett. 1998. Effects of bacterial antibiotic production on rhizosphere microbial communities from a culture-independent perspective. Appl. Environ. Microbiol. 64:5020-5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romanowski, G., M. G. Lorenz, and W. Wackernagel. 1993. Use of polymerase chain reaction and electroporation of Escherichia coli to monitor the persistence of extracellular plasmid DNA introduced into natural soils. Appl. Environ. Microbiol. 59:3438-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwieger, F., T. Dammann-Kalinowski, U. Dresing, W. Selbitschka, J. C. Munch, A. Pühler, M. Keller, and C. C. Tebbe. 2000. Field lysimeter investigation with luciferase-gene (luc)-tagged Sinorhizobium meliloti strains to evaluate the ecological significance of soil inoculation and a recA-mutation. Soil Biol. Biochem. 32:859-868. [Google Scholar]
- 40.Schwieger, F., and C. C. Tebbe. 2000. Effect of field inoculation with Sinorhizobium meliloti L33 on the composition of bacterial communities in rhizospheres of a target plant (Medicago sativa) and a non-target plant (Chenopodium album)—linking of 16S rRNA gene-based single-strand conformation polymorphism community profiles to the diversity of cultivated bacteria. Appl. Environ. Microbiol. 66:3556-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selbitschka, W., U. Dresing, M. Hagen, S. Niemann, and A. Pühler. 1995. A biological containment system for Rhizobium meliloti based on the use of recombination-deficient (recA) strains. FEMS Microbiol. Ecol. 16:223-232. [Google Scholar]
- 42.Seldin, L., A. S. Rosado, D. W. da Cruz, A. Nobrega, J. D. van Elsas, and E. Paiva. 1998. Comparison of Paenibacillus azotofixans strains isolated from rhizoplane, rhizosphere, and non-root-associated soil from maize planted in two different Brazilian soils. Appl. Environ. Microbiol. 64:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and H. K. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Streeter, J. G. 1994. Failure of inoculant rhizobia to overcome the dominance of indigenous strains for nodule formation. Can. J. Microbiol. 40:513-522. [Google Scholar]
- 46.Toro, N. 1996. Nodulation competitiveness in the Rhizobium-legume symbiosis. World J. Microb. Biol. 12:157-162. [DOI] [PubMed] [Google Scholar]
- 47.Triplett, E. W., B. T. Breil, and G. A. Splitter. 1994. Expression of tfx and sensitivity to the rhizobial peptide antibiotic trifolitoxin in a taxonomically distinct group of α-proteobacteria including the animal pathogen Brucella abortus. Appl. Environ. Microbiol. 60:4163-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Triplett, E. W., and M. J. Sadowsky. 1992. Genetics of competition for nodulation of legumes. Annu. Rev. Microbiol. 46:399-428. [DOI] [PubMed] [Google Scholar]
- 49.van Dillewijn, P., M. J. Soto, P. J. Villadas, and N. Toro. 2001. Construction and environmental release of a Sinorhizobium meliloti strain more competitive for alfalfa nodulation. Appl. Environ Microbiol. 67:3860-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viallard, V., I. Poirier, B. Cournoyer, J. Haurat, S. Wiebkin, K. Ophel-Keller, and J. Balandreau. 1998. Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of [Pseudomonas] phenazinium, [Pseudomonas] pyrrocinia and [Pseudomonas] glathei as Burkholderia. Int. J. Syst. Bacteriol. 48:549-563. [DOI] [PubMed] [Google Scholar]
- 51.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 52.Wieland, G., R. Neumann, and H. Backhaus. 2001. Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl. Environ. Microbiol. 67:5849-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]