Abstract
Two filamentous fungi with different phenotypes were isolated from crushed healthy spores or perforated dead spores of the arbuscular mycorrhizal fungus (AMF) Scutellospora castanea. Based on comparative sequence analysis of 5.8S ribosomal DNA and internal transcribed spacer fragments, one isolate, obtained from perforated dead spores only, was assigned to the genus Nectria, and the second, obtained from both healthy and dead spores, was assigned to Leptosphaeria, a genus that also contains pathogens of plants in the Brassicaceae. PCR and randomly amplified polymorphic DNA-PCR analyses, however, did not indicate similarities between pathogens and the isolate. The presence of the two isolates in both healthy spores and perforated dead spores of S. castanea was finally confirmed by transmission electron microscopy by using distinctive characteristics of the isolates and S. castanea. The role of this fungus in S. castanea spores remains unclear, but the results serve as a strong warning that sequences obtained from apparently healthy AMF spores cannot be presumed to be of glomalean origin and that this could present problems for studies on AMF genes.
The order Glomales (class Zygomycota) comprises arbuscular mycorrhizal fungi (AMF) that are able to form mutualistic symbioses with roots of approximately 60% of all plant species (25). AMF are important for plants because they assist plants in absorbing nutrients, especially phosphate (14), and have a protective role against plant-pathogenic fungi (10, 17). It has also been shown that the diversity of AMF is an important determinant of plant diversity and productivity (26, 27, 28). AMF are obligate biotrophs. Their hyphae are coenocytic, and asexual spores form on the termini of hyphae (8). These spores contain hundreds to thousands of nuclei per spore (1, 11, 29). In the species Scutellospora castanea, one spore contains approximately 800 nuclei (11). The spores are the only form by which individual species can be identified on the basis of morphological characteristics (33).
Because of the symbiotic function of AMF, there is great interest in identifying AMF genes that are involved in the establishment and functioning of the symbiosis. However, at present, very few genes other than rRNA genes have been studied in these fungi. Several studies have shown that genetic diversity of the internal transcribed spacer (ITS) exists among and within single spores (9, 16, 22). Six different ITS1-5.8S-ITS2 types (T1 to T6) from S. castanea were reported (8). The 5.8S sequences published by Hijri et al. (9) were subsequently used in a phylogenetic analysis of Glomales and other fungi (20). This analysis showed that the highly divergent sequences T1, T3, T5, and T6 clustered within the ascomycetes (with Phoma-Leptosphaeria as the closest relatives to T1 and T3). Other studies have also reported sequences of rRNA genes obtained from glomalean spores that clustered in the ascomycetes in phylogenetic analyses (4, 12, 18). The question therefore arises as to what is the true origin of these sequences. Other researchers have suggested that these sequences are the result of fungal contaminants on the AMF spore wall (24). Three possibilities were suggested by Redecker et al. (20): (i) that these sequences were from contaminating fungi on the surface of the AMF spores, (ii) that they were sequences from ascomycetes which were living inside S. castanea spores, and (iii) that they originated from the S. castanea genome. The first two possibilities were seen as the most likely explanations (20).
In this study, we have addressed the question of whether ascomycete fungi live inside healthy S. castanea spores or only on the surface of the spores. This is an important question, because if other fungi are able to live inside healthy AMF, then they pose a serious problem for studies of AMF genes. This is not problematic for studies of AMF ribosomal genes because so much information on these sequences exists in the databases, allowing identification of any potential nonglomalean sequences. It would, however, be a serious problem for the study of other glomalean genes, for which much less information is present in the databases that allows verification of the sequences as glomalean or nonglomalean. Because the databases contain a strong bias for ascomycete sequences compared to fungi in the zygomycetes and basidiomycetes, it is likely that sequences of genes found in AMF would cluster in the ascomycetes in a phylogenetic analysis. In this study, we attempted to isolate fungi from inside S. castanea spores and to check whether ribosomal DNA sequences of these fungi matched those already found in AMF spores. Electron microscopy was also used to demonstrate that fungi lived inside healthy spores of S. castanea that had just germinated.
MATERIALS AND METHODS
Isolation of fungi from inside S. castanea spores.
The AMF S. castanea (BEG1) was provided by the Banque Européenne des Glomales (from the Institut National de la Recherche Agronomique, Dijon, France). Spores of S. castanea were recovered from pot cultures of Allium porrum L. by wet sieving. The freshly sieved spores were observed under a binocular microscope and sorted into two groups according to color and appearance of the spore contents. One group of spores was chestnut in color, had a shiny surface, and contained clearly visible lipid globules (Fig. 1a and b, spore H). Spores having these characteristics were assumed to be healthy spores, since a preliminary test showed that a high proportion of these spores were able to germinate, and they are termed healthy spores below. The second group of spores had contents with a cloudy white appearance (Fig. 1a and b, spore C). We assumed that these were dead, parasitized spores that were not viable, as we have never observed them germinating in previous experiments. These spores are termed cloudy spores below.
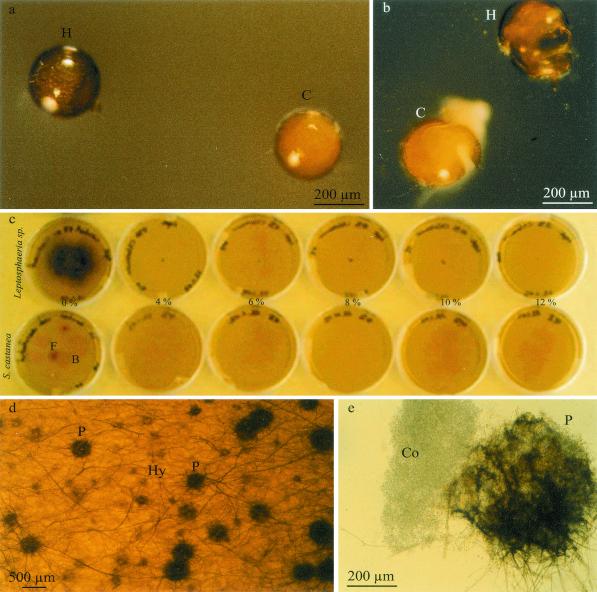
FIG. 1.
(a) Spores of S. castanea observed in incident light under a binocular microscope. C, cloudy spores; H, healthy spores with vacuolate content. (b) Crushed healthy and cloudy spores. (c) Control for surface sterilization. V8 agar medium plates contain either a Leptosphaeria sp. suspension or 10 spores of S. castanea per plate. Five sterilization solutions were used, consisting of 4% to 12% chloramin T, 0.02% streptomycin, and 0.01% gentamicin. Sterile distilled water (0%) was used as a control. The Leptosphaeria sp. only grew in the control treatment (0%). L, Leptosphaeria sp. suspension; F, fungal colony; B, bacterial colony. (d and e) Leptosphaeria sp. isolated from healthy S. castanea spores. The mycelium comprises septate yellow-brown hyphae (Hy), and the fungus also forms black pycnidia (P) which contain conidia (Co).
After sorting, all spores were rinsed several times with distilled water, surface sterilized with 4% chloramin T-0.02% streptomycin-0.01% gentamicin for 15 min, and then rinsed again at least three times with autoclaved distilled water. As a control to check that surface sterilization treatment killed any fungal propagules that were on the surface, several solutions with different concentrations (4, 6, 8, 10, and 12%) of chloramin T were used to wash a suspension of Leptosphaeria sp. which contained pycnidia, conidia, and mycelium. Surface-sterilized cloudy and healthy spores and the Leptosphaeria sp. suspension were placed onto V8 juice agar medium containing 0.2% CaCO3, 1% agar, and 75 μg of ampicillin per ml. Cloudy and healthy spores were placed separately on plates. Sixty-seven plates and 417 spores were used in total, and each plate contained 4 to 12 spores. Cloudy and healthy spores on half of the plates were crushed on the medium with sterilized forceps. Plates were incubated at 25°C for 3 weeks and checked every day for outgrowths of fungi. The crushed spore treatment was performed to see if any fungi could be isolated from inside the AMF spores and whether there was a higher probability of isolating fungi from crushed rather than intact AMF spores. Isolated fungi were grown in a V8 agar medium containing 75 μg of ampicillin per ml.
Characterization of isolated fungi by using molecular techniques.
DNA of the fungi isolated from S. castanea spores was extracted from mycelium obtained from the fungi after cultivation in pure culture in V8 liquid medium. DNA was extracted using a DNeasy Plant minikit (Qiagen). The ITS regions of rRNA genes were amplified by PCR using the universal ITS primers ITS1 and ITS4 (35). PCR products were cloned into pGEM-T vector (Promega) and sequenced with an ABI 377 DNA sequencer (ABI; Perkin-Elmer).
PCR was performed to compare one fungus that was isolated from healthy S. castanea spores with different Leptosphaeria maculans isolates, using the pathogenicity group-specific primers A1-A3 and A2-A3 (both specific for aggressive isolates and giving 300- and 100-bp fragments, respectively) and the NA1-3-NA2-1 primers (which are specific to nonaggressive isolates and give rise to a 100-bp fragment) (30, 31).
Random amplified polymorphic DNA (RAPD)-PCR was also performed on seven isolates of Phoma lingam (anamorph of L. maculans). These comprise one aggressive and one nonaggressive isolate, P. lingam Thlaspi, P. lingam Sisymbrium, P. lingam Erysimum, P. lingam Lepidium, and P. lingam PhTrZgg. DNA samples from Phoma nigrificans, Sclerotinia sclerotiorum, Cylindrosporium concentricum, and Verticillium dahliae were also used for the RAPD analysis. We used the decamer primer 17 (23, 30, 31, 32), which generates several polymorphic RAPD bands.
Transmission electron microscopy (TEM) of spores of S. castanea.
Cloudy spores of S. castanea and healthy spores that had just germinated (germination was carried out in 0.6% water-agar in petri dishes at 25°C in the dark for approximately 3 weeks) were fixed for 24 h at room temperature on a rotator in a solution of either 4.5% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2) or 2% (vol/vol) paraformaldehyde-2% (vol/vol) glutaraldehyde-3 mM CaCl2 in 0.1 M sodium cacodylate (pH 7.2) (cacodylate buffer). Each solution contained approximately 0.1% (wt/vol) Brij 35 (Sigma). The spores were then placed in fresh fixative without Brij 35 for 2 days and were rotated at room temperature.
After several rinses with cacodylate buffer, the spores were placed in 2% sucrose in cacodylate buffer for 40 min, followed by a postfixation in 1% (vol/vol) osmium tetroxide in cacodylate buffer for 1 h. Spores were then precontrasted for 40 min in 2% (wt/vol) uranyl acetate in 10% ethanol at room temperature and dehydrated through a graded ethanol series (30 to 50% on ice and then 70 to 100% at −25°C, with 20 min per step). Spores were embedded in LR White medium grade resin with 0.5% benzoin methyl ether as follows: one night in 1:3 resin-ethanol, 3 h in 2:2 resin-ethanol, one night in 3:1 resin-ethanol, and finally in pure resin at −25°C. Polymerization was performed at −20°C under UV irradiation for 24 h, followed by 24 h at room temperature under UV irradiation.
The resin blocks were cut with a diatome diamond knife on a Reichert Ultracut S microtome. Fine sections of 70 to 80 μm were placed on Formvar-coated copper grids and contrasted with a 2% aqueous solution of uranyl acetate for 6 min followed by lead citrate for 6 min. The grids were examined with a Philips Biotwin CM100 (Lab6 filament) transmission electron microscope.
Nucleotide sequence accession numbers.
The ITS sequences of the two fungi that were isolated from S. castanea spores have been deposited in the EMBL data library under accession numbers AJ317958 (Leptosphaeria-like fungus) and AJ317959 (Nectria-like fungus).
RESULTS AND DISCUSSION
Isolation and molecular identification of fungi from S. castanea spores.
A low frequency (1.2%) of intact surface-sterilized healthy spores gave rise to some fungal colonies. This was probably due to the breaking of some spores during the treatment, because the use of 4% (or more) chloramin T, 0.02% streptomycin, and 0.01% gentamicin killed the fungi isolated from AMF (Fig. 1c). In contrast, 6.7% of fungi grew out from intact surface-sterilized cloudy spores, because cloudy spores are heavily infected by microorganisms and their walls were perforated (as shown later by electron microscopy). Two filamentous fungi with different morphology grew out from surface-sterilized crushed spores; 19 and 25% of crushed healthy and cloudy spores, respectively, gave fungal colonies. One fungus with septate white hyphae was isolated only from cloudy spores. This fungus grew out of S. castanea spores after 2 days of incubation on a V8 agar medium and had morphology similar to that of Nectria spp. Another fungus showing morphological similarity to L. maculans was isolated from crushed healthy spores after a week (Fig. 1d and e). This fungus was also observed in crushed cloudy spores. However, the Nectria-like fungus was never observed in crushed healthy spores.
The ITS sequence of the Nectria-like fungus that was isolated from cloudy spores was 566 bp in length. A BLAST search showed that it had 98% identity with ITS and 5.8S sequences from Nectria haematococca. The ITS sequence from the Leptosphaeria-like fungus, isolated from healthy spores, was 576 bp in length and showed 97% identity to the previously published sequences T1 and T3, which were previously shown to phylogenetically fit into the Leptosphaeria group (20).
Other microorganisms, such as bacterium-like organisms, are known to live inside some healthy AMF (2). However, little is known about fungi living inside healthy AMF, although there are reports of fungi that parasitize or colonize dead AMF spores. It has already been reported that intracellular bacterial endosymbionts exist in spores of six species of the family Gigasporaceae, including S. castanea (3). Hyperparasites have also been isolated from spores of the AMF Glomus epigaeum and Glomus fasciculatum and have been identified as Anguillospora pseudolongissima (hyphomycetes) and the ascomycete Humicola fuscoatra (5). Another hyperparasite, identified as Phlyctochytrium sp. (Chytridiomycota), was isolated from G. fasciculatum (5) and Glomus macrocarpum (21). All of these hyperparasites are known to be saprophytes in the soil. They were isolated from spores of supposedly pure cultures of AMF that were maintained on plants in the greenhouse. An unidentified microorganism, which clearly has a cell wall and cytoplasmic membrane, was also observed in the hyphae of a mycorrhizal fungus by electron microscopy (19). The wall of the microorganism was in a close contact with the cytoplasm of the mycorrhizal fungus.
TEM of microorganisms living inside S. castanea spores.
TEM was performed on cloudy spores of S. castanea, on healthy germinated spores, and on the two filamentous fungi grown in pure culture that had been isolated from S. castanea. The fungus that was isolated from cloudy spores and was identified as a Nectria sp. possessed a hyphal wall with two layers (an external electron-dense layer and an internal electron- transparent layer), a nucleus with diffuse chromatin, mitochondria, and electron-transparent vacuoles of different sizes (Fig. 2a and b).
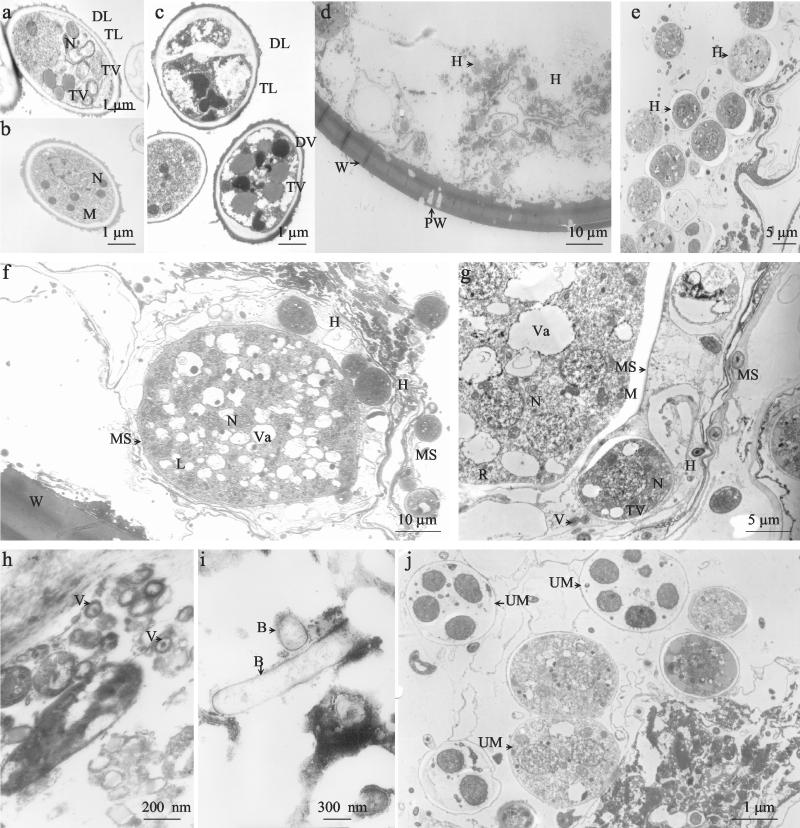
FIG. 2.
Electron micrographs showing similarities between the isolated fungi and fungal structures inside the S. castanea spores and also the presence of other microorganisms. (a and b) Ultrastructural features of the Nectria sp. isolated from cloudy spores of S. castanea. (c) Leptosphaeria sp. isolated from healthy spores of S. castanea. (d and e) Ultrastructure of cloudy nonviable spores. The spore wall contains many holes, and there are many hyphae inside the spore. (f and g) Healthy S. castanea spores. Many nuclei are present, surrounded by dense cytoplasm which is filled with ribosomes, mitochondria, lipids, membrane systems, and vacuoles containing electron-dense bodies. Beside the living structures of S. castanea spores, fungal structures with the same ultrastructural features as Leptosphaeria sp. are present. (h) Healthy spores of S. castanea harboring virus-like particles. (i) Bacterium-like organisms are also present in healthy spores of S. castanea. (j) Other eucaryotic microorganisms that we were not able to identify were also observed inside S. castanea. B, bacterium-like organisms; DL, electron-dense layer; DV, electron-dense vacuole; H, hyphae; L, lipids; M, mitochondria; MS, membrane system; N, nucleus; PW, perforated wall; R, ribosomes; TL, electron-transparent layer; TV, electron-transparent vacuole; UM, unidentified microorganisms; V, virus-like particle; Va, vacuole; W, spore wall.
The Leptosphaeria-like fungus isolated from healthy spores (Fig. 2c) also exhibited a hyphal wall of two layers. The external layer was thick and electron dense. The contents of some vacuoles were condensed to a dark electron-dense form. Some other vacuoles were large and electron transparent.
TEM performed on cloudy S. castanea spores (Fig. 2d) showed a spore wall which is formed by four recognizable layers with a thickness of approximately 8 μm in total. The wall observed in fresh spores of this species was thicker (34) than that observed by TEM, and this could be the result of the fixation and dehydration procedures. Cloudy spores exhibited a perforated wall as has previously been reported for parasitized spores. Many hyphae were observed inside the spores, and these had an ultrastructural similarity to those of the Nectria sp. that was isolated from cloudy spores of S. castanea (Fig. 2e).
Figure 2f and g show ultrastructural features of typical healthy S. castanea spores, which contain many nuclei surrounded by dense cytoplasm filled with ribosomes, mitochondria, lipids, membrane systems, and vacuoles containing electron-dense bodies. Fungal structures with the same ultrastructural features as Leptosphaeria spp. (two layers of the hyphal wall, electron-dense vacuoles, and an electron-dense nucleolus) are present inside these healthy spores.
Figure 2h shows that healthy spore of S. castanea harbor virus-like particles. These virus-like particles have ultrastructural similarity to those previously observed in nematodes by Dollet et al. (6). As previously described for AMF, bacterium-like organisms (2, 3) were also observed in healthy S. castanea spores (Fig. 2i). Figure 2j shows the presence of other eucaryotic microorganisms; some of them have a vacuole-like structure with a single wall layer and the inside is dark around bodies. These unidentified microorganisms are probably not able to grow in the V8 agar medium used in this study.
The nuclear diameters of both the Leptosphaeria sp. and N. haematococca were approximately 1.24 (± 0.2) μm. This is considerably smaller than the S. castanea nuclear diameter, which is between 3 and 5 μm (13, 15).
The finding of a Nectria sp. in spores that appear to be old, with cloudy contents, was not surprising, since such spores of many different AMF species can be found in field soils with such contents and nearly always have degraded perforated walls (I. R. Sanders, unpublished observation). From the results of this study, we cannot be sure of the relationship between the Nectria sp. and S. castanea. The Nectria sp. may be a parasite or colonizer of old, dead spores. We cannot say for certain whether spores colonized by the Nectria sp. are already dead or nonviable, because the fungus grew out onto the agar plate much faster than the time necessary for S. castanea spores to germinate.
The finding of a Leptosphaeria sp. inside healthy spores of S. castanea is interesting. The fact that the isolated Leptosphaeria sp. contained sequences very closely matching those of T1 and T3 (9, 20) makes it very likely that the previously reported sequences T1 and T3 also came from a fungus in the Leptosphaeria genus living inside healthy S. castanea spores rather than as contaminants on the surface of the spores as previously suggested (24). The further observation of fungal hyphae inside S. castanea spores with a morphology similar to that of the isolated Leptosphaeria sp. also supports this hypothesis.
We present evidence that the Leptosphaeria sp. isolate lives in healthy spores of S. castanea. As far as we know, this is the first report showing a fungus living inside healthy AMF spores. It is not possible to determine the relationship between the Leptosphaeria sp. and S. castanea. The relationship could be parasitic, mutualistic, or commensal. Future experiments need to be performed to find out whether the Leptosphaeria sp. has any effect on S. castanea fitness or on symbiotic function.
Comparison of the Leptosphaeria strain originating from S. castanea with other known Leptosphaeria isolates.
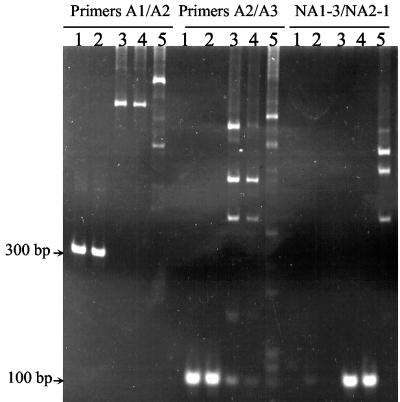
We used PCR to compare the Leptosphaeria strain that was isolated from healthy S. castanea spores with different strains of aggressive and nonaggressive L. maculans isolates. Figure 3 shows that the Leptosphaeria strain that was isolated from S. castanea is different from its relatives.
FIG. 3.
Gel electrophoresis of PCR products carried out on genomic DNAs of different Leptosphaeria isolates with primer pairs A1-A3, A2-A3, and NA1-3-NA2-1. Lanes 1 and 2, L. maculans aggressive isolate; lanes 3 and 4, L. maculans nonaggressive isolate; lane 5, Leptosphaeria sp. isolated from S. castanea spores.
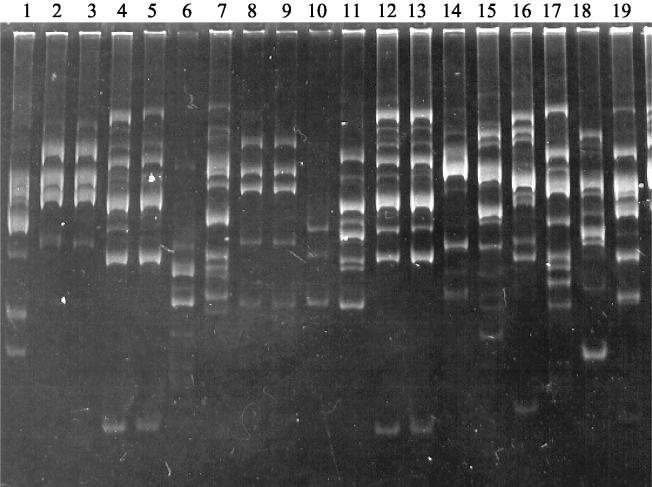
Because Leptosphaeria is known to be a pathogen of Brassicaceae, we performed RAPD-PCR with different ascomycete fungi which are known to be pathogens of Brassicaceae (including several Leptosphaeria isolates) and with the Leptosphaeria sp. isolated from S. castanea. The banding patterns produced by the RAPD-PCR for the different fungi were compared with the profiles for the Leptosphaeria sp. that was isolated from S. castanea (Fig. 4). The banding pattern produced by the Leptosphaeria sp. that was isolated from S. castanea spores was different from those produced by other Leptosphaeria isolates or other pathogenic fungi of Brassicaceae. The observed differences cannot be correlated to any particular functions.
FIG. 4.
Banding patterns produced from RAPD-PCR amplification with primer 17 on DNA. Lanes: 1, Leptosphaeria sp. isolated from S. castanea spores; 2, 3, 8, and 9, P. lingam aggressive isolate; 4, 5, 12, and 13, P. lingam nonaggressive isolate; 6, P. lingam PhTrZgg; 7, P. nigrificans; 10 and 11, P. lingam Thlaspi; 14, P. lingam Sisymbrium; 15, P. lingam Erysimum; 16, P. lingam Lepidium; 17, C. concentricum; 18, V. dahliae; 19, S. sclerotiorum.
The finding of a fungus living inside healthy AMF spores may be of considerable biological interest. Interestingly, some strains of L. maculans are known pathogens of plants in the family Brassicaceae. Because of the growth habit of AMF that allows hyphae from one AMF to connect roots of different host plants (both intra- and interspecifically), AMF hyphae could potentially act as a vector for dispersal of fungi from plant to plant. Our results are particularly intriguing, however, because the plants on which Leptosphaeria can be pathogenic are a group of plants that are usually nonmycorrhizal (25). One obvious hypothesis to test is whether the fungi carried by AMF result in an improved fitness of mycorrhizal plants over their nonmycorrhizal competitors that is mediated by pathogenic fungi: in other words, whether AMF-isolated fungi act as pathogens to non-AMF host plants. Francis and Read (7) have shown that the presence of AMF hyphae in the rhizospheres of nonmycorrhizal plant species can inhibit the growth of the nonmycorrhizal plant species, and this could be a potential mechanism for that effect.
Since inocula of some AMF species are sold commercially for the enhancement of crop quality and productivity, the presence of other microorganisms in AMF inocula could be seen as problematic. We therefore stress here that there is no evidence from the results of our experiments that these fungi that we have isolated from S. castanea would be likely to have a negative effect on the growth of plant species that are normally mycorrhizal.
Conclusions.
The presence of other fungi in AMF is not problematic for studies of AMF ribosomal genes, because so much information on these sequences exists in the databases from all fungal groups, allowing identification of any probable nonglomalean sequences. However, it is a very serious problem for the study of other glomalean genes, where much less information is available in the DNA sequence databases. There is a high bias in the DNA sequence databases towards ascomycete and basidiomycete DNA sequences, with much less data existing for zygomycetes. Therefore, new sequences of genes found in AMF are much more likely to show similarities to sequences of genes already found in ascomycetes or basidiomycetes than to Glomales.
We have described here one fungal species that lives inside healthy S. castanea spores. We are not claiming in this study that Leptosphaeria spp. live inside all AMF, but our results demonstrate that symptomless infections of apparently healthy AMF spores are indeed possible. Considering the number of molecular studies of DNA sequences from AMF spores that have also revealed Ascomycetes-like sequences, it is entirely possible that other fungi also have the ability to colonize AMF. Therefore, the search for glomalean functional genes should certainly take into consideration the presence of other fungi that could live inside AMF spores and hyphae. The presence of ascomycete gene sequences should not be discounted as being due solely to contaminations on the surface of spores as has been assumed by other workers. Future research will show whether the presence of such fungi also plays an important role in the functioning of AMF or their interaction with plants.
Acknowledgments
This work was supported by grants 3100-050481.97 and 631-058108.99 from the Swiss National Science Foundation (SNSF). I.R.S. was in receipt of a professorial fellowship from the SNSF, which is gratefully acknowledged.
We thank Thomas Boller and Andres Wiemken for allowing part of this work to be carried out in the Botanical Institute, Basel, Switzerland, and Paola Bonfante for advice on interpretation of electron micrographs.
REFERENCES
- 1.Bécard, G., and P. E. Pfeffer. 1993. Status of nuclear division in arbuscular mycorrhizal fungi during in vitro development. Protoplasma 194:62-68. [Google Scholar]
- 2.Bianciotto, V., C. Bandi, D. Minerdi, M. Sironi, H. V. Tichy, and P. Bonfante. 1996. An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 62:3005-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianciotto, V., E. Lumini, L. Lanfranco, D. Minerdi, P. Bonfante, and S. Perotto. 2000. Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae. Appl. Environ. Microbiol. 66:4503-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clapp, J. P., A. H. Fitter, and J. P. Young. 1999. Ribosomal small subunit sequence variation within spores of an arbuscular mycorrhizal fungus, Scutellospora sp. Mol. Ecol. 8:915-921. [DOI] [PubMed] [Google Scholar]
- 5.Daniels, B. A., and J. A. Menge. 1980. Hyperparasite of vesicular-arbuscular mycorrhizal fungi. Phytopathology 70:584-588. [Google Scholar]
- 6.Dollet, M., S. Marche, D. Gardani, E. Muller, and T. Baltz. 1996. Virus of plant trypanosomes (Phytomonas spp.), p. 227-236. In M. Nicole and V. Gianinazzi-Pearson (ed.), Histology, ultrastructure and molecular cytology of plant microorganism interactions. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Francis, R., and D. J. Read. 1995. Mutualism and antagonism in the mycorrhizal symbiosis with special reference to impacts on plant community structure. Can. J. Bot. 73:1301-1309. [Google Scholar]
- 8.Gerdemann, J. W., and J.M. Trappe. 1974. The Endogonaceae in the Pacific Northwest. Mycol. Mem. 5:1-76. [Google Scholar]
- 9.Hijri, M., M. Hosny, D. van Tuinen, and H. Dulieu. 1999. Intraspecific ITS polymorphism in Scutellospora castanea (Glomales, Zygomycota) is structured within multinucleate spores. Fungal Genet. Biol. 26:141-151. [DOI] [PubMed] [Google Scholar]
- 10.Hooker, J. E., M. Jaizme-Vega, and D. Atkinson. 1994. Biocontrol of plant pathogens using arbuscular mycorrhizal fungi, p. 191-200. In S. Gianinazzi and H. Schüepp (ed.), Impact of arbuscular mycorrhizas on sustainable agriculture and natural ecosystems. Birkhäuser Verlag, Basel, Switzerland.
- 11.Hosny, M., V., Gianinazzi-Pearson, and H. Dulieu. 1998. Nuclear DNA contents of eleven fungal species in Glomales. Genome 41:422-428. [Google Scholar]
- 12.Hosny, M., M. Hijri, E. Passerieux, and H. Dulieu. 1999. rDNA units are highly polymorphic in Scutellospora castanea (Glomales, Zygomycetes). Gene 226:61-71. [DOI] [PubMed] [Google Scholar]
- 13.Hosny, M. 1997. Ph.D. thesis. University of Burgundy, Dijon, France.
- 14.Jakobsen, I. 1995. Transport of phosphorus and carbon in VA mycorrhizas, p. 297-324. In A. Varma and B. Hock (ed.), Mycorrhiza. Springer-Verlag, Berlin, Germany.
- 15.Kuhn, G., M. Hijri, and I. R. Sanders. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature 414:745-748. [DOI] [PubMed]
- 16.Lanfranco, L., M. Delpero, and P. Bonfante. 1999. Intrasporal variability of ribosomal sequences in the endomycorrhizal fungus Gigaspora margarita. Mol. Ecol. 8:37-45. [DOI] [PubMed] [Google Scholar]
- 17.Nesheim, O. N., and M. B. Linn. 1969. Deleterious effect of certain fungitoxicants on the formation of mycorrhizae on corn by Endogone fasciculata and corn root development. Phytopathology 59:297-300. [Google Scholar]
- 18.Pringle, A., J. M. Moncalvo, and R. Vilgalys. 2000. High levels of variation in ribosomal DNA sequences within and among spores of a nature population of the arbuscular mycorrhizal fungus Acaulospora colossica. Mycologia 92:259-268. [Google Scholar]
- 19.Protsenko, M. A. 1975. Microorganism in the hyphae of mycorrhiza-forming fungus. Mikrobiologiya 44:1121-1124. [PubMed] [Google Scholar]
- 20.Redecker, D., M. Hijri, H. Dulieu, and I. R. Sanders. 1999. Phylogenetic analysis of a dataset of fungal 5.8S rDNA sequences shows that highly divergent copies of internal transcribed spacers reported from Scutellospora castanea are of ascomycete origin. Fungal Genet. Biol. 28:238-244. [DOI] [PubMed] [Google Scholar]
- 21.Ross, J. P., and R. Ruttencutter. 1977. Population dynamics of two vesicular-arbuscular endomycorhizal fungi and the role of hyperparasitic fungi. Phytopathology 67:490-496. [Google Scholar]
- 22.Sanders, I. R., M. Alt, K. Groppe, T. Boller, and A. Wiemken. 1995. Identification of ribosomal DNA polymorphisms among and within spores of the Glomales: application to studies on the genetic diversity of arbuscular mycorrhizal fungal communities. New Phytol. 130:419-427. [Google Scholar]
- 23.Schleier, S., K. Voigt, and J. Woestemeyer. 1997. RAPD-based molecular diagnosis of mixed fungal infection on oilseed rape (Brassica napus): evidence for genus- and species-specific sequences in the fungal genomes. J. Phytopathol. 145:81-87. [Google Scholar]
- 24.Schüβler, A., H. Gehrig, D. Schwarzott, and C. Walker. 2001. Analysis of partial Glomales SSU rRNA gene sequences: implications for primer design and phylogeny. Mycol. Res. 105:5-15. [Google Scholar]
- 25.Smith, S. E., and D. J. Read. 1997. The mycorrhizal symbiosis. Academic Press, Oxford, United Kingdom.
- 26.Streitwolf-Engel, R., M. G. A. van der Heijden, A. Wiemken, and I. R. Sanders. 2001. The ecological significance of arbuscular mycorrhizal fungal effects on clonal reproduction in plants. Ecology 82:2846-2859. [Google Scholar]
- 27.van der Heijden, M. G. A., J. N. Klironomos, M. Ursic, P. Moutoglis, R. Streitwolf-Engel, T. Boller, A. Wiemken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69-72. [Google Scholar]
- 28.van der Heijden, M. G. A., T. Boller, A. Wiemken, and I. R. Sanders. 1998. Differential responses of plants species to arbuscular mycorrhizal fungal species: are they potential determinants of plant community structure? Ecology 79:2082-2091. [Google Scholar]
- 29.Viera, A., and M. G. Glenn. 1990. DNA content of vesicular-arbuscular mycorrhizal fungal spores. Mycology 82:263-267. [Google Scholar]
- 30.Voigt, K., M. Jedryczka, and J. Wöstemeyer. 2001. Strain typing of Polish Leptosphaeria maculans isolates supports at the genetic level the multi-species concept of aggressive and non-aggressive strains. Microbiol. Res. 156:169-177. [DOI] [PubMed] [Google Scholar]
- 31.Voigt, K., S. Schleier, and J. Wöstemeyer. 1998. RAPD-based molecular probe for the blackleg fungus Leptosphaeria maculans (Phoma lingam). Evidence for pathogenicity group-specific sequences in the fungal genomes. J. Phytopathol. 146:567-576. [Google Scholar]
- 32.Voigt, K., E. Cigelnik, and K. O'Donnell. 1999. Phylogeny and PCR identification of clinically important zygomycetes based on nuclear ribosomal DNA sequence data. J. Clin. Microbiol. 37:3957-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, C. 1992. Systematics and taxonomy of the arbuscular endomycorrhizal fungi (Glomales): a possible way forward. Agronomie 12:887-898. [Google Scholar]
- 34.Walker, C., V. Gianinazzi-Pearson, and H. Marrion-Espinasse. 1993. Scutellospora castanea, a newly described arbuscular mycorrhizal fungus. Mycology 14:279-286. [Google Scholar]
- 35.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sminsk, and T. J. White (ed.), PCR protocols, a guide to methods and applications. Academic Press, San Diego, Calif.