Abstract
While Paenibacillus polymyxa strain Pw-2 has been identified as an endophyte of lodgepole pine (M. Shishido, B. M. Loeb, and C. P. Chanway, Can. J. Microbiol. 41:707-713, 1995), P. polymyxa strain L6 has not, a distinction that could be explained by the differential abilities of these isolates to form spores, rather than the differential abilities to colonize the interior tissues of lodgepole pine. Chemical disinfection was used to destroy bacteria on the root exterior, but bacterial endospores are known for their ability to withstand chemical disinfection, and strain Pw-2 was found to produce 300 to 11,000 times more germinating endospores than strain L6 under the experimental conditions used by Shishido et al. (Can. J. Microbiol. 41:707-713, 1995). Attempts to identify strain Pw-2 within lodgepole pine root tissues by using confocal microscopy techniques failed. We discuss the possibility that spore-forming bacteria can be mistakenly identified as endophytes when culture-based methods alone are used.
Bacterial endophytes within plants are the focus of much recent interest, as their location within plants places them in a strong position to affect plant nutrition (17, 23), pollutant catabolism (20), stress or defense responses (23, 24), and invading pathogens (5, 22). Work within the last decade has identified abundant and diverse populations of bacterial endophytes in many plants, including potato (9, 22), corn (7, 13), cotton (13, 14), and cucumber (12). Endophytic bacteria are usually identified as such on the basis of their culture from chemically surface-disinfected root segments (7, 14) or macerates (12, 19, 26).
Problems with reliance upon chemical disinfection for identifying bacterial endophytes.
A bacterial cell or spore that adheres to a root surface and is not removed or killed by chemical disinfection can be mistaken for a true endophyte. Bacterial endospores can resist a variety of harsh treatments (6), including many of those used in disinfection protocols. Some root mucilage is difficult or impossible to remove from the root surface, even with sonication (7a). Thus, it is possible for bacterial cells and spores in such mucilage to adhere to the root surface even when the root is rinsed multiple times with liquids, as during chemical disinfection. Spores adhering to the root surface that survive chemical disinfection could germinate to form colonies when root segments or macerates are plated on nutrient agar. These colonies would be indistinguishable from those formed by bacteria located within the root.
Rhizobacteria isolated from surface-sterilized shoot tissues have been identified as systemic endophytes based on the assumption that the bacteria must have spread from the root to the shoot (13, 21, 25). However, bacteria on seed coats or in soils may colonize the exterior of the shoot during the emergence of the radicle from the seed coat or of the seedling from the soil (16). Isolation of a bacterial species that is resistant to chemical disinfection from surface-disinfected shoot tissue will demonstrate that the bacterium is an endophyte only when (i) care is taken to thoroughly remove any spores that might adhere to the shoot surface or (ii) the seedling has not been grown from a seed coated with bacteria or a seed germinated in soil containing bacteria.
Prior identification via culture-based methods of Paenibacillus polymyxa strain Pw-2, but not strain L6, as an endophyte.
P. polymyxa Pw-2 and P. polymyxa L6 are rhizobacteria that are able to improve the growth of inoculated lodgepole pine, Douglas fir, and spruce (10, 19). P. polymyxa Pw-2 was first identified as an endophyte of lodgepole pine on the basis of its consistent recovery from chemically surface-disinfected, macerated root tissues (19), an observation repeated in subsequent investigations (3). While consistent, the recovery rate of strain Pw-2 from surface-disinfected pine roots has always been relatively low, sometimes as little as 10 cells per root (3). P. polymyxa L6 was not considered to be an endophyte of lodgepole pine, as it was never recovered from the interior of surface-disinfected roots (3, 19).
Both strain L6 and strain Pw-2 form endospores. If strain Pw-2 produces a greater number of spores in the rhizosphere than strain L6, or forms spores more likely to germinate under experimental conditions than those of L6, it is possible that Pw-2 has been identified as an endophyte because it forms these spores and not because it can colonize the interior of lodgepole pine roots.
Problems with prior microscopic observations of P. polymyxa Pw-2.
In lodgepole pine shoots, ovoid objects were observed in shoot vascular tissues and identified as fluorescein isothiocyanate (FITC)-immunolabeled cells of a Pw-2 derivative (18), despite the fact that they were quite large (5 to 7 μm long). The vacuoles of phloem parenchyma cells in conifers contain phenolic compounds that tend to form circular or ovoid deposits that fall within the 5- to 7-μm size range and autofluoresce brightly at the same wavelengths as FITC when under similar excitation wavelengths (8). It is possible that the ovoid objects observed by Shishido et al. (19) may have been autofluorescing plant components, such as phenolic deposits, rather than oversized bacteria.
Experimental methods.
To verify that P. polymyxa Pw-2 is an endophyte of lodgepole pine, direct microscopic evaluation of bacterial colonization within root tissues was combined with evaluations of the spore-forming ability of each strain in the lodgepole pine rhizosphere. The latter approach was designed to demonstrate whether it is possible for P. polymyxa Pw-2, but not P. polymyxa L6, to be mistaken for an endophyte of lodgepole pine under the experimental conditions used.
For all experiments, seedlings were grown from treated seeds in sealed tubes containing sterilized nursery mix (Sunshine 4 mix; Fisons Horticulture, Inc., Vancouver, Canada), and bacteria were cultured, washed, and resuspended in cold buffer at standardized concentrations (108 CFU/ml) as described by Bent et al. (4). Three seed treatments were used: strain L6, strain Pw-2, and a sterile buffer. At least 30 seeds were treated and sown per treatment.
Confocal microscopy.
Microscopic evaluations of root interior colonization of inoculated lodgepole pines by P. polymyxa Pw-2 and L6 were conducted in two separate experiments. In the first, performed 7 weeks after inoculation and with two to three seedlings per treatment, cross sections were prepared from five different and randomly chosen areas of the roots ranging from the root tip to the root base. In the second, conducted 13 weeks after inoculation and with two seedlings per treatment, cross sections were prepared from (i) an area about 1 cm from the root tip and (ii) an area of the root near the middle or the base of the root only. In each case, 10 to 20 cross sections were prepared per sampled area on each root. Preparation of root sections for microscopy was conducted as described elsewhere (2) with the exception that cross sections instead of segments were prepared. Cross sections were made by slicing formaldehyde-fixed and rinsed roots in a droplet of 0.1 M PO4 (pH 7.4) buffer with a new razor blade. The preparation procedure can be summarized as follows: each section was exposed to polyclonal mouse antibodies that recognized either strain Pw-2 or strain L6 (as appropriate), rinsed, and then exposed to secondary anti-mouse monoclonal antibodies labeled with FITC. Appropriate microscopy controls to identify nonspecific antibody binding or autofluorescence were also included, as described elsewhere (2). Sections were mounted in 80% glycerol containing an antifade reagent (2.5% [wt/vol] 1,4-diazabicyclo [2.2.2] octane [DABCO]) on glass slides and kept at 4°C in the dark until viewed. Slides were viewed using the preprogrammed COMOS red and green channel settings of a Bio-Rad MRC 600 confocal scanning laser microscope, with the channel settings and the collection and manipulation of digital data performed as described previously (2).
Spore formation.
The extent of spore formation by P. polymyxa Pw-2 and L6 in the rhizosphere of lodgepole pines was determined for five seedlings sampled from each treatment after 6, 9, and 12 weeks of incubation (15 seedlings in total were sampled per treatment in each experiment). Roots from each seedling were aseptically removed and washed for 20 min in an aliquot of KP buffer containing 0.01% Tween 20, as described by Bent et al. (4). Two 1-ml samples of each root wash were placed in separate, sterile 2-ml microcentrifuge tubes. One of the tubes was placed in a 55°C water bath for 30 min. This treatment was sufficient to kill 100% of a 108-CFU/ml suspension of Pseudomonas fluorescens strain M20 cells (data not shown). The other tube was kept on ice until used. Unheated and heat-treated root washes were diluted and plated on one-half-strength tryptic soy agar, and the plates were incubated for 2 days at 25°C. These conditions were identical to those used previously to evaluate root interior colonization by strains L6 and Pw-2 (3). Extension of the period of incubation up to 1 week for plates devoid of colonies or with very few colonies did not result in the formation of new colonies (data not shown). Colonies were counted and representative samples of colonies from each plate were subjected to strain verification tests by toothpicking colonies onto a variety of differential or selective media as described by Bent and Chanway (3). The entire spore formation experiment was conducted three times. Data from each experiment were pooled, and the mean CFU of cells recovered per root before and after heat treatment, after 6, 9, or 12 weeks of incubation, was determined with analysis of variance and Duncan's mean separation procedures by using SAS v. 6.1 software and an error rate at a P of < 0.05.
Recovery of bacteria from roots.
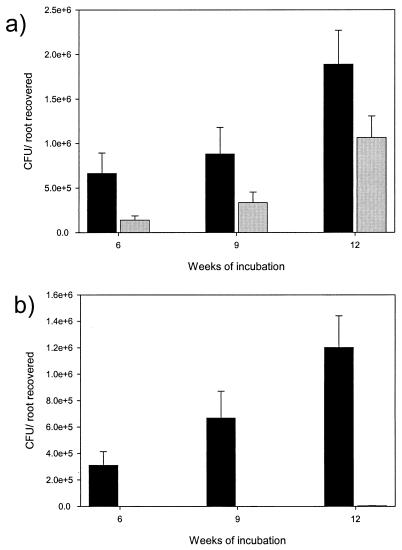
P. polymyxa strains L6 and Pw-2 were recovered from exterior washes of roots inoculated with L6 and Pw-2, respectively (Fig. 1). No bacteria were recovered from uninoculated roots, even when plates were incubated for 7 days (data not shown). Before heat treatment, about twice as many Pw-2 colonies than L6 colonies were observed (Fig. 1a), a significant difference (P < 0.05) that confirms prior observations (4).
FIG. 1.
Recovery of P. polymyxa strains L6 and Pw-2 from washes of lodgepole pine seedling roots before (a) and after (b) heat treatment (55°C, 30 min), determined for roots after 6, 9, or 12 weeks of incubation. Standard errors are indicated by bars. Black bars represent data for strain Pw2; gray bars represent data for strain L6. The average CFU of L6 recovered per root after heat treatment (±standard error) for weeks 6, 9, and 12 were 12 (±9), 520 (±380), and 4,200 (±2,000), respectively.
After heat treatment, colonies formed by strain Pw-2 outnumbered strain L6 colonies 300- to 11,000-fold (Fig. 1b) throughout the experiment. The recovery of strain L6 after heat treatment was minimal. The recovery of L6 from surface-disinfected roots was not observed by Shishido et al. (19) or Bent and Chanway (3), but this may have been due to the experimental methods used in those studies (exposure to sterilants rather than heat and the use of higher dilutions that will not detect low concentrations of bacteria).
The fact that strain Pw-2 formed more colonies than strain L6 after heat treatment may be due to (i) greater spore production by Pw-2 and/or (ii) a greater ability of strain Pw-2 spores to germinate under the experimental conditions used. Either way, it is possible that strain Pw-2 was recovered previously from chemically surface-disinfected, macerated lodgepole pine roots while strain L6 was not (3, 19) because of the greater ability of Pw-2 to form spores that will germinate under the experimental conditions (Fig. 1) and not because Pw-2 actually colonizes the root interior of lodgepole pine.
Microscopic observations of root colonization.
Neither P. polymyxa Pw-2 nor L6 was observed within lodgepole pine root tissues, although both organisms were observed in these experiments to colonize the root exterior (data not shown). No bacterial cells were observed either in the root interior or the exterior of uninoculated roots (data not shown). Similar observations of colonization of the root exterior by strains L6 and Pw-2 were made previously (2). In the absence of a bacterium verified to be an endophyte of lodgepole pine, the applicability of the method we used to locate bacterial endophytes is difficult to assess. Electron microscopy has been used to identify bacterial endophytes in angiosperms (11, 15) and could be employed to determine whether P. polymyxa strain Pw-2 is an endophyte of lodgepole pine.
The present work has implications for the use of culture-based methods in identifying other spore-forming bacteria as endophytes. Whenever a spore-forming organism is identified as an endophyte of a particular plant species, care must be taken to ensure that this identification cannot be explained by its spore-forming ability. Experimental results should be interpreted with this in mind, and the endophytic status of any spore-forming bacterium in plant tissues should be verified through direct methods where possible.
Acknowledgments
Funding for E. Bent was provided by an NSERC PGS B scholarship. Lodgepole pine seeds were donated by the British Columbia Ministry of Forests Tree Seed Centre, Surrey, British Columbia, Canada.
REFERENCES
- 1.Bent, E. 2000. The effect of other rhizosphere microorganisms on the ability of Paenibacillus spp. to promote the growth of lodgepole pine [Pinus contorta var. latifolia (Dougl. Engelm.)]. Ph.D. thesis. University of British Columbia, Vancouver, British Columbia, Canada.
- 2.Bent, E., C. Breuil, S. Enebak, and C. P. Chanway. 2002. Surface colonization of lodgepole pine (Pinus contorta var. latifolia [Dougl. Engelm.]) roots by Pseudomonas fluorescens and Paenibacillus polymyxa under gnotobiotic conditions. Plant Soil 24:187-196. [Google Scholar]
- 3.Bent, E., and C. P. Chanway. 1998. The growth-promoting effects of a bacterial endophyte on lodgepole pine are partially inhibited by the presence of other rhizobacteria. Can. J. Microbiol. 44:980-988. [Google Scholar]
- 4.Bent, E., S. Tuzun, S. Enebak, and C. P. Chanway. 2001. Alterations in plant growth and in root hormone levels of lodgepole pines inoculated with rhizobacteria. Can. J. Microbiol. 46:793-800. [DOI] [PubMed] [Google Scholar]
- 5.Downing, K. J., and J. A. Thomson. 2000. Introduction of the Serratia marcesens chiA gene into an endophytic Pseudomonas fluorescens for the biocontrol of phytopathogenic fungi. Can. J. Microbiol. 46:363-369. [DOI] [PubMed] [Google Scholar]
- 6.Fairhead, H., B. Setlow, W. M. Waites, and P. Setlow. 1994. Small, acid-soluble proteins bound to DNA protect Bacillus subtilis spores from being killed by freeze-drying. Appl. Environ. Microbiol. 60:2647-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher, P. J., O. Petrini, and H. M. Lappin-Scott. 1992. The distribution of some fungal and bacterial endophytes in maize (Zea mays L.). New Phytol. 122:299-305. [DOI] [PubMed] [Google Scholar]
- 7a.Foster, R. C. 1986. The ultrastructure of the rhizoplane and rhizosphere. Annu. Rev. Phytopathol. 24:211-234. [Google Scholar]
- 8.Franceschi, V. R., T. Krekling, A. A. Berryman, and E. Christiansen. 1998. Specialized phloem parenchyma cells in Norway spruce (Pinaceae) bark are an important site of defense reactions. Am. J. Bot. 85:601-615. [PubMed] [Google Scholar]
- 9.Garbeva, P., L. S. van Overbeek, J. W. L. van Vuurde, and J. D. van Elsas. 2001. Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16s rDNA based PCR fragments. Microb. Ecol. 41:369-383. [DOI] [PubMed] [Google Scholar]
- 10.Holl, F. B., and C. P. Chanway. 1992. Rhizosphere colonization and seedling growth promotion of lodgepole pine by Bacillus polymyxa. Can. J. Microbiol. 38:303-308. [Google Scholar]
- 11.Jacobs, M. J., W. M. Bugbee, and D. A. Gabrielson. 1985. Enumeration, location and characterization of endophytic bacteria within sugar beet roots. Can. J. Bot. 63:1262-1265. [Google Scholar]
- 12.Mahafee, W. F., and J. W. Kloepper. 1997. Bacterial communities of the rhizosphere and endorhiza associated with field-grown cucumber plants inoculated with a plant growth-promoting rhizobacterium or its genetically modified derivative. Can. J. Microbiol. 43:344-353. [DOI] [PubMed] [Google Scholar]
- 13.McInroy, J. A., and J. W. Kloepper. 1995. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil 173:337-342. [Google Scholar]
- 14.Misaghi, I. J., and C. R. Donndelinger. 1990. Endophytic bacteria in symptom-free cotton plants. Phytopathology 80:808-811. [Google Scholar]
- 15.M'Piga, P., R. R. Belanger, T. C. Paulitz, and N. Benhamou. 1997. Increased resistance to Fusarium oxysporum f. sp. radicis lycopersici in tomato plants treated with the endophytic bacterium Pseudomonas fluorescens strain 63-28. Physiol. Mol. Plant Pathol. 50:301-320. [Google Scholar]
- 16.Raaijmakers, J. M., M. Leeman, M. M. P. van Oorshot, I. van der Sluis, B. Schippers, and P. A. H. M. Bakker. 1995. Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology 85:1075-1081. [Google Scholar]
- 17.Sevilla, M., R. H. Burris, N. Gunapala, and C. Kennedy. 2001. Comparison of benefit to sugarcane plant growth and 15N2 incorporation following inoculation of sterile plants with Acetobacter diazotrophicus wild-type and Nif-mutant strains. Mol. Plant-Microbe Interact. 14:358-366. [DOI] [PubMed] [Google Scholar]
- 18.Shishido, M., C. Breuil, and C. P. Chanway. 1999. Endophytic colonization of spruce by plant growth-promoting rhizobacteria. FEMS Microbiol. Ecol. 29:191-196. [Google Scholar]
- 19.Shishido, M., B. M. Loeb, and C. P. Chanway. 1995. External and internal root colonization of lodgepole pine seedlings by two growth-promoting Bacillus strains originated from different root microsites. Can. J. Microbiol. 41:707-713. [Google Scholar]
- 20.Siciliano, S. D., N. Fortin, A. Mihoc, G. Wisse, S. Labelle, D. Beaumier, D. Ouellette, R. Roy, L. G. Whyte, M. K. Banks, P. Schwab, K. Lee, and C. W. Greer. 2001. Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl. Environ. Microbiol. 67:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturz, A. V., and B. R. Christie. 1995. Endophytic bacterial systems governing red clover growth and development. Ann. Appl. Biol. 126:285-290. [Google Scholar]
- 22.Sturz, A. V., B. R. Christie, B. G. Matheson, W. J. Arsenault, and N. A. Buchanan. 1999. Endophytic bacterial communities in the periderm of potato tubers and their potential to improve resistance to soil-borne pathogens. Plant Pathol. 48:360-369. [Google Scholar]
- 23.Sturz, A. V., B. R. Christie, and J. Nowak. 2000. Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit. Rev. Plant Sci. 19:1-30. [Google Scholar]
- 24.Sturz, A. V., and J. Nowak. 2000. Endophytic communities of rhizobacteria and the strategies required to create yield enhancing associations with crops. J. Appl. Soil Ecol. 15:183-190. [Google Scholar]
- 25.Tester, C. F. 1992. Influence of a genetically modified endophytic bacterium on composition and decomposition of corn residue. Soil Biol. Biochem. 24:1107-1112. [Google Scholar]
- 26.Van Peer, R., H. L. M. Punte, L. A. De Weger, and B. Schippers. 1990. Characterization of root surface and endorhizosphere pseudomonads in relation to their colonization of roots. Appl. Environ. Microbiol. 56:2462-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]