Abstract
Sphingomonas paucimobilis SYK-6 degrades ferulic acid to vanillin, and it is further metabolized through the protocatechuate 4,5-cleavage pathway. We obtained a Tn5 mutant of SYK-6, FA2, which was able to grow on vanillic acid but not on ferulic acid. A cosmid which complemented the growth deficiency of FA2 on ferulic acid was isolated. The 5.2-kb BamHI-EcoRI fragment in this cosmid conferred the transformation activity of ferulic acid to vanillin on Escherichia coli host cells. A sequencing analysis revealed the genes ferB and ferA in this fragment; these genes consist of 852- and 2,127-bp open reading frames, respectively. The deduced amino acid sequence of ferB showed 40 to 48% identity with that of the feruloyl-coenzyme A (CoA) hydratase/lyase genes of Pseudomonas and Amycolatopsis ferulic acid degraders. On the other hand, the deduced amino acid sequence of ferA showed no significant similarity to the feruloyl-CoA synthetase genes of other ferulic acid degraders. However, the deduced amino acid sequence of ferA did show 31% identity with pimeloyl-CoA synthetase of Pseudomonas mendocina 35, which has been classified as a new superfamily of acyl-CoA synthetase (ADP forming) with succinyl-CoA synthetase (L. B. Sánchez, M. Y. Galperin, and M. Müller, J. Biol. Chem. 275:5794-5803, 2000). On the basis of the enzyme activity of E. coli carrying each of these genes, ferA and ferB were shown to encode a feruloyl-CoA synthetase and feruloyl-CoA hydratase/lyase, respectively. p-coumaric acid, caffeic acid, and sinapinic acid were converted to their corresponding benzaldehyde derivatives by the cell extract containing FerA and FerB, thereby indicating their broad substrate specificities. We found a ferB homolog, ferB2, upstream of a 5-carboxyvanillic acid decarboxylase gene (ligW) involved in the degradation of 5,5′-dehydrodivanillic acid. The deduced amino acid sequence of ferB2 showed 49% identity with ferB, and its gene product showed feruloyl-CoA hydratase/lyase activity with a substrate specificity similar to that of FerB. Insertional inactivation of each fer gene in S. paucimobilis SYK-6 suggested that the ferA gene is essential and that ferB and ferB2 genes are involved in ferulic acid degradation.
Lignin is the most abundant aromatic substance in the biosphere, and therefore its utilization could potentially be advantageous in many areas. One present method for making use of lignin is to convert it into useful chemical materials by the implementation of microbial lignin degradation systems. It is believed that lignin degradation is initiated by white rot fungi, which secrete extracellular degradation enzymes such as lignin peroxidase, manganese peroxidase, and laccase (8). The resulting low-molecular-weight lignin is further degraded and mineralized by bacteria (34). Bacterial lignin degradation systems consist of many unique and specific enzymes with the ability to catalyze the production of various useful compounds (12). Due their productivity, bacterial enzyme systems are expected to serve as useful tools for the conversion of lignin into intermediate metabolites.
Sphingomonas paucimobilis SYK-6 has the ability to degrade a wide variety of dimeric lignin compounds, including β-aryl ether, biphenyl, phenylcoumarane, diarylpropane, and pinoresinol (11, 12). We have already characterized the enzyme genes involved in the degradation of β-aryl ether and biphenyl (12, 20, 21). These dimeric lignin compounds are degraded to vanillic acid or syringic acid, and they are further degraded through the protocatechuate 4,5-cleavage pathway (9, 13, 14, 17).
In this study we focused on the degradation of ferulic acid, which is the precursor of lignin biosynthesis. Cinnamic acid derivatives, including ferulic acid and p-coumaric acid, play important roles in the cross-linking of the cell walls of various grasses and represent up to 1.5% of the weight of their cell walls. Cinnamic acid derivatives are therefore expected to become popular as an abundant class of bioresources.
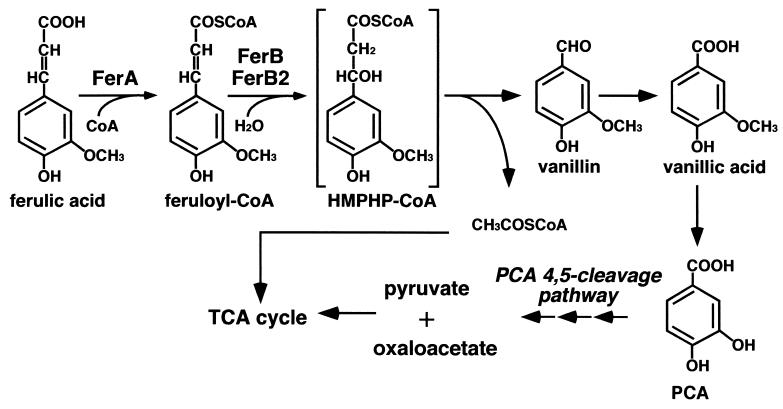
Two types of ferulic acid side chain cleavage have been reported for a number of microorganisms (24). One type is catalyzed by nonoxidative decarboxylase, which eliminates one carbon from the ferulic acid side chain, resulting in the formation of 4-hydroxy-3-methoxystyrene. The other type is characterized by the elimination of two carbons from the ferulic acid side chain, and this latter type can produce vanillin, a valuable flavor compound. For this reason the enzymes and the genes involved in the transformation of ferulic acid to vanillin have been the focus of much attention. Two enzymes involved in the side chain cleavage reaction have been reported for Pseudomonas fluorescens AN103 (7), Pseudomonas sp. strain HR199 (18, 23), Pseudomonas putida WCS358 (33), and Amycolatopsis sp. strain HR167 (1). The feruloyl-coenzyme A (CoA) synthetase catalyzes the transfer of CoA to the carboxyl group of ferulic acid, which then forms feruloyl-CoA in the presence of ATP and Mg2+ as cofactors. The resulting feruloyl-CoA is degraded by the feruloyl-CoA hydratase/lyase, which hydrates feruloyl-CoA to form 4-hydroxy-3-methoxyphenyl-β-hydroxypropionyl-CoA and cleaves 4-hydroxy-3-methoxyphenyl-β-hydroxypropionyl-CoA to produce vanillin and acetyl-CoA (Fig. 1). Although the catabolic pathway of ferulic acid is well characterized, little is known about the diversity of the degradation genes.
FIG. 1.
Ferulic acid catabolic pathway of S. paucimobilis SYK-6. Feruloyl-CoA synthetase (FerA) and feruloyl-CoA hydratases/lyases (FerB and FerB2) catalyze the side chain cleavage of ferulic acid to give vanillin and acetyl-CoA. Vanillin is converted to pyruvate and oxaloacetate through the protocatechuate (PCA) 4,5-cleavage pathway.
In this study we isolated a novel type of the feruloyl-CoA synthetase gene and two feruloyl-CoA hydratase/lyase genes from S. paucimobilis SYK-6. The substrate specificities of the gene products and the role of each gene in ferulic acid degradation by SYK-6 were characterized.
(This study was presented in part at the 100th General Meeting of American Society for Microbiology, Los Angeles, Calif., 2000).
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. S. paucimobilis SYK-6 was grown at 30°C in W minimal medium (20) containing 10 mM vanillic acid or ferulic acid or in Luria-Bertani (LB) medium (Bacto Tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 5 g/liter).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| S. paucimobilis | ||
| SYK-6 | Wild type; Nalr Smr | 11 |
| FA2 | A Tn5 mutant of SYK-6, vanillic acid+ and ferulic acid deficient | This study |
| FAK | Mutant derivative of SYK-6; Kmr gene insertion mutant of ferA; Nalr Smr Kmr | This study |
| FBK | Mutant derivative of SYK-6; Kmr gene insertion mutant of ferB; Nalr Smr Kmr | This study |
| FB2K | Mutant derivative of SYK-6; Kmr gene insertion mutant of ferB2; Nalr Smr Kmr | This study |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thiΔ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | 35 |
| S17-1 | recA; harboring the tra genes of plasmid RP4 in the chromosome, proA thi | 31 |
| HB101 | supE44 hsdS20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 4 |
| Plasmids | ||
| pSUP5011 | pBR325::Tn5-mob Apr Kmr Cmr | 31 |
| pVK100 | Broad-host-range cosmid vector, Kmr Tetr | 5 |
| pRK2013 | Kmr Tra+ Mob+ | 6 |
| pTS1210 | Broad-host-range vector, pSa ori pBR ori, Kmr Ampr | T. Nakazawa |
| pTS1210MCS | pTS1210 with multicloning site from pUC19 into EcoRI and HindIII sites | This study |
| pBluescript II KS(+) and SK(+) | Cloning vectors; Apr | 30 |
| pUC18 | Cloning vector; Apr | 35 |
| pUC4K | Apr Kmr | 32 |
| pK19mobsacB | oriT sacB Kmr | 29 |
| pKYO2 | pVK100 with an approx 20-kb fragment carrying ferBA | This study |
| pBAB3 | SK(+) with a 5.2-kb BamHI-EcoRI fragment of pKYO2 | This study |
| pBAB19 | SK(+) carrying the same fragment as pBAB3 in the opposite direction | This study |
| pKB4 | KS(+) carrying the same fragment as pBAB3 in the opposite direction | This study |
| pKH128 | Deletion derivative of pBAB3 | This study |
| pKHR201, pKHR128, pKHR126, pKHR213 | Deletion derivatives of pKB4 | This study |
| pKHR201E | pKHR201; a 2.7-kb EcoRI fragment in the insert was deleted | This study |
| pAH41 | KS(+) with a 1.2-kb DNA fragment carrying ferB2 | This study |
| pUCBam | pUC18 with a 5.2-kb BamHI-EcoRI fragment from pBAB3 | This study |
| pUCBamKm | pUCBam with insertion of the Kmr gene from pUC4K into an ApaI site | This study |
| pSCFA | pK19mobsacB with a 6.5-kb BamHI fragment from pUCBamKm | This study |
| pXESK | SK(+) with a 1.0-kb XhoI-EcoRI fragment from pKHR201 | This study |
| pXESKKm | pXESK with insertion of the Kmr gene from pUC4K into a SalI site | This study |
| pSCFB | pK19mobsacB with a 2.3-kb KpnI-EcoRI fragment from pXESKKm | This study |
| pKS7E2 | KS(+) with a 7.0-kb EcoRI fragment carrying ligW and ferB2 | 22 |
| pKS7E2Km | pKS7E2 with insertion of the Kmr gene from pUC4K into a BstXI site | This study |
| pSCFB2 | pK19mobsacB with an 8.3-kb EcoRI fragment from pKS7E2Km | This study |
| pBABX | pTS1210MCS with a 4.0-kb XhoI-EcoRI fragment carrying ferA | This study |
Chemicals.
Cinnamic acid derivatives, including ferulic acid, caffeic acid, sinapinic acid, cinnamic acid, p-coumaric acid, o-coumaric acid, m-coumaric acid, 3-methoxycinnamic acid, and 4-methoxycinnamic acid, were purchased from Tokyo Kasei Kogyo Co. (Tokyo, Japan). Benzaldehyde derivatives and other chemicals were purchased from Tokyo Kasei Kogyo Co., Wako Pure Chemical Industries (Osaka, Japan), or Sigma Chemical Company (St. Louis, Mo.)
Analysis of the metabolites.
S. paucimobilis SYK-6 and its disruption mutants were grown to an optical density at 600 nm (OD600) of 1.0 in LB medium at 30°C. Cells were washed twice with W medium and suspended to an OD600 of 0.2 in 10 ml of the same medium. After the addition of ferulic acid to a final concentration of 10 mM, the mixtures were shaken at 30°C. A portion of the cultures (200 μl) was collected every 3 h from 0 to 48 h and were acidified with hydrochloric acid to a pH value of 1. The metabolites were extracted with 200 μl of ethyl acetate, and then the extract was dried in vacuo and was trimethylsilylated. The resultant samples were analyzed by gas chromatography-mass spectrometry analysis (GC-MS) by using a model 5971A (Hewlett-Packard Co., Palo Alto, Calif.) with an Ultra-2 capillary column (50 m by 0.2 mm; Hewlett-Packard Co.). The analytical conditions for GC-MS were the same as those described previously (14).
Isolation of a Tn5 insertion mutant and cloning of the genes.
Tn5 insertion mutants of S. paucimobilis SYK-6 were generated by using pSUP5011, which was transferred from Escherichia coli S17-1 to S. paucimobilis SYK-6 by conjugation. A mutant which grows on vanillic acid but not on ferulic acid was selected in the following way. Tn5 insertion mutants of SYK-6 were grown in LB to an OD600 of 1.0 at 30°C. Cells were then washed twice with W medium and suspended to an OD600 of 0.2 in 10 ml of the same medium. After addition of ferulic acid to a final concentration of 10 mM the culture was shaken at 30°C. To enrich cultures for target mutants unable to grow with ferulic acid by the method of pencillin screening, ampicillin (200 mg/liter) and d-cycloserine (100 mg/liter) were added to the culture when the OD600 reached 0.4, and the culture was then incubated for 48 h. The resultant cells were inoculated on W medium containing either ferulic acid or vanillic acid, and then a mutant was selected.
A gene library of SYK-6 constructed with pVK100 as a vector was introduced into a mutant strain by conjugation. The resulting transconjugants were plated on W medium containing 10 mM ferulic acid.
Preparation of cell extracts.
E. coli JM109 harboring each plasmid was grown in 200 ml of LB medium containing 100 mg of ampicillin/liter. When the OD600 of the culture reached 0.5, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. After 5 h of incubation, cells were harvested by centrifugation and ruptured by passage though a French pressure cell in 100 mM Tris-HCl buffer (pH 7.5). The cell lysate was centrifuged at 15,000 × g, and the supernatant was collected. Streptomycin was added to a final concentration of 1% to the supernatant, and the resultant supernatant was incubated on ice for 10 min and centrifuged at 15,000 × g for 15 min to remove nucleic acids. The protein concentration was measured by a protein assay kit with bovine serum albumin as the standard (Bio-Rad, Hercules, Calif.).
SDS-PAGE and N-terminal amino acid sequencing.
The expression of the enzymes was examined by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE). To determine the N-terminal amino acid sequence the cell extract was subjected to SDS-PAGE and was electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad). The enzyme band was cut out and analyzed on a PPSQ-21 protein sequencer (Shimadzu Co., Kyoto, Japan).
Enzyme assay.
Substrate was added to a final concentration of 500 μM to 1 ml of 100 mM Tris-HCl buffer (pH 7.5) containing 0.2 mM CoA, 2.5 mM MgSO4, 2.5 mM ATP, and crude extract of E. coli JM109 harboring pKHR201 carrying the ferBA genes (400 μg of protein). After incubation for 3 min at 25°C, the mixture was extracted and analyzed by GC-MS as described above. The ferulic acid transformation activity (FerA activity) was estimated by the decrease in the amount of the substrate. The total activity of FerA and FerB was estimated by the increase in the amount of product generated by the successive reactions catalyzed by FerA and FerB.
To examine the metal ion dependency of FerA, EDTA or a metal salt solution containing either MgSO4 · 7H2O, Na2SO4 · 10H2O, FeSO4 · 7H2O, Fe2(SO4)3 · nH2O, CuSO4 · 5H2O, CaSO4 · 2H2O, CoSO4 · 7H2O, ZnSO4 · 7H2O, or MnSO4 · 4H2O was added to a final concentration of 5 mM for EDTA or 2.5 mM for the metal salt solution to 1 ml of the reaction mixture. Ferulic acid was added to a final concentration of 500 μM to initiate the reaction, and feruloyl-CoA synthetase activity (FerA activity) was determined by measuring the absorbance at 352 nm derived from the generation of feruloyl-CoA with a DU-7500 spectrophotometer (Beckman, Fullerton, Calif.).
DNA manipulation and nucleotide sequencing.
DNA manipulations were carried out essentially as described elsewhere (2, 26). A Kilosequence kit (Takara Shuzo Co., Ltd., Kyoto, Japan) was used to construct a series of deletion derivatives. The nucleotide sequences were determined by the dideoxy termination method (28) with an ALFexpress DNA sequencer (Pharmacia Biotech, Milwaukee, Wis.). A Sanger reaction was carried out by using a Thermosequenase fluorescence-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Sequencing analysis and homology alignment were carried out with the GeneWorks programs (IntelliGenetics, Inc., Mountain View, Calif.). Multiple sequence alignment was produced by using the program CLUSTAL W, and the phylogenetic tree was inferred from the alignments by using the neighbor-joining method (25). Graphics for phylogenetic trees were produced by using the TreeView program (19). The DDBJ databases were used for searching homologous proteins. Southern hybridization analysis was done with the DIG System (Roche Molecular Biochemicals, Mannheim, Germany).
Insertional inactivation of the ferulic acid catabolic genes.
To disrupt each ferA, ferB, and ferB2 in SYK-6, the kanamycin resistance gene from pUC4K was inserted in each of the three genes by the gene replacement technique. The fer gene disrupted plasmids by using pK19mobsacB as a vector, and pSCFA, pSCFB, and pSCFB2 were introduced into E. coli S17-1 and then introduced into SYK-6 by conjugation. The kanamycin-resistant transformants were selected on an LB agar plate containing 50 mg of kanamycin/liter and 25 mg of nalidixic acid/liter. They were cultured for 12 h in LB liquid medium containing 10% sucrose. The candidates for mutants were isolated on an LB agar plate containing 10% sucrose and kanamycin in order to select the cells in which the sacB-containing vector portion was deleted by a double crossover.
To examine the disruption of each gene, Southern hybridization analysis was carried out. The total DNA of the candidates for ferA, ferB, and ferB2 mutants were digested with EcoRI, BamHI and EcoRI, and SalI, respectively. The 1.3-kb SalI fragment with kanamycin resistance, the 3.0-kb EcoRI fragment carrying ferA, the 1.0-kb XhoI-EcoRI fragment carrying ferB, and the 2.6-kb SalI fragment carrying ferB2 were labeled with the DIG System and were used as probes.
Nucleotide sequence accession numbers.
The nucleotide sequences of ferBA and ferB2 have been deposited in the DDBJ, EMBL, and GenBank sequence databases under accession number nos. AB072376 and AB072377, respectively.
RESULTS
Cloning of the ferulic acid catabolic genes.
To determine the ferulic acid catabolic pathway, S. paucimobilis SYK-6 was grown in 10 ml of W medium containing 10 mM ferulic acid for 12 h. The culture was acidified and extracted with ethyl acetate. GC-MS analysis of the trimethylsilyl (TMS) derivative of the metabolite showed the production of a compound with a retention time of 23.4 min (data not shown). Its retention time and the mass spectrum corresponded to those of authentic TMS-vanillic acid. This result suggests that the two carbons of ferulic acid side chain were eliminated by SYK-6.
Tn5 mutagenesis with pSUP5011 was used to obtain the insertion mutant of SYK-6, which was unable to degrade ferulic acid. Tn5 mutants of SYK-6 were subsequently screened for the ability to grow on vanillic acid, but not on ferulic acid, as the sole carbon and energy source. A mutant strain, designated FA2, was obtained. GC-MS analysis indicated that this strain completely lost the ability to degrade ferulic acid. FA2 was used for a complementation test to isolate a wild-type copy of the mutated gene(s). The pVK100 cosmid library carrying the partial SalI-digested fragments of SYK-6 total DNA was introduced from E. coli HB101 to FA2 by triparental mating. Four cosmids, each containing approximately 20- to 30-kb DNA fragments, were obtained as they complemented the growth deficiency of FA2 on ferulic acid. A subcloning experiment of a cosmid, pKYO2, indicated that a 5.2-kb BamHI-EcoRI fragment included in all the cosmids was responsible for this complementation.
We identified the reaction product of ferulic acid incubated with the cell extract of E. coli JM109 harboring pBAB3 carrying the 5.2-kb BamHI-EcoRI fragment by GC-MS. A gas chromatogram of the TMS derivatives of the reaction product showed a peak with a retention time of 16.2 min. The retention time and mass spectrum of this compound corresponded to those of a TMS derivative of authentic vanillin (data not shown). Gasson et al. (7) and Overhage et al. (18) have reported that two enzymes, feruloyl-CoA synthetase and feruloyl-CoA hydratase/lyase, are involved in the sequential reactions for the conversion of ferulic acid to vanillin in P. fluorescens AN103 and Pseudomonas sp. strain HR199, respectively. Feruloyl-CoA synthetase catalyzes the conversion of ferulic acid to feruloyl-CoA, which has an absorption maximum at approximately 350 nm at pH 7.5, in the presence of CoA, ATP, and Mg2+. We confirmed the increase in absorbance at 352 nm of the reaction mixture with 100 mM Tris-HCl buffer (pH 7.5) containing ferulic acid and the cell extract of E. coli harboring pBAB3 only in the presence of CoA, ATP, and Mg2+. These results suggested that the 5.2-kb BamHI-EcoRI fragment encodes both feruloyl-CoA synthetase and feruloyl-CoA hydratase/lyase.
Nucleotide sequence of the feruloyl-CoA synthetase and feruloyl-CoA hydratase/lyase genes.
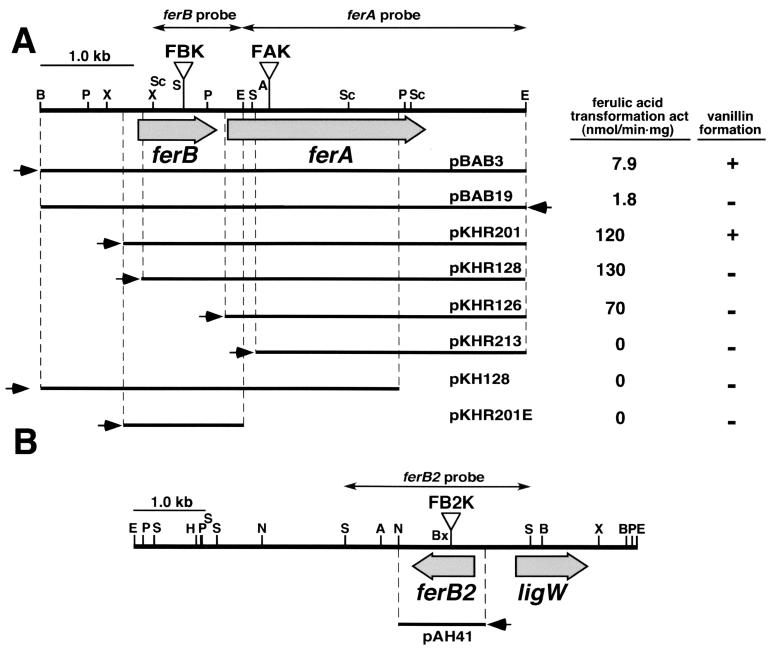
A series of subclones of the 5.2-kb BamHI-EcoRI fragment was generated by using restriction enzymes and exonuclease III. In order to map the region involved in ferulic acid degradation, the ferulic acid transformation activity and vanillin formation ability from ferulic acid were determined in the cell extract of each E. coli JM109 harboring a deletion plasmid (Fig. 2A). Deletion of the 1.2-kb region from the 5′ end of the 5.2-kb BamHI-EcoRI fragment resulted in a deficiency of vanillin formation ability (pKHR128). However, this mutant retained the ferulic acid transformation activity. Further deletion (pKHR213) resulted in the complete loss of the ferulic acid transformation activity. E. coli harboring pBAB19, which carries the same fragment as pBAB3 but in the opposite direction, showed no vanillin formation activity and showed 23% of the ferulic acid transformation activity of the recombinant containing pBAB3. These results suggest that there are feruloyl-CoA synthetase and feruloyl-CoA hydratase/lyase genes carried by pKHR201. The direction of the transcription of these genes is the same as that of the lac promoter in pKHR201, and the feruloyl-CoA hydratase/lyase gene is located upstream of the feruloyl-CoA synthetase gene.
FIG. 2.
Deletion analysis of the 5.2-kb BamHI-EcoRI fragment carrying ferBA (A) and the restriction map of the 7.0-kb EcoRI fragment carrying ferB2 and ligW (B). The ferulic acid transformation activity and the ability to generate vanillin from ferulic acid of E. coli JM109 harboring each plasmid are presented on the right-hand side of panel A. Small arrows indicate the direction of transcription from the lac promoter. Triangles indicate the positions of the Kmr gene insertion of the ferA mutant (FAK), ferB mutant (FBK), and ferB2 mutant (FB2K). Double-headed arrows indicate the probes used for Southern hybridization analysis of the fer gene mutants. The following genes (and their products) were used: ferA, feruloyl-CoA synthetase; ferB and ferB2, feruloyl-CoA hydratase/lyase; ligW, 5-carboxyvanillic acid decarboxylase. Abbreviations: A, ApaI; B, BamHI; Bx, BstXI; E, EcoRI; H, HindIII; N, NotI; P, PstI; S, SalI; Sc, SacI; X, XhoI.
The nucleotide sequence of the insert DNA of pKHR201 was determined, and two open reading frames (ORFs) consisting of 852 and 2,127 bp, designated ferB and ferA, respectively, were found (Fig. 2A). A homology search of the DDBJ database revealed that the deduced amino acid sequence of ferB showed 40 to 48% identity with those of the feruloyl-CoA hydratase/lyase genes of the ferulic acid degraders P. fluorescens AN103 (ORFA) (7), Pseudomonas sp. strain HR199 (Ech) (18), P. putida WCS358 (Fca) (33), and Amycolatopsis sp. strain HR167 (1). On the other hand, the deduced amino acid sequence of ferA showed no significant similarities with those of the feruloyl-CoA synthetase genes of Pseudomonas sp. HR199 and Amycolatopsis sp. strain HR167. In addition, the FerA amino acid sequence did show 31% identity with pimeloyl-CoA synthetase of Pseudomonas mendocina 35, which is involved in biotin synthesis (3).
Identification of the ferB homolog.
Recently we characterized the 5-carboxyvanillic acid decarboxylase gene (ligW) of S. paucimobilis SYK-6 (22), which is involved in the degradation of a lignin-related biphenyl compound, 5,5′-dehydrodivanillic acid (20, 21). The nucleotide sequencing of the 7.0-kb EcoRI fragment of pKS7E2 carrying ligW revealed a divergent transcribed ORF of 831 bp, which was located 575 bp upstream of ligW (Fig. 2B). The deduced amino acid sequence of this ORF revealed a 49% identity with that of ferB and was designated as ferB2.
Expression of ferA, ferB, and ferB2 in E. coli.
The ferA gene expression induced by IPTG in E. coli JM109 was examined. A 70-kDa protein was observed by SDS-PAGE of the cell extracts from E. coli that harbored pKHR201 carrying ferBA and that harbored pKHR126 carrying ferA (data not shown). The size of the protein was in good agreement with the value calculated from the deduced amino acid sequence of ferA (73,778 Da). N-terminal amino acid sequencing of the 70-kDa protein revealed the sequence TVEAGVRPQAGARDINRL, which corresponded to the deduced N-terminal amino acid sequence of ferA, except for the first Met, which appeared to be processed.
The ferB and ferB2 gene expressions induced by IPTG in E. coli JM109 were examined with pKHR201 and pKHR201E carrying ferB and with pAH41 carrying ferB2, respectively. SDS-PAGE showed the expression of 34- and 32-kDa proteins in E. coli carrying ferB and ferB2, respectively (data not shown). These sizes are in good agreement with the value calculated from the deduced amino acid sequences of ferB (32,463 Da) and ferB2 (30,828 Da). N-terminal amino acid sequencing of these proteins was carried out. Only the sequence for a 32-kDa protein expressed in E. coli harboring pAH41 was determined: SDELTXETVXXTLDDGIA. This sequence corresponded to the deduced amino acid sequence of ferB2. The first Met of FerB2 also appeared to be processed.
Substrate specificities of FerA, FerB, and FerB2.
The feruloyl-CoA synthetase activity of E. coli harboring pKHR126 was detected only in the presence of 0.2 mM CoA, 2.5 mM Mg2+, and 2.5 mM ATP. Replacement of Mg2+ in the reaction mixture with 2.5 mM of Mn2+ or Co2+ also showed 94 or 39% of activity obtained with 2.5 mM Mg2+, respectively. However, FerA had no activity in the presence of Cu2+, Fe2+, Fe3+, Zn2+, and Ca2+.
To determine the substrate specificity of FerA and FerB, 0.5 mM cinnamic acid derivatives were incubated with the cell extract of E. coli JM109 harboring pKHR201 carrying ferBA in the presence of CoA, ATP, and Mg2+. The amount of the substrate remained, and the corresponding benzaldehyde formed was estimated by GC-MS. FerA transformed ferulic acid, p-coumaric acid, caffeic acid, and sinapinic acid (3,5-dimethoxy-4-hydroxycinnamic acid) at a similar rate, and 4-methoxycinnamic acid was also transformed at half the rate of ferulic acid transformation (Table 2). Formation of the FerB reaction products, 4-hydroxybenzaldehyde, protocatechualdehyde, and syringaldehyde, was confirmed in the reaction mixtures from p-coumaric acid, caffeic acid, and sinapinic acid, respectively. The formation rates of these benzaldehydes from cinnamic acid derivatives were at almost the same level (approximately 20 nmol/min·mg of protein). However, the formation of 4-methoxybenzaldehyde from 4-methoxycinnamic acid was not observed. The substrate specificity of FerB2 was also determined by the incubation of cinnamic acid derivatives with the cell extracts of E. coli harboring pAH41 carrying ferB2 and E. coli containing pKHR126 carrying ferA. Formation of vanillin, p-hydroxybenzaldehyde, protocatechualdehyde, and syringaldehyde from the corresponding cinnamic acid derivatives were found, thus suggesting a substrate specificity of FerB2 similar to that of FerB.
TABLE 2.
Substrate specificity of FerAaa
| Substrate | Transformation activity ± SD (nmol/min·mg) |
|---|---|
| Ferulic acid | 120 ± 14 |
| Caffeic acid | 99 ± 17 |
| Sinapinic acid | 130 ± 25 |
| Cinnamic acid | 0 |
| p-Coumaric acid | 110 ± 23 |
| o-Coumaric acid | 0 |
| m-Coumaric acid | 0 |
| 3-Methoxycinnamic acid | 0 |
| 4-Methoxycinnamic acid | 58 ± 6 |
Cinnamic acid derivatives (0.5 mM) were incubated with the cell extract of E. coli JM109 harboring pKHR201 carrying ferBA in the presence of 0.2 mM CoA, 2.5 mM ATP, and 2.5 mM Mg2+. E. coli JM109 harboring pBluescript II KS(+) showed no transformation activity toward the substrates tested.
Characterization of the insertion mutants of the ferA, ferB, and ferB2 genes.
In order to examine the actual roles of the ferA, ferB, and ferB2 genes in the ferulic acid catabolism of S. paucimobilis SYK-6, these genes were inactivated by the insertion of kanamycin resistance genes by using the gene replacement technique based on homologous recombination (Fig. 2). The plasmids pSCFA, pSCFB, and pSCFB2, which contain the disrupted ferA, ferB, and ferB2 genes, respectively, were constructed (Table 1) and were introduced into SYK-6. The disruption of each gene was confirmed by Southern hybridization analysis by using each fer gene (Fig. 2) and the kanamycin resistance gene as probes (data not shown). The resulting ferA mutant (FAK) and ferB mutant (FBK) were unable to grow on ferulic acid, although they were able to grow on vanillic acid. However, it was found that FBK also lost the transformation activity of ferulic acid. This may have been due to a polar effect caused by the insertion of a kanamycin resistance gene in ferB. Introduction of pBABX carrying ferA in pTS1210MCS into FAK and FBK restored the growth of both strains on ferulic acid. These results indicate that the ferA gene is essential to the growth of S. paucimobilis SYK-6 on ferulic acid. In contrast, the ferB gene appears not to be essential to such growth. On the other hand, the disruption of the ferB2 gene did not affect the growth of the mutant (FB2K) on ferulic acid.
DISCUSSION
For some Pseudomonas strains and a gram-positive Amycolatopsis strain, ferulic acid has been shown to degrade to vanillin by the consecutive enzyme reactions catalyzed by feruloyl-CoA synthetase and feruloyl-CoA hydratase/lyase (1, 7, 18, 33). S. paucimobilis SYK-6 has similar yet different enzyme systems for the degradation of ferulic acid. We found the ferBA genes, which restored the growth deficiency of the Tn5 mutant of SYK-6, FA2, on ferulic acid; we also found the ferB homolog, ferB2, which was located upstream of ligW. The gene organization of ferB and ferA and the short intergenic distance (120 bp) suggest that these two genes constitute an operon. This gene organization is the same as that of Amycolatopsis sp. strain HR167 (1). The organization of these genes differs from that of Pseudomonas sp. strain HR199. The feruloyl-CoA synthetase gene (fcs) and feruloyl-CoA hydratase/lyase gene (ech) are separately localized in HR199 (18). However, ferB2 was located on a different locus in SYK-6. We determined the nucleotide sequence of the 7.0-kb EcoRI fragment carrying ferB2 and ligW in order to demonstrate the existence of another feruloyl-CoA synthetase gene; however, no enzyme gene related to acyl-CoA synthetase was found (data not shown).
Three findings led to the conclusion that the ferA gene encodes feruloyl-CoA synthetase. (i) The reaction product of ferulic acid catalyzed by FerA showed a specific absorption at 352 nm (pH 7.5), which is a spectral feature of feruloyl-CoA (7). When the reaction was carried out in 100 mM Tris-HCl buffer (pH 8.5), the reaction mixture became yellowish in color and revealed an absorption at 400 nm (data not shown). This result is in good agreement with the properties of feruloyl-CoA (15). (ii) The feruloyl-CoA synthetase activity was observed only when CoA, ATP, and Mg2+ were present. (iii) The deduced amino acid sequence of ferA was similar to that of an acyl-CoA synthetase gene, the pimeloyl-CoA synthetase gene (pauA) of P. mendocina 35 (3). The successful cloning of the feruloyl-CoA synthetase gene (fcs) was reported for Pseudomonas sp. strain HR199 (18) and Amycolatopsis sp. strain HR167 (1). The deduced amino acid sequence of the fcs genes showed similarity with those of the AMP-forming acyl-CoA synthetase genes. Interestingly, the deduced amino acid sequence of ferA did not show significant similarity to those of fcs and other AMP-forming acyl-CoA synthetase genes. Recently, Sánchez et al. (27) proposed a new superfamily of nucleoside diphosphate (NDP)-forming acyl-CoA synthetases. This superfamily contains pimeloyl-CoA synthetase, acetyl-CoA synthetase (ADP forming), succinyl-CoA synthetase, (both ADP and GDP forming), malyl-CoA synthetase, and ATP citrate lyase. BLAST searches revealed that the N-terminal (amino acid positions 1 to 532) and C-terminal (amino acid positions 457 to 709) amino acid sequences of FerA had 29% identity in a 457-amino-acid overlap and 37% identity in a 222-amino-acid overlap with the α-subunit (AcdA) and β-subunit (AcdB), respectively, of Pyrococcus furiosus acetyl-CoA synthetase (ADP forming) (16). These results suggest that FerA is an ADP-forming acyl-CoA synthetase and that the origin of ferA differs completely from that of the fcs genes of HR199 and HR167. In the case of NDP-forming acyl-CoA synthetases, a His residue of the enzyme is phosphorylated by nucleoside triphosphates to release NDP. This phosphoryl group transfers to the substrate, and then the phosphoryl group is replaced by CoA. On the basis of the alignment of the amino acid sequence of these enzymes with that of the α-subunit of succinyl-CoA synthetase of E. coli, which has been biochemically characterized, the His residue to be phosphorylated in the reaction was implicated. The amino acid sequence alignment of FerA with ADP-forming acyl-CoA synthetases suggested that His270 of FerA was phosphorylated during the reaction.
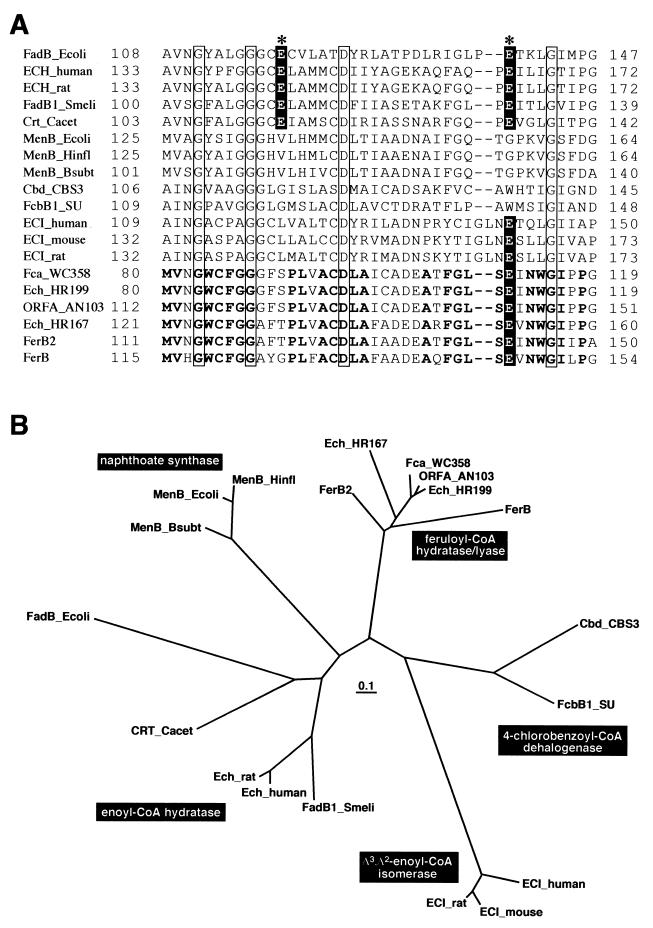
For the following reasons it was concluded that the ferB and ferB2 genes encode feruloyl-CoA hydratase/lyase. (i) Vanillin was produced from ferulic acid by the addition of FerB or FerB2 to the reaction mixture containing FerA. (ii) The deduced amino acid sequences of ferB and ferB2 were similar to those of the feruloyl-CoA hydratase/lyase genes of P. fluorescens AN103, P. putida WCS358, Pseudomonas sp. strain HR199, and Amycolatopsis sp. strain HR167. Gasson et al. (7) and Overhage et al. (18) reported that ORFA and Ech belong to a superfamily that includes enoyl-CoA hydratase, which is involved in fatty acid β-oxidation; this superfamily also includes a number of other proteins that catalyze, or are assumed to catalyze, the related reactions of CoA thiolesters. The two catalytic Glu residues have been implicated in the α-subunit (FadB) of the multienzyme complex of fatty acid oxidation from E. coli, which shows enoyl-CoA hydratase activity (10). The protonated Glu139 of FadB transfers a proton to the α-carbon of 2-trans-enoyl-CoA, and the deprotonated Glu119 attracts a proton from water, the oxygen of which launches a nucleophilic attack on the β-carbon of the substrate. Glu119 and Glu139 are well conserved in enoyl-CoA hydratase (Fig. 3A). As indicated by Gasson et al. (7), only a Glu residue corresponding to Glu139 of E. coli FadB is conserved among all of the feruloyl-CoA hydratases/lyases, including FerB (Glu146) and FerB2 (Glu142). These Glu residues may involve the transfer of a proton to the substrate in the feruloyl-CoA hydratase/lyase family. The phylogenetic tree of this superfamily indicated that FerB, FerB2, ORFA, Ech, and Fca consist of a subgroup of feruloyl-CoA hydratase/lyase (Fig. 3B). However, both FerB and FerB2 are rather distant from Pseudomonas and Amycolatopsis feruloyl-CoA hydratases/lyases.
FIG.3.
Comparison of the amino acid sequences of FerB and FerB2 with those of the enoyl-CoA hydratase/isomerase superfamily. (A) Partial amino acid sequence alignment of the region including the catalytic sites of E. coli FadB with the corresponding regions of the enoyl-CoA hydratase/isomerase superfamily. The N-terminal domain of FadB (α-subunit of the multienzyme complex of E. coli) showed enoyl-CoA hydratase and Δ3,Δ2-enoyl-CoA isomerase activities. Two Glu residues, indicated by asterisks, have been implicated as the catalytic sites for enoyl-CoA hydratase (10). Glu residues aligned with these catalytic Glu are offset by a black background. Amino acids conserved among all of the sequences are shown in squares. Boldface roman type indicates the conserved residues among all of the feruloyl-CoA hydratases/lyases. (B) Phylogenetic tree of FerB and FerB2 with the enoyl-CoA hydratase/isomerase superfamily. The scale corresponds to a genetic distance of 0.1 substitution per position (10% difference). Accession numbers for the sequences are as follows: FadB_Ecoli (to residue 285), P21177; ECH_human, short-chain enoyl-CoA hydratase of human mitochondria (D13900-1); ECH_rat, enoyl-CoA hydratase precursor of rat mitochondria (S06477); FadB1_Smeli, enoyl-CoA hydratase of Sinorhizobium meliloti (L39265); Crt_Cacet, 3-hydroxybutyryl-CoA dehydratase of Clostridium acetobutylicum ATCC824 (P52046); MenB_Ecoli, naphthoate synthase of E. coli (P27290); MenB_Hinfl, naphthoate synthase of Haemophilus influenzae Rd (U32777); MenB_Bsubt, naphthoate synthase of Bacillus subtilis (F69656); Cbd_CBS3, 4-chlorobenzoate dehalogenase of Pseudomonas sp. CBS-3 (A42560); FcbB1_SU, 4-chlorobenzoyl-CoA dehalogenase of Arthrobacter sp. SU (M93187); ECI_human, Δ3,Δ2-enoyl-CoA isomerase of human (L24774); ECI_mouse, Δ3,Δ2-enoyl-CoA isomerase of mouse (Z14049); ECI_rat, Δ3,Δ2-enoyl-CoA isomerase of rat (X61184); Fca_WCS358, ferulic acid hydratase of P. putida WCS358 (Y14772); Ech_HR199, enoyl-CoA hydratase of Pseudomonas sp. HR199 (Y11520); ORFA_AN103, p-hydroxycinnamoyl-CoA hydratase/lyase of P. fluorescens AN103 (Y13067); Ech_HR167, enoyl-CoA hydratase of Amycolatopsis sp. HR167 (AJ290449); FerB2, feruloyl-CoA hydratase/lyase of S. paucimobilis SYK-6; FerB, feruloyl-CoA hydratase/lyase of S. paucimobilis SYK-6.
There are few reports discussing the substrate specificity of the feruloyl-CoA synthetase for various cinnamic acid derivatives. Only one example of this enzyme is available for P. putida (36). This enzyme showed activity in the presence of ferulic acid, p-coumaric acid, and caffeic acid. In addition to these substrates, sinapinic acid, which had previously not been examined in P. putida, was converted to syringaldehyde by FerA and either FerB or FerB2 enzyme mixtures. Furthermore, a decrease in the amount of 4-methoxycinnamic acid by the FerA reaction was observed; however, the FerB reaction product, 4-methoxybenzaldehyde, was not detected. On the other hand, 3-methoxycinnamic acid and cinnamic acid did not serve as the substrate for FerA. These results suggest that certain functional groups, such as hydroxyl and methoxyl, at the para position of cinnamic acid derivatives are important for FerA enzyme activity. Viewed from the point of view of utilization of these enzymes for the biochemical conversion of cinnamic acid derivatives, the broad substrate spectra of SYK-6 feruloyl-CoA synthetase and feruloyl-CoA hydratase/lyase are potentially advantageous.
In order to elucidate the roles of these fer genes, each fer gene was disrupted by insertion of a kanamycin resistance marker gene. This technique revealed that the ferA gene was essential to ferulic acid degradation in this particular strain. On the other hand, ferB disruption also led to growth deficiency among the cells on ferulic acid. However, this growth deficiency was restored by the introduction of pTS1210MCS carrying the ferA gene, thereby indicating that the ferB gene was not essential to the growth on ferulic acid. The ferB2 gene is most likely expressed in the wild-type strain during the degradation of ferulic acid, and its gene product potentially complements feruloyl-CoA hydratase/lyase activity in the ferB mutant (FBK).
Acknowledgments
We thank T. Nakazawa for providing pTS1210 and S. Kajita for helpful discussion.
REFERENCES
- 1.Achterholt, S., H. Priefert, and A. Steinbuchel. 2000. Identification of Amycolatopsis sp. strain HR167 genes, involved in the bioconversion of ferulic acid to vanillin. Appl. Microbiol. Biotechnol. 54:799-807. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Binieda, A., M. Fuhrmann, B. Lehner, C. Rey-Berthod, S. Frutiger-Hughes, G. Hughes, and N. M. Shaw. 1999. Purification, characterization, DNA sequence and cloning of a pimeloyl-CoA synthetase from Pseudomonas mendocina 35. Biochem. J. 15:793-801. [PMC free article] [PubMed] [Google Scholar]
- 4.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 5.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figurshi, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasson, M. J., Y. Kitamura, W. R. McLauchlan, A. Narbad, A. J. Parr, E. L. H. Parsons, J. Payne, M. J. C. Rhodes, and N. J. Walton. 1998. Metabolism of ferulic acid to vanillin. J. Biol. Chem. 273:4163-4170. [DOI] [PubMed] [Google Scholar]
- 8.Gold, M. H., and M. Alic. 1993. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol. Rev. 57:605-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara, H., E. Masai, Y. Katayama, and M. Fukuda. 2000. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6950-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, X.-Y., and S.-Y. Yang. 1997. Glutamate-119 of the large α-subunit is the catalytic base in the hydration of 2-trans-enoyl-coenzyme A catalyzed by the multienzyme complex of fatty acid oxidation from Escherichia coli. Biochemistry 36:11044-11049. [DOI] [PubMed] [Google Scholar]
- 11.Katayama, Y., S. Nishikawa, M. Nakamura, K. Yano, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1987. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi 33:77-79. [Google Scholar]
- 12.Masai, E., Y. Katayama, S. Nishikawa, and M. Fukuda. 1999. Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J. Ind. Microbiol. Biotechnol. 23:364-373. [DOI] [PubMed] [Google Scholar]
- 13.Masai, E., K. Momose, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 2000. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6651-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitra, A., Y. Kitamura, M. J. Gasson, A. Narbad, A. J. Parr, J. Payne, M. J. C. Rhodes, C. Sewter, and N. J. Walton. 1999. 4-hydroxycinnamoyl-CoA hydratase/lyase (HCHL), an enzyme of phenylpropanoid chain cleavage from Pseudomonas. Arch. Biochem. Biophys. 365:10-16. [DOI] [PubMed] [Google Scholar]
- 16.Musfeldt, M., M. Selig, and P. Schönheit. 1999. Acetyl coenzyme A synthetase (ADP forming) from the hyperthermophilic Archaeon Pyrococcus furiosus: identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J. Bacteriol. 181:5885-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noda, Y., S. Nishikawa, K. Shiozuka, H. Kadokura, H. Nakajima, K. Yoda, Y. Katayama, N. Morohoshi, T. Haraguchi, and M. Yamasaki. 1990. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 172:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overhage, J., H. Priefert, and A. Steinbüchel. 1999. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65:4837-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 20.Peng, X., T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara, and M. Fukuda. 1998. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng, X., E. Masai, Y. Katayama, and M. Fukuda. 1999. Characterization of the meta-cleavage compound hydrolase gene involved in degradation of the lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 65:2789-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng, X., E. Masai, H. Kitayama, K. Harada, Y. Katayama, and M. Fukuda. 2002. Characterization of the 5-carboxyvanillate decarboxylase gene and its role in lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 68:4407-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Priefert, H., J. Rabenhorst, and A. Steinbüchel. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 179:2595-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosazza, J. P., Z. Huang, L. Dostal, T. Volm, and B. Rousseau. 1995. Review: biocatalytic transformations of ferulic acid: an abundant aromatic natural product. J. Ind. Microbiol. 15:457-471. [DOI] [PubMed] [Google Scholar]
- 25.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Sánchez, L. B., M. Y. Galperin, and M. Müller. 2000. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of Acyl-CoA synthetase (nucleotide diphosphate-forming). J. Biol. Chem. 275:5794-5803. [DOI] [PubMed] [Google Scholar]
- 28.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schäfer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 30.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. λ ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-mob transposon. Mol. Gen. Genet. 196:413-420. [DOI] [PubMed] [Google Scholar]
- 32.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venturi, V., F. Zennaro, G. Degrassi, B. C. Okeke, and C. V. Bruschi. 1998. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 144:965-973. [DOI] [PubMed] [Google Scholar]
- 34.Vicuña, R. 1988. Bacterial degradation of lignin. Enzyme Microbiol. Technol. 10:646-655. [Google Scholar]
- 35.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 36.Zenk, M. H., B. Ulbrich, J. Busse, and J. Stöckigt. 1980. Procedure for the enzymatic synthesis and isolation of cinnamoyl-CoA thioesters using a bacterial system. Anal. Biochem. 101:182-187. [DOI] [PubMed] [Google Scholar]