Abstract
We constructed a novel cell-surface display system, using as a new type of cell-wall anchor 3,297 or 4,341 bp of the 3′ region of the FLO1 gene (FS or FL gene, respectively), which encodes the flocculation functional domain of Flo1p. In this system, the N terminus of the target protein was fused to the FS or FL protein and the fusion proteins were expressed under the control of the inducible promoter UPR-ICL (5′ upstream region of the isocitrate lyase of Candida tropicalis). Using this new system, recombinant lipase with a pro sequence from Rhizopus oryzae (rProROL), which has its active site near the C terminus, was displayed on the cell surface. Cell-surface display of the FSProROL and FLProROL fusion proteins was confirmed by immunofluorescence microscopy and immunoblotting. Lipase activity reached 145 IU/liter (61.3 IU/g [dry cell weight]) on the surface of the yeast cells, which successfully catalyzed the methanolysis reaction. Using these whole-cell biocatalysts, methylesters synthesized from triglyceride and methanol reached 78.3% after 72 h of reaction. To our knowledge, this is the first example of cell-surface display of lipase with high activity. Interestingly, the yeast cells displaying the FLProROL protein showed strong flocculation, even though the glycosylphosphatidylinositol anchor attachment signal and cell-membrane-anchoring region of Flo1p had been deleted from this gene. The cell-surface display system based on FL thus endows the yeast strain with both novel enzyme display and strong flocculation ability.
Cell-surface display of heterologous proteins by using microorganisms has been widely utilized in various areas (2, 6, 23). For instance, cell-surface display of enzymes such as glucoamylase and cellulase on bacteria (4, 5, 12) and yeast (16, 17, 18) has been found effective for the preparation of whole-cell biocatalysts. Antigenic proteins have also been displayed on the surface of yeast cells for the development of vaccines (20, 21, 29). Among microorganisms, yeast cells are suitable because of their safety, simplicity of genetic manipulation, and rigidity of cell-wall structure (11).
In the most widely used yeast-based cell-surface display system, the gene encoding the target protein with the secretion signal is fused with the gene encoding the C-terminal half of α-agglutinin and containing the putative glycosylphosphatidylinositol (GPI) anchor attachment signal sequence. However, the activity of enzymes whose active site is spatially near to the C terminus (1) may be inhibited by fusion with an anchor protein, as in the results of a previous study, in which Rhizopus oryzae lipase (ROL) activity was strongly inhibited by fusion with a GPI anchor protein (30).
For surface display of such enzymes as ROL in active form, we have developed a new system, based on the FLO1 gene encoding a lectin-like cell-wall protein (Flo1p) in Saccharomyces cerevisiae. Flo1p is composed of several domains: the secretion signal domain, the flocculation functional domain, the GPI anchor attachment signal domain, and the membrane-anchoring domain (15, 26, 31). The Flo1p flocculation functional domain, thought to be located near the N terminus, recognizes and adheres noncovalently to cell-wall components such as α-mannan carbohydrates, causing reversible aggregation of cells into flocs (3, 15, 31). We have therefore developed a new cell-surface display system, consisting of the flocculation functional domain of Flo1p with secretion signal and insertion sites for the target protein. In this system, the N termini of target proteins such as ROL (i.e., with a pro sequence [ProROL]), are fused to the Flo1p flocculation functional domain.
In the present study, the applicability of yeast cells displaying active lipase to biodiesel production in a solvent-free and water-containing system was studied. Biodiesel fuel refers to methylesters (MEs) synthesized from natural triglycerides and methanol (8, 10). Since biodiesel is a clean fuel (28) and can be produced from waste oil, the development of an efficient biodiesel fuel production process using lipase (19, 22) is considered of great importance to help overcome environmental problems by utilizing renewable nonpetroleum sources of fuel.
MATERIALS AND METHODS
Strains and media.
The FLO1 gene was cloned from S. cerevisiae ATCC 60715 (MATa FLO8 his4 leu2 STA1). The ProROL gene was cloned from R. oryzae IFO4697. S. cerevisiae MT8-1 (MATa ade2 his3 leu2 trp1 ura3) was used for production of Flo1-ProROL fusion proteins (24). Yeasts were grown in 100 ml of complete medium (1% yeast extract, 2% peptone, 2% glucose) or selective medium (SD: 0.67% yeast nitrogen base, supplemented with appropriate amino acids and nucleotides, and 2% glucose unless otherwise noted) in a 500-ml shake flask shaken at 150 strokes/min at 30°C. For the plate medium, 2% agar was added.
Construction of FLO1-ProROL surface expression vectors.
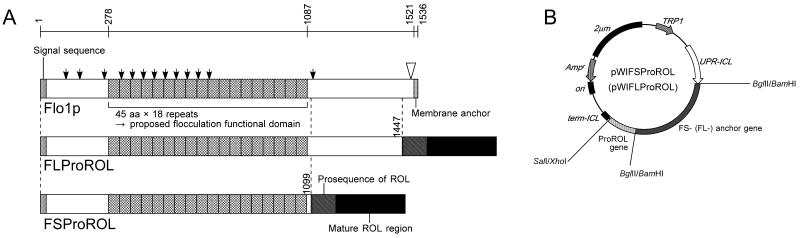
Figure 1 shows the newly developed cell-surface display systems using the flocculation functional domain of Flo1p (FS and FL). To investigate the effect of the length of Flo1p on expression, the genes encoding amino acids 1 to 1099 (FS) and 1 to 1417 (FL) of the flocculation functional region of Flo1p were used. For efficient expression of the fusion genes of FSProROL and FLProROL on the yeast cell surface, the plasmids pWIFSProROL and pWIFLProROL (Fig. 1) were constructed. Both plasmids have the secretion signal sequence, and the fusion gene of the flocculation functional regions of the FLO1 gene and the ProROL gene was expressed under the control of the 5′ upstream region of the isocitrate lyase of the Candida tropicalis (UPR-ICL) gene. UPR-ICL-mediated transcription is strongly induced by either glucose exhaustion or a nonfermentable carbon source such as ethanol or acetate (9, 27). Neither constructed plasmid possesses the GPI anchor attachment signal on the C terminus of the FLO1 gene.
FIG. 1.
(A) Structural features of Flo1p (31), FLProROL, and FSProROL. Arrowheads indicate possible N-glycosylation sites. Open arrowhead indicates the possible GPI attachment site (amino acid 1514). Numbers written vertically indicate lengths of proteins. The 45-amino-acid repeats comprise the proposed flocculation functional domain. (B) Yeast expression plasmids pWIFSProROL and pWIFLProROL.
To amplify the FS gene from S. cerevisiae ATCC 60715 chromosomal DNA, the following two oligonucleotides were used as primers: FSBam (5′-ACATGGATCCATGACAATGCCTCATCGCTATATGTTTTTG-3′) and FS-rvBgl (5′-GATAGATCTGGTGATTTGTCCTGAAGATGATGATGACAAA-3′). The FL gene was also amplified using FSBam and Flo1Xp (5′-AATGCCTCGAGTTAAATAATTGCCAGCAATAAGGACGCAATGAAG-3′). PCR was carried out using KOD Plus polymerase (Toyobo Co. Ltd., Osaka, Japan). Amplified fragments were ligated into the multicopy expression plasmid pWI3 using the following procedure. pWI3, containing the UPR-ICL promoter, was digested with BglII. Subsequently, the FS and FL fragments, digested with BamHI and BglII, were inserted into the digested plasmid, and the resulting plasmids were named pWIFS and pWIFL, respectively. The ProROL gene was amplified from R. oryzae IFO4697 chromosomal DNA using two synthesized primers, ICs (5′-CTCCGGATCCATGGTTCCTGTTTCTGGTAAATCTGGATCT-3′) and ROLrvSalI (5′-CGATGTCGACTTACAAACAGCTTCC-3′). The ProROL gene fragment, digested with BglII and SalI, was ligated into pWIFS and pWIFL and digested with BglII and XhoI, and the resulting plasmids were named pWIFSProROL and pWIFLProROL, respectively.
Yeast transformation.
The expression plasmids prepared as described above were transformed into S. cerevisiae cells using YEASTMAKER (Clontech Laboratories Inc., Palo Alto, Calif.) according to the protocol specified by the supplier. The transformants were selected by plating and incubating for 2 days on an SD medium plate.
Cultivation.
Transformants harboring the plasmids for cell-surface expression of FS- and FLProROL were precultivated in SD medium at 30°C for 30 h (optical density at 600 nm [OD600] > 1.5) and used as starters to inoculate 100 ml of SDC medium (SD medium containing 2% Casamino Acids) in a 500-ml shaking flask to give an initial OD600 of 0.03. Initial concentration of glucose was 0.5%.
Measurement of lipase activity on yeast cells.
The hydrolytic activity of lipase in culture broth and yeast cells was measured with Lipase Kit S (Dainippon Pharmaceutical Co., Osaka, Japan) according to the protocol specified by the supplier and the resulting values were expressed in international units (IU). One unit of lipase activity was defined as the amount of enzyme catalyzing the formation of 1 μmol of 2,3-dimercaptopropan-1-ol from 2,3-dimercaptopropan-1-ol tributyl ester per min. Lipase activity on the yeast cell surface was measured as follows. Yeast cells harvested from the culture broth were washed and resuspended in distilled water with vigorous agitation using a vortex. The cell concentration was determined by measuring dried-cell weight, and an appropriate amount of the suspension was used for lipase activity assay.
Immunofluorescence microscopy.
Immunofluorescence microscopy was carried out as reported previously (30). Immunostaining was performed as follows. The rabbit polyclonal anti-ROL antiserum was raised against recombinant ROL produced by S. cerevisiae (25) and used as the primary antibody. The antibody was preincubated with cells harboring a control plasmid (pWI3) to prevent nonspecific binding to the yeast cell. Cells were washed with 10 mM potassium phosphate buffer, pH 7.2, containing 150 mM sodium chloride (PBS). The cells and the pretreated antibody were then incubated with 2% bovine serum albumin at room temperature for 1.5 h. After the cells had been washed with PBS, the second antibody, fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (IgG) (Molecular Probes, Eugene, Oreg.), was diluted to 1:300 and allowed to react with the cells at room temperature for 1 h. The cells were then washed with PBS and observed with a fluorescence microscope.
Isolation of cell wall and protein extraction.
Cell wall fractions were separated according to the previously reported method (11), with minor modifications. Cells were harvested by centrifugation at 3,000 × g and washed with iced buffer (10 mM Tris-HCl [pH 7.8], 1 mM phenylmethylsulfonyl fluoride). The cells, buffer, and glass beads (0.5 mm diameter) were mixed at a ratio of 1:2:1 (wet weight/vol/wt) in a microcentrifugation tube and agitated vigorously with a vortex mixer at maximum speed for 5 min at 0°C. The cell wall fraction was recovered by centrifugation of the homogenate at 1,000 × g for 5 min and washed with the same buffer. Sodium dodecyl sulfate (SDS) extraction and subsequent glucanase extraction were carried out according to the previously reported methods (3).
Endo Hf treatment.
To remove N-glycosylated carbohydrates from glycoproteins, endoglycosidase treatment was conducted using Endo Hf (New England BioLabs, Beverly, Mass.) according to the protocol specified by the supplier. That is, 20 μg of proteins, extracted from yeast cell wall by SDS, was denatured in 0.5% SDS and 1% β-mercaptoethanol by 10 min of boiling and then incubated for 1 h at 37°C with 1/10 volume of 0.5 M sodium citrate (pH 5.5) and 2.5 μl of Endo Hf.
Western blot analysis of rProROL.
Using a 10% (wt/vol) gel, SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as described previously. The proteins separated on the gel were electroblotted on polyvinylidene difluoride membrane (Millipore Co., Boston, Mass.) and allowed to react with primary rabbit anti-ROL IgG antibodies (30) and secondary goat anti-rabbit IgG alkaline phosphatase-conjugated antibodies (Promega Co., Madison, Wis.). The membrane was then stained with nitroblue tetrazolium (Promega Co.) and 5-bromo-4-chloro-3-indolylphosphate (BCIP; Promega Co.) according to the protocol specified by the supplier. The staining solution was prepared by adding 66 μl of nitroblue tetrazolium and 33 μl of BCIP sequentially to 10 ml of alkaline phosphatase buffer (100 mM Tris-HCl [pH 9.0] containing 150 mM NaCl and 1 mM MgCl2).
Flocculation measurement.
Flocculation ability was measured according to previously reported methods (3). Yeast cells were deflocculated by washing with 50 mM sodium citrate (pH 3.0)-5 mM EDTA buffer twice and then suspended, giving a concentration of 108 cells/ml (5 OD units) in the same buffer. A total of 0.1 ml of distilled water or 1 M CaCl2 solution (20 mM final concentration to induce flocculation) was added to 4.9 ml of deflocculated cell suspension in a 10-ml test tube. The tubes were agitated at a rate of 50 oscillations per min for 5 min and then left standing vertically. After 1 min, 0.2 ml of suspension was collected from just below the meniscus of each tube and mixed with 1 ml of 0.25 M EDTA (pH 8.0) and the OD600 was measured. From the OD600 values of deflocculated and flocculated cell suspensions (ODdefloc and ODfloc, respectively), the ability to induce flocculation (i.e., the percentage of cells flocculated) was calculated using the following equation: ability to induce flocculation (percent) = (ODdefloc − ODfloc)/ODdefloc× 100. Percent values of 100 and 0 indicate complete cell flocculation and dispersion, respectively. Flocculation ability was also recorded photographically. Culture medium, cultivated in a shake flask for 150 h, was transferred to a test tube and shaken quickly and photographed immediately and after 5 s.
Methanolysis reaction using whole-cell biocatalysts.
The methanolysis reaction was performed as follows. A total of 100 mg (dry weight) of FSProROL or FLProROL surface-displaying yeast cells was suspended in 0.5 ml of 0.1 M acetate buffer (pH 7.0) and used as a catalyst. The yeast cell suspension was added to a mixture of soybean oil and methanol (9.65 g/0.35 g = 1 mol/1 mol). The molar amount of soybean oil was calculated using apparent molecular weight values calculated from the fatty acid composition (7). The reaction was carried out in a 30-ml screw vial at 37°C at 150 oscillations per min. Methanol (0.35 g [1 molar equivalent]) was added twice into the reaction mixture, after 24- and 50-h reactions.
Amount of ME produced by methanolysis reaction was measured using a capillary gas chromatograph GC-18A apparatus (Shimadzu Co., Kyoto, Japan) connected to a DB-5 capillary column according to the method previously reported (8), with minor modification. Aliquots of 150 μl were taken from the reaction mixture and centrifuged at 19,000 × g to obtain the upper layer, of which 80 μl was mixed with 20 μl of tricaprylin in a 10-ml bottle, to which were then added a specified amount of sodium sulfate as dehydrogenating agent and 3.0 ml of hexane. A 1.0-μl aliquot of the treated sample was subjected to gas chromatography to quantify the ME content.
RESULTS
Immunofluorescence microscopy.
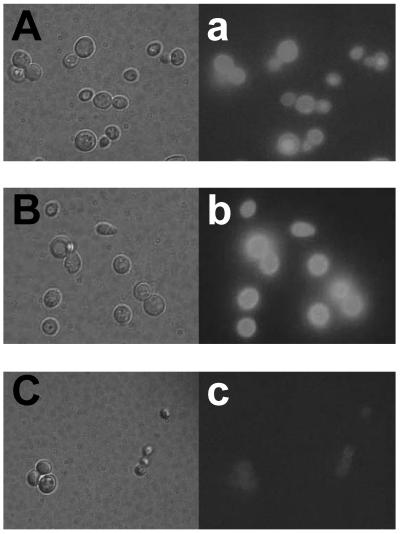
Immunofluorescent labeling of cells was performed sequentially with anti-ROL IgG and fluorescein isothiocyanate-conjugated goat IgG. The green fluorescence of the immunostained FSProROL and FLProROL fusion proteins was clearly observed outlining both the yeast MT8-1 cells harboring pWIFSProROL (MT8-1/pWIFSProROL) (Fig. 2A and a) and those harboring pWIFLProROL (MT8-1/pWIFLProROL) (Fig. 2B and b). In contrast, the yeast cells harboring the control plasmid pWI3 were not immunostained (Fig. 2C and c). This confirmed that the FSProROL and FLProROL fusion proteins were anchored on the yeast cell surface.
FIG. 2.
Microscopy of immunofluorescence-labeled yeast cells. Differential interference contrast micrographs (panels A, B, and C) and fluorescence micrographs (panels a, b, and c) are shown. Panels A and a, S. cerevisiae MT8-1/pWIFSProROL; panels B and b, S. cerevisiae MT8-1/pWIFLProROL; panels C and c, S. cerevisiae MT8-1/pWI3 (control).
Enzyme activity.
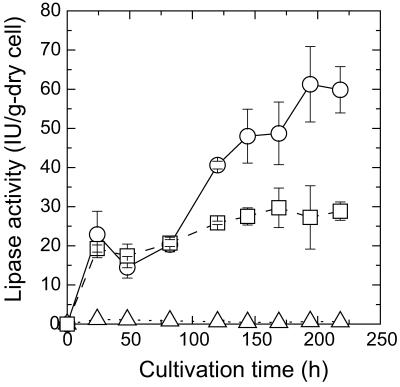
Expression experiments with the FSProROL and FLProROL fusion genes were performed by flask cultivation. Figure 3 shows the time course of lipase activity in whole cells of MT8-1/pWIFSProROL and MT8-1/pWIFLProROL. In both transformants, the lipase activity of yeast whole cells started increasing at 24 h, when the glucose in the medium was exhausted (data not shown). Lipase activity reached 61.3 IU/g (dry cell weight) in MT8-1/pWIFSProROL after 194 h of cultivation and 29.7 IU/g (dry cell weight) in MT8-1/pWIFLProROL after 169 h of cultivation. On the other hand, detectable lipase activity was not observed in the culture medium of MT8-1/pWIFSProROL or MT8-1/pWIFLProROL. These results confirmed that the produced FSProROL and FLProROL fusion proteins were not secreted into the culture media and were mostly immobilized on the yeast cells.
FIG. 3.
Time courses of lipase activity in cell pellets during cultivation of S. cerevisiae MT8-1/pWIFSProROL (open circle), MT8-1/pWIFLProROL (open square), and MT8-1/pWI3 (open triangle [control]). Vertical axis indicates lipase activity in cell pellet per dry cell weight.
Localization of FSProROL and FLProROL fusion proteins.
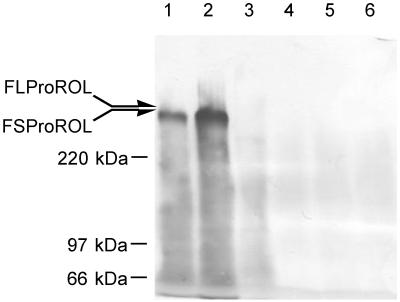
The localization of the FSProROL and FLProROL proteins and their association with the cell wall were examined. Cells were disrupted to obtain the cell wall, and FSProROL and FLProROL were extracted using a two-step procedure. First, proteins bound noncovalently or bound via disulfide bridges were extracted from the cell wall with hot SDS. Following this, proteins covalently bound to cell walls were further extracted by digestion with laminarinase (β-1,3-glucanase). These extracts were treated with Endo Hf. Then, they were applied to SDS-PAGE and blotted onto polyvinylidene difluoride membrane and FSProROL and FLProROL were immunostained. As shown in Fig. 4, both FSProROL and FLProROL were mainly observed in SDS-extracted fractions (lanes 1 and 2) with little or no signal observed in glucanase-extracted fractions (lanes 4 and 6). This result indicates that FSProROL and FLProROL were noncovalently attached to the cell wall. Since the molecular masses of these fusion proteins seemed rather higher than the calculated values (FSProROL, 156 kDa; FLProROL, 191 kDa), they were thought to have been produced in highly o-glycosylated form.
FIG. 4.
Immunoblotting of extracts from S. cerevisiae MT8-1/pWIFSProROL (lanes 1 and 4), MT8-1/pWIFLProROL (lanes 2 and 5), and MT8-1/pWI3 (lanes 3 and 6 [controls]) cell walls. Lanes 1 to 3, SDS-extracted fractions; lanes 4 to 6, glucanase-extracted fractions. All fractions were treated with Endo Hf prior to analysis.
Flocculation profile of yeast cells displaying FSProROL and FLProROL fusion proteins.
Figure 5 shows the flocculation ability of yeast cells displaying the FSProROL and FLProROL fusion proteins and wild-type nonflocculent yeast cells. Interestingly, the yeast cells harboring pWIFLProROL at the late stage of cultivation seemed more flocculent than the wild-type flocculent strain S. cerevisiae ATCC 60715 (data not shown), even though the FLProROL fusion protein produced had neither a GPI anchor attachment site nor a membrane-anchoring domain (Fig. 1). Meanwhile, yeast cells displaying FSProROL seemed to flocculate slightly on flask cultivation, although distinct ability was not detected by flocculation ability measurement (Fig. 5). The level of surface display of the FL protein was just enough to obtain sufficient flocculation ability. This result leads to the conclusion that the hydrophobic C-terminal region of Flo1p (amino acids 1448 to 1536) is not necessary for flocculation but the C-terminal region of the FL anchor (consisting of amino acids 1100 to 1447) is.
FIG. 5.
Photographs of flocculating yeast during agitation (A, B, and C) and after sedimentation for 5 s (a, b and c). Panels A and a, S. cerevisiae MT8-1/pWIFSProROL; panels B and b, S. cerevisiae MT8-1/pWIFLProROL; panels C and c, S. cerevisiae MT8-1/pWI3 (control). Ability of different constructs to trigger flocculation is indicated by +++ (full flocculation), + (slight flocculation), or − (no flocculation). Values in parentheses represent percentages of cells that settled after 1 min.
Methanolysis reaction using yeast cells displaying ProROL.
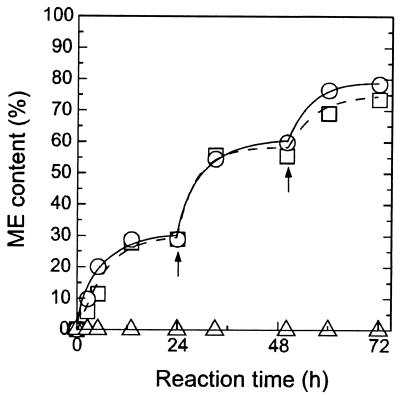
In a methanolysis reaction using plant oil, reaction substrates such as methanol and triglyceride can easily access surface-displayed lipase. The yeast cells displaying FSProROL and FLProROL were therefore used for biodiesel production without any permeabilizing treatment. Figure 6 shows the time course of ME content in the methanolysis reaction. In yeast cells displaying FSProROL and FLProROL, ME content after 72 h of reaction reached 78.6% and 73.5%, respectively, with a two-step addition of methanol. Further, the initial reaction rate, defined as ME production rate, reached 3.2 g of ME/liter · min when 150 mg of lyophilized FSProROL-displaying cells were used (data not shown), a level as high as that for ROL in enzyme solution (8).
FIG. 6.
Time course of methanolysis reaction using yeast whole cells. Percentages of ME weight in reaction mixture are plotted against reaction time. ME content was determined as the ratio of the weight of ME to that in the total oil phase in the reaction mixture. Arrows show times of addition of 0.35 g of methanol. Symbols: open circle, S. cerevisiae MT8-1/pWIFSProROL; open square, MT8-1/pWIFLProROL; open triangle, MT8-1/pWI3 (control).
DISCUSSION
In the present study, we successfully developed a novel surface-display system utilizing the flocculation functional domain of Flo1p. This protein is thought to produce cell adhesion via noncovalent interaction of its flocculation functional domain with the mannan chain of the cell wall. We noted this adhesive ability and attempted to utilize the relevant domain in a novel surface display system. As described in Fig. 1, in the new cell-surface display system the FS and FL proteins, which consisted of amino acids 1 to 1099 and 1 to 1447, respectively, of Flo1p, were fused to the N terminus of the ProROL protein. This system is expected to be effective for N-terminal immobilization of target proteins whose catalytic site is near the C terminus. Since both of the surface-displayed proteins, FSProROL and FLProROL, were extracted with hot SDS (Fig. 4), these fusion proteins are probably immobilized noncovalently to the cell wall component. In conventional GPI anchor systems, which have been the most commonly used in cell-surface display, the GPI attachment signal has to be fused to the C terminus of the target proteins. It is therefore very difficult to display ProROL on the yeast cell surface using GPI anchor systems, and no report has been published detailing cell-surface display of lipase with sufficiently high activity; only 4.1 IU/g (dry cell weight) of lipase activity is achieved with the GPI anchor system (30). In our new surface display system using FL and FS proteins, lipase activity reached 61.3 IU/g (dry cell weight), indicating effectiveness in the display of enzymes with active sites located near the C terminus.
It is also very interesting that S. cerevisiae MT8-1/pWIFLProROL showed stronger flocculation than the wild-type flocculent yeast S. diastaticus ATCC 60715, despite MT8-1 being a nonflocculent strain (Fig. 5a). The mechanism of flocculation is thought to consist of lectin-like proteins, fixed covalently at the C terminus by the GPI anchor, interacting with the mannan chains of another cell via the N-terminal domain (15, 26, 31). Recently, Bony et al. reported similarly that the removal of the hydrophobic C-terminal region was observed to prevent the anchoring of Flo1p at the cell surface and that the truncated Flo1p was mostly secreted into the culture medium (3). However, our findings reveal that Flo1p, a lectin-like protein, does not need to be fixed to the yeast cell surface via the C-terminal anchoring region for surface display and flocculation to occur. The mechanism of this flocculation phenomenon is under investigation.
Yeast cells displaying high lipase activity on their surface were used as whole-cell biocatalysts for methanolysis reaction in a solvent-free system. With cell surface-displayed ProROL, substrate molecules could easily access ProROL and no treatment was needed to catalyze methanolysis reaction. Since the initial reaction rate of FSProROL-displaying cells was as high as that of soluble ROL, the displayed FSProROL may have the same accessibility to substrates as free enzymes. There is almost no barrier to diffusion of substrates and products in cell-surface display systems, whereas in whole-cell biocatalyst containing intracellular ROL, various permeabilization treatments such as freeze-thawing and air drying are necessary to improve reactivity (14). For the industrial bioconversion process, lipases immobilized on the cell surface are more cost effective and convenient. These whole-cell biocatalysts are prepared by simple cultivation and recovered easily. Moreover, MT8-1/pWIFLProROL with strong flocculation ability was spontaneously immobilized within porous support particles (13) during cultivation (data not shown). The stability of lipase-displaying yeast whole-cell biocatalyst, which is important for industrial applications, is presently under investigation, and details will be described elsewhere.
Our novel FS-anchoring system allows N-terminal immobilization of targeted proteins in cell suspension form, while the FL-anchoring systems additionally give strong flocculation. Each of these novel surface display systems will be suited to a different range of applications.
REFERENCES
- 1.Beer, H. D., G. Wohlfahrt, J. E. McCarthy, D. Schomburg, and R. D. Schmid. 1996. Analysis of the catalytic mechanism of a fungal lipase using computer-aided design and structural mutants. Protein Eng. 9:507-517. [DOI] [PubMed] [Google Scholar]
- 2.Boder, E. T., and K. D. Wittrup. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553-557. [DOI] [PubMed] [Google Scholar]
- 3.Bony, M., D. Thines-Sempoux, P. Barre, and B. Blondin. 1997. Localization and cell surface anchoring of the Saccharomyces cerevisiae flocculation protein Flo1p. J. Bacteriol. 179:4929-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francisco, J. A., C. F. Earhart, and G. Georgiou. 1992. Transport and anchoring of β-lactamase to the external surface of Escherichia coli. Proc. Natl. Acad. Sci. USA 89:2713-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francisco, J. A., C. Stathopoulos, R. A. Warren, D. G. Kilburn, and G. Georgiou. 1993. Specific adhesion and hydrolysis of cellulose by intact Escherichia coli expressing surface anchored cellulase or cellulose binding domains. Bio/Technology 11:491-495. [DOI] [PubMed] [Google Scholar]
- 6.Georgiou, G., H. L. Poetschke, C. Stathopoulos, and, J. A. Francisco. 1993. Practical applications of engineering gram-negative bacterial cell surfaces. Trends Biotechnol. 11:6-10. [DOI] [PubMed] [Google Scholar]
- 7.Gunstone, F. D., J. L. Harwood, and F. B. Padley. 1994. The lipid handbook, 2nd ed., p. 97-101. Chapman & Hall, London, United Kingdom.
- 8.Kaieda, M., T. Samukawa, T. Matsumoto, K. Ban, Kondo, Y. Shimada, H. Noda, F. Nomoto, K. Ohtuka, E. Izumoto, and A. H. Fukuda. 1999. Biodiesel fuel production from plant oil catalyzed by Rhizopus oryzae lipase in a water-containing system without an organic solvent. J. Biosci. Bioeng. 88:627-631. [DOI] [PubMed] [Google Scholar]
- 9.Kanai, T., H. Atomi, K. Umemura, H. Ueno, Y. Teranishi, M. Ueda, and A. Tanaka. 1996. A novel heterologous gene expression system in Saccharomyces cerevisiae using the isocitrate lyase gene promoter from Candida tropicalis. Appl. Microbiol. Biotechnol. 44:759-765. [DOI] [PubMed] [Google Scholar]
- 10.Linco, Y. Y., M. Lamsa, X. Wu, E. Uosukainen, J. Seppala, and P. Linko. 1998. Biodegradable products by lipase biocatalysis. J. Biotechnol. 66:41-50. [DOI] [PubMed] [Google Scholar]
- 11.Lipke, P. N., and R. Ovalle. 1998. Cell wall architecture in yeast: new structure and new challenges. J. Bacteriol. 180:3735-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Little, M., P. Fuchs, F. Breitling, and S. Dubel. 1993. Bacterial surface presentation of proteins and peptides: an alternative to phage technology? Trends Biotechnol. 11:3-5. [DOI] [PubMed] [Google Scholar]
- 13.Liu, Y., A. Kondo, H. Ohkawa, N. Shiota, and H. Fukuda. 1998. Bioconversion using immobilized recombinant flocculent yeast cells carrying a fused enzyme gene in an “intelligent” bioreactor. Biochem. Eng. J. 2:229-235. [Google Scholar]
- 14.Matsumoto, T., S. Takahashi, M. Kaieda, M. Ueda, A. Tanaka, H. Fukuda, and A. Kondo. 2001. Yeast whole-cell biocatalyst constructed by intracellular overproduction of Rhizopus oryzae lipase is applicable to biodiesel fuel production. Appl. Microbiol. Biotechnol. 57:515-520. [DOI] [PubMed] [Google Scholar]
- 15.Miki, B. L., N. H. Poon, A. P. James, and V. L. Seligy. 1982. Possible mechanism for flocculation interactions governed by gene FLO1 in Saccharomyces cerevisiae. J. Bacteriol. 150:878-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murai, T., M. Ueda, H. Atomi, Y. Shibasaki, N. Kamasawa, M. Osumi, T. Kawaguchi, M. Arai, and A. Tanaka. 1997. Genetic immobilization of cellulase on the cell surface of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 48:499-503. [DOI] [PubMed] [Google Scholar]
- 17.Murai, T., M. Ueda, T. Kawaguchi, M. Arai, and A. Tanaka. 1998. Assimilation of cellooligosaccharides by a cell surface-engineered yeast expressing β-glucosidase and carboxymethylcellulase from Aspergillus aculeatus. Appl. Environ. Microbiol. 64:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murai, T., M. Ueda, M. Yamamura, H. Atomi, Y. Shibasaki, N. Kamasawa, M. Osumi, T. Amachi, and A. Tanaka. 1997. Construction of a starch-utilizing yeast by cell surface engineering. Appl. Environ. Microbiol. 63:1362-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson, L. A., T. A. Foglia, and W. N. Marmer. 1996. Lipase-catalyzed production of biodiesel. J. Assoc. Off. Anal. Chem. 73:1191-1195. [Google Scholar]
- 20.Schreuder, M. P., C. Deen, W. J. A. Boersma, P. H. Pouwels, and F. M. Klis. 1996. Yeast expressing hepatitis B virus surface antigen determinants on its surface: implications for a possible oral vaccine. Vaccine 14:383-388. [DOI] [PubMed] [Google Scholar]
- 21.Schreuder, P. S., A. T. A. Mooren, H. Y. Toschka, C. T. Verrips, and F. M. Klis. 1996. Immobilizing proteins on the surface of yeast cells. Trends Biotechnol. 14:115-120. [DOI] [PubMed] [Google Scholar]
- 22.Shimada, Y., Y. Watanabe, T. Samukawa, A. Sugihara, H. Noda, H. Fukuda, and Y. Tominaga. 1999. Conversion of plant oil to biodiesel using immobilized Candida antarctica lipase. J. Assoc. Off. Anal. Chem. 76:789-793. [Google Scholar]
- 23.Stahl, S., and M. Uhlen. 1997. Bacterial surface display: trends and progress. Trends Biotechnol. 15:185-192. [DOI] [PubMed] [Google Scholar]
- 24.Tajima, M., Y. Nogi, and T. Fukasawa. 1985. Primary structure of the Saccharomyces cerevisiae GAL7 gene. Yeast 1:67-77. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi, S., M. Ueda, H. Atomi, H. D. Beer, U. T. Bornscheuer, R. D. Schmid, and A. Tanaka. 1998. Extracellular production of active Rhizopus oryzae lipase by Saccharomyces cerevisiae. J. Ferment. Bioeng. 86:164-168. [Google Scholar]
- 26.Teunissen, A. W., E. Holub, J. van der Hucht, J. A. van den Berg, and H. Y. Steensma. 1993. Sequence of the open reading frame of the FLO1 gene from Saccharomyces cerevisiae. Yeast 9:423-427. [DOI] [PubMed] [Google Scholar]
- 27.Umemura, K., H. Atomi, T. Kanai, Y. Teranishi, M. Ueda, and A. Tanaka. 1995. A novel promoter, derived from the isocitrate lyase gene of Candida tropicalis, inducible with acetate in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 43:489-492. [PubMed] [Google Scholar]
- 28.Varese, R., and M. Varese. 1996. Methyl ester biodiesel: opportunity or necessity? INFORM 7:816-824. [Google Scholar]
- 29.Walker, R. I. 1994. New strategies for using mucosal vaccination to achieve more effective immunization. Vaccine 12:387-400. [DOI] [PubMed] [Google Scholar]
- 30.Washida, M., S. Takahashi, M. Ueda, and A. Tanaka. 2001. Spacer-mediated display of active lipase on the yeast cell surface. Appl. Microbiol. Biotechnol. 56:681-686. [DOI] [PubMed] [Google Scholar]
- 31.Watari, J., Y. Takata, M. Ogawa, H. Sahara, S. Koshino, M. L. Onnela, U. Airaksinen, R. Jaatinen, M. Penttila, and S. Keranen. 1994. Molecular cloning and analysis of the yeast flocculation gene FLO1. Yeast 10:211-225. [DOI] [PubMed] [Google Scholar]