Abstract
The objective of this study was to evaluate the effect of human gut-derived lactic acid bacteria and bifidobacteria on cholesterol levels in vitro. Continuous cultures inoculated with fecal material from healthy human volunteers with media supplemented with cholesterol and bile acids were used to enrich for potential cholesterol assimilators among the indigenous bacterial populations. Seven potential probiotics were found: Lactobacillus fermentum strains F53 and KC5b, Bifidobacterium infantis ATCC 15697, Streptococcus bovis ATCC 43143, Enterococcus durans DSM 20633, Enterococcus gallinarum, and Enterococcus faecalis. A comparative evaluation regarding the in vitro cholesterol reduction abilities of these strains along with commercial probiotics was undertaken. The degree of acid and bile tolerance of strains was also evaluated. The human isolate L. fermentum KC5b was able to maintain viability for 2 h at pH 2 and to grow in a medium with 4,000 mg of bile acids per liter. This strain was also able to remove a maximum of 14.8 mg of cholesterol per g (dry weight) of cells from the culture medium and therefore was regarded as a candidate probiotic.
Since Shaper et al. (18) and later Mann (12) observed that men from the tribes of Samburu and Masai warriors in Africa showed a reduction in serum cholesterol levels after consumption of large amounts of milk fermented with a wild Lactobacillus strain, there has been considerable interest in the beneficial effects of fermented milk products containing lactobacilli and/or bifidobacteria on human lipid metabolism. Several human studies have suggested a moderate cholesterol-lowering action of dairy products fermented with a certain strain(s) of probiotic bacteria (1, 2, 10, 17; G. Schaarmann, J. Schneider, A. Zorn, C. Vilser, and G. Jahreis, Am. J. Clin. Nutr. 73[Suppl.]:496S, 2001). However, the role of fermented milk products as hypocholesterolemic agents in humans is still equivocal, as the clinical studies performed have given variable data and no firm conclusions can be drawn (6, 23). From several in vitro studies a number of mechanisms have been proposed for the purported cholesterol-lowering action of probiotic bacteria (7, 9, 11, 13, 21, 22, 24). These include physiological actions of the end products of short-chain fatty acid fermentation (especially propionate), cholesterol assimilation by the bacteria, cholesterol binding to the bacterial cell wall, and enzymatic deconjugation of bile acids. These hypotheses need to be confirmed in animal and human studies, and the exact mechanism(s) of action of probiotic bacteria on cholesterol reduction remains unclear.
In this study, we evaluated the in vitro cholesterol-lowering effects of strains of lactic acid bacteria and bifidobacteria of human origin and compared them to those of commercial probiotic strains. Further, we have investigated the ability of the cultures to tolerate acid and bile concentrations typically found in the upper gastrointestinal tract of humans. We hope this study provides further background and new, improved strains for the understanding of the purported action of probiotic bacteria on cholesterol levels.
Source and maintenance of bacterial strains.
The origins of the strains used in this study are shown in Table 1. The human isolates of Lactobacillus fermentum (F53 and KC5b), Enterococcus gallinarum, Enterococcus faecalis, and Lactobacillus brevis (NR1C1684) have not been deposited in a culture collection and will be provided upon request. Stock cultures were maintained at −70°C on Microbank cryovials (Pro-Lab Diagnostics). Working cultures were maintained and subcultured in MRS-THIO broth (MRS broth [Oxoid] supplemented with 0.2% [wt/vol] sodium thioglycolate [Sigma]). Prior to assay, strains were serially transferred three times in broth and incubated anaerobically (10% H2, 10% CO2, and 80% N2) at 37°C for 24 h. Seed cultures of each strain were taken at the end of the exponential phase of growth at cell densities of ca. 109 CFU/ml. All the bacterial strains used in this study were identified by PCR techniques (34 cycles of amplification of DNA by PCR) followed by sequencing of the bacterial 16S rRNA gene (data not shown). Approximately 500 nucleotides proximal to the 5′ end of the rRNA were sequenced with an ABI PRISM dRhodamine terminator cycle sequencing kit with AmpliTaq DNA polymerase FS (PE Applied Biosystems, Inc.) and an automatic DNA sequencer (model 373A; PE Applied Biosystems, Inc.). Screening of the isolates was carried out using the reverse primers pD* (GTATTACCGCGGCTGCTG) and γ (ACTGCTGCCTCCCGTAGGAG). Generated sequences were compared to those available in the GenBank/EMBL (European Bioinformatics Institute) database by using the program WU-Blast2 (Washington University), in order to ascertain their closest phylogenetic relatives (Table 1).
TABLE 1.
Origin and source of bacterial strainsa
| Bacterial strainb | Source | Origin |
|---|---|---|
| Lactobacillus fermentum F53 | FMSUc | Human feces |
| Lactobacillus fermentum KC5b | FMSUc | Human feces |
| Bifidobacterium infantis ATCC 15697 | FMSUc | Human feces |
| Streptococcus bovis ATCC 43143 | FMSUc | Human feces |
| Enterococcus durans DSM 20633 | FMSUc | Human feces |
| Enterococcus gallinarum | FMSUc | Human feces |
| Enterococcus faecalis | FMSUc | Human feces |
| Lactobacillus brevis NR1C1684 | FMSUd | Human feces |
| Enterococcus durans ATCC 59607 | FMSUd | Human feces |
| Lactobacillus pentosus (A), (B) | St. Ivel | NA |
| Lactobacillus reuteri DSM 20016T | St. Ivel | NA |
| Lactobacillus reuteri JCM 1112 | St. Ivel | NA |
| Lactobacillus plantarum NDV | Novartise | NA |
| Bifidobacterium infantis ATCC 15697 | Danone | Bio Danone yoghurt |
| Lactobacillus casei | Danone | Actimel FM drink |
| Lactobacillus delbrueckii JCM 1002 | Danone | Actimel FM drink |
| Lactobacillus casei | Yakult | Yakult FM drink |
| Lactobacillus acidophilus johnsonii | Nestlé | LC1 FM drink |
| Lactobacillus crispatus ATCC 33820 | Nestlé | LC1 FM drink |
Abbreviations: FMSU, Food Microbial Sciences Unit (The University of Reading); NA, not available; FM, fermented milk.
Identification based on sequence homology of the 16S rRNA bacterial gene.
Strains isolated at steady state from continuous cultures enriched with cholesterol (100 mg liter−1) and bile acids (0.2 and 0.4% [wt/vol] oxgall); data not shown.
Strains isolated from 24-h static batch cultures with the same medium used in the chemostats; data not shown.
Rifampin-resistant, wild-type strain obtained from Novartis (Basel, Switzerland).
Assimilation of cholesterol.
The ability of cultures to assimilate cholesterol was determined by a modification of the method described by Danielson et al. (5). The test medium used for screening cultures for cholesterol uptake was sterile modified MRS broth (mMRS) supplemented with oxgall and a water-soluble form of cholesterol (polyoxyethanyl-cholesteryl sebacate; Sigma). The composition of the mMRS broth was (grams per liter unless otherwise indicated): peptone (Oxoid), 10.0; Lab-Lemco powder (Oxoid), 8.0; yeast extract (Oxoid), 4.0; Tween 80, 1 ml liter−1; K2HPO4, 2.0; triammonium citrate, 2.0; sodium acetate, 3.0; MgSO4 · 7H2O, 0.2; MnSO4 · H2O, 0.04; sodium thioglycolate, 2.0; and glucose, 10.0. Two different bile concentrations were used, 0.2 and 0.4% (wt/vol) oxgall (OXOID), to mimic approximate levels in the intestinal tract (19). The final cholesterol concentration in the medium was ca. 100 mg liter−1. All chemicals were obtained from Merck unless otherwise stated. The seed culture was added at a 1% (vol/vol) inoculum size to Hungate tubes containing 9.9-ml aliquots of the test medium. The cultures were statically fermented for 12 h at 37°C under anaerobic conditions. Bacterial growth was monitored hourly by measuring the optical density of the culture broth at 650 nm. Each strain was tested in three different runs and each time in triplicate. Uninoculated sterile broth was used as the control. Following incubation, bacterial cells were removed by centrifugation (2000 × g, 10 min, 4°C), and the spent broth and uninoculated control broths were then assayed for their cholesterol content. The dry weight of the cultures was determined after drying the centrifuged cells to a constant weight in an 80°C oven. Strains were compared for cholesterol assimilation in terms of their specific cholesterol uptake after the 12-h incubation period, according to the following equation:
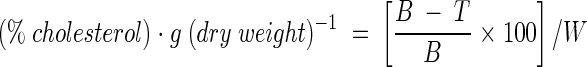 |
(1) |
where B is cholesterol content in the uninoculated control (milligrams liter−1), T is cholesterol in the culture medium (milligrams liter−1), and W is cells (dry weight [grams]) after 12 h of incubation.
Cholesterol assay.
The cholesterol in the spent broths was first extracted by the procedure described by Gilliland et al. (7). The total cholesterol content of the evaporated residues was then determined by the enzymatic assay described by Salè et al. (15).
Bile tolerance.
All strains were evaluated for rapidity of growth in a broth medium with and without bile acids. Overnight cultures were inoculated (1% [vol/vol]) into mMRS broth and mMRS broth containing 0.2 and 0.4% (wt/vol) oxgall and incubated anaerobically at 37°C for 12 h. Cultures were monitored hourly for growth spectrophotometrically at 650 nm. Comparison of cultures was based on their growth rates in each broth. The experiments were repeated three times in triplicate.
Acid tolerance.
Overnight cultures of strains were inoculated (10% [vol/vol]) into MRS broth (Oxoid) previously adjusted to pH 2.0 with HCl. The mixtures were incubated anaerobically at 37oC for 2 h. One-milliliter samples were taken at various times (0, 15, 30, 45, 60, and 120 min), serially 10-fold diluted in anaerobic diluent (half-strength peptone water plus 0.5 g of l-cysteine HCl liter−1, pH 7.0), and plated in triplicate onto MRS agar (Oxoid). The plates were incubated at 37°C for 24 h under anaerobic conditions before enumeration. The experiments were repeated three times.
Statistical analysis.
Cholesterol assimilation and acid and bile tolerance were analyzed by the one-way analysis of variance procedure of SPSS to determine whether significant (P < 0.05) variation occurred among means in each experiment (8). The least significant difference (Bonferroni t test) was used to determine which means differed significantly.
The standard deviation on the specific cholesterol uptake was estimated from the standard deviations on the cholesterol assimilations and the standard error of the dry weight determinations by the law of error propagation in arithmetic operations.
Cholesterol reduction from the culture media.
The amount of cholesterol assimilated during 12 h of anaerobic growth at 37°C (Table 2) revealed a wide variation among strains as well as between trials for the same strain. All strains examined were able to assimilate cholesterol to some extent with the exception of Lactobacillus crispatus ATCC 33820 and Enterococcus durans ATCC 59607, which did not grow well in the media used. The amounts of cholesterol assimilated by the cultures ranged from 0.09 to 29.73 mg/g of cells (0.4 to 47% of the cholesterol in the media). No statistically significant differences were found between the cultures tested with regard to their specific cholesterol uptake rate (P > 0.05), due to the high variability of data. In the majority of cases, uptake of cholesterol was higher in the medium with 0.4% (wt/vol) oxgall, although this was significant only (P < 0.05) for Lactobacillus casei Shirota. Cultures of L. brevis NR1C1684 and E. faecalis assimilated more than 1.5 times the average of the other strains in the media with 0.2 and 0.4% (wt/vol) oxgall, respectively. However, this effect was not statistically significant (P > 0.05).
TABLE 2.
Cholesterol specific uptake during 12 h of anaerobic growth in mMRS broth supplemented with bile acids and 100 mg of cholesterol per litera
| Strain | Cholesterol uptake (mg of cholesterol g [dry wt])
|
|||||
|---|---|---|---|---|---|---|
| 0.2% oxgall
|
0.4% oxgall
|
|||||
| Run 1 | Run 2 | Run 3 | Run 1 | Run 2 | Run 3 | |
| L. brevis NR1C1684 | 8.6 (0.5) | 19 (1) | 3.9 (0.6) | 1.6 (0.1) | 8.0 (0.8) | 13.2 (0.7) |
| L. pentosus (A) | 9.4 (0.5) | 0.88 (0.05) | 0.10 (0.03) | 6.9 (0.4) | 16.6 (0.9) | 15.3 (0.8) |
| L. plantarum NDV | 0.92 (0.07) | 6.9 (0.4) | 0.02 (0.04) | 5.5 (0.3) | 11.1 (0.6) | 16.4 (0.9) |
| L. reuteri JCM 1112 | 0.09 (0.08) | 10.6 (0.6) | 4.8 (0.3) | 10.4 (0.5) | 12.5 (0.7) | 11.3 (0.6) |
| L. casei Shirota | 5.5 (0.3) | 6.0 (0.3) | 1.14 (0.07) | 8.1 (0.4) | 8.7 (0.5) | 9.5 (0.5) |
| L. casei | 2.9 (0.2) | 3.7 (0.2) | 0.40 (0.05) | 14.1 (0.7) | 3.0 (0.2) | 8.5 (0.4) |
| L. acidophilus johnsonii | 3.0 (0.2) | 0.22 (0.02) | 1.47 (0.08) | 3.3 (0.2) | 0.94 (0.3) | 11.7 (0.6) |
| L. delbrueckii JCM 1002 | 4.9 (0.3) | 3.8 (0.2) | 6.7 (0.4) | 13.7 (0.7) | 2.5 (0.2) | 13.9 (0.7) |
| L. fermentum F53 | 0.39 (0.06) | 6.2 (0.3) | 0.77 (0.05) | 12.9 (0.7) | 6.9 (0.4) | 8.8 (0.5) |
| L. fermentum KC5B | 8.3 (0.4) | 2.4 (0.1) | 2.4 (0.1) | 14.8 (0.8) | 5.3 (0.3) | 4.1 (0.2) |
| S. thermophilus DSM 20617 | 3.7 (0.1) | ND | ND | 6.1 (0.2) | ND | ND |
| S. bovis ATCC 43143 | 2.47 (0.09) | 6.4 (0.2) | 1.55 (0.07) | 6.6 (0.2) | 6.7 (0.2) | 20.0 (0.7) |
| L. pentosus (B) | 2.4 (0.1) | 10.5 (0.6) | 2.6 (0.2) | 6.5 (0.3) | 22 (1) | 26 (1) |
| L. reuteri DSM 20016T | 2.1 (0.1) | 0.84 (0.07) | 0.64 (0.05) | 5.2 (0.3) | 23 (1) | 10.7 (0.6) |
| E. durans DSM 20633 | 6.2 (0.4) | 0.66 (0.07) | 0.45 (0.05) | 4.8 (0.3) | 26 (1) | 19 (1) |
| E. gallinarum | 7.1 (0.4) | 0.30 (0.09) | 0.28 (0.07) | 8.3 (0.5) | 27 (1) | 19 (1) |
| E. faecalis | 6.5 (0.4) | 5.3 (0.3) | 3.6 (0.2) | 1.50 (0.09) | 30 (1) | 26 (2) |
| B. infantis ATCC 15697 | 1.31 (0.05) | 3.9 (0.1) | 4.7 (0.2) | 9.6 (0.3) | 7.3 (0.3) | 4.3 (0.2) |
| B. infantis D86184 | 4.3 (0.4) | 2.0 (0.2) | 6.1 (0.3) | ND | ND | ND |
Results are shown as means (standard deviations) (n = 3). L. crispatus ATCC 33820 and E. durans ATCC 59607 did not grow in the test media used. ND, no growth observed.
Growth in the presence of bile salts.
Table 3 shows the effect of bile salts on the growth of the strains arranged in order of decreasing bile tolerance, as determined by a lowering of the growth rate in the medium with 0.4% bile acids. Significant variations existed among the cultures with regard to their ability to grow in mMRS broth and in mMRS broth supplemented with bile acids (P < 0.01). L. crispatus ATCC 33820 was less bile tolerant than all the other strains tested, not being able to grow in the presence of 0.2% bile acids. In a comparison of all the other strains, L. casei Shirota and Bifidobacterium infantis ATCC 15697 were the most sensitive to bile acid, with significantly lower growth rates in the medium with 0.4% (wt/vol) bile acids than in the absence of bile (P < 0.05). Lactobacillus delbrueckii JCM 1002, E. gallinarum, and L. fermentum strains F53 and KC5b showed some sensibility to bile acids; however, the differences found in the growth rate were not significant (P > 0.05). The remaining strains showed no significant differences in their growth in the media with and without bile acids.
TABLE 3.
Effect of bile acids on the growth rate of strains in mMRS brotha
| Strain | Growth rate (h−1) with oxgall concn (% [wt/vol]):
|
||
|---|---|---|---|
| 0 | 0.2 | 0.4 | |
| E. durans DSM 20633 | 1.2 (0.2)bdf, x | 1.4 (0.2)b, x | 1.5 (0.4)b, x |
| L. acidophilus johnsonii | 0.8 (0.2)ab, x | 1.0 (0.3)bce, x | 1.0 (0.4)ab, x |
| L. plantarum NDV | 0.64 (0.03)ac, x | 0.8 (0.1)acde, x | 0.70 (0.03)ac, x |
| E. durans ATCC 59607 | 1.2 (0.2)bdf, x | 1.4 (0.4)be, x | 1.2 (0.5)bce, x |
| E. faecalis | 1.3 (0.2)bd, x | 1.3 (0.1)bd, x | 1.3 (0.3)bc, x |
| S. thermophilus DSM 20617 | 0.7 (0.3)ace, x | 0.7 (0.1)ac, x | 0.71 (0.06)ac, x |
| L. casei | 0.50 (0.03)ac, x | 0.45 (0.03)ac, x | 0.5 (0.1)a, x |
| S. bovis ATCC 43143 | 1.5 (0.1)d, x | 1.6 (0.1)b, x | 1.5 (0.2)bd, x |
| L. brevis NR1C1684 | 0.45 (0.02)a, x | 0.39 (0.05)a, x | 0.42 (0.07)a, x |
| L. pentosus (B) | 0.75 (0.07)aef, x | 0.7 (0.1)ac, x | 0.7 (0.1)ac, x |
| L. reuteri DSM 20016T | 0.75 (0.06)aef, x | 0.69 (0.05)acd, x | 0.66 (0.01)ac, x |
| L. reuteri JCM 1112 | 0.9 (0.2)bcf, x | 0.8 (0.1)acde, x | 0.80 (0.07)acd, x |
| L. pentosus (A) | 0.64 (0.03)ac, x | 0.62 (0.08)ac, x | 0.53 (0.09)a, x |
| L. casei Shirota | 0.54 (0.02)ac, x | 0.40 (0.02)ae, y | 0.41 (0.02) a, y |
| L. fermentum F53 | 0.6 (0.2)ac, x | 0.4 (0.1)ac, x | 0.48 (0.06)a, x |
| L. fermentum KC5b | 0.81 (0.09)aef, x | 0.5 (0.3)ac, x | 0.6 (0.2)ac, x |
| L. delbrueckii JCM 1002 | 0.82 (0.09)ab, x | 0.5 (0.3)ac, x | 0.63 (0.07)ae, x |
| E. gallinarum | 1.14 (0.04)bde, x | 1.0 (0.1)bc, x | 0.89 (0.09)ab, x |
| B. infantis ATCC 15697 | 1.4 (0.2)d, x | 1.3 (0.2)bd, x | 0.98 (0.03)ab, y |
| L. crispatus ATCC 33820 | 0.42 (0.07)a | ND | ND |
Results are shown as means (standard deviations) (n = 3). Means in the same column with different letters (a to f) differ (P < 0.05). Means in the same row with different letters (x and y) differ (P < 0.05). Cultures were incubated anaerobically at 37°C for 12 h. ND, no growth observed.
Acid tolerance of cultures.
The effects of acidity on the viability of strains are presented in Table 4 arranged in order of decreasing acid tolerance. Lactobacillus pentosus (B) and Streptococcus thermophilus DSM 20617 were the most acid sensitive of all strains tested, losing viability in less than 15 min at pH 2.0. The total CFU of L. casei Shirota, E. gallinarum, L. brevis NR1C1684, B. infantis ATCC 15697, and Streptococcus bovis ATCC 43143 significantly decreased (P < 0.01) after 15 min in medium at pH 2.0. This decrease ranged from 1.7 to 4.5 log cycles. The viability of L. casei and L. crispatus ATCC 33820 at pH 2.0 significantly decreased (ca. 1 log cycle, P < 0.01) after 30 min, and after 60 min these strains showed a 3- to 4-log-cycle decrease in counts. L. fermentum KC5b, L. delbrueckii JCM 1002, and Lactobacillus acidophilus johnsonii were the most acid-tolerant strains, retaining around 100% viability for up to 2 h at pH 2.0. No significant differences were found between the viability after 2 h for L. fermentum KC5b and L. acidophilus johnsonii (P > 0.05). Although the latter showed a small but significant decrease in viability after 30 min at pH 2.0 (0.4 log cycles, P = 0.042), this was transient, and the viable counts recovered after that time. The maintenance of viability of L. delbrueckii JCM 1002 after 2 h at pH 2.0 was significantly lower than that of L. fermentum KC5b (P = 0.037) but not than that of L. acidophilus johnsonii (P > 0.05).
TABLE 4.
Effect of pH 2.0 on viability of lactic acid bacteria and bifidobacteriaa
| Strain | Viable count (log CFU/ml) at time (min):
|
|||||
|---|---|---|---|---|---|---|
| 0 | 15 | 30 | 45 | 60 | 120 | |
| L. fermentum KC5b | 8.30 (0.03) | 8.25 (0.04) | 8.29 (0.09) | 8.16 (0.01) | 8.17 (0.01) | 8.12 (0.02)* |
| L. acidophilus johnsonii | 8.1 (0.1) | 7.98 (0.05) | 7.6 (0.1)** | 7.9 (0.2) | 8.0 (0.1) | 7.8 (0.1) |
| L. delbrueckii JCM 1002 | 8.50 (0.04) | 8.65 (0.02) | 8.28 (0.03) | 8.28 (0.01) | 8.15 (0.01) | 8.02 (0.02)* |
| E. faecalis | 8.46 (0.03) | 8.45 (0.04) | 8.37 (0.02) | 8.2 (0.1) | 7.83 (0.02)* | 5.26 (0.09)* |
| L. fermentum F53 | 8.20 (0.08) | 8.52 (0.04) | 8.50 (0.02) | 8.16 (0.01) | 7.22 (0.03)* | 5.73 (0.03)* |
| E. durans DSM 20633 | 7.7 (0.1) | 8.8 (0.2) | 7.78 (0.05) | 7.64 (0.06) | 6.92 (0.02)* | ND |
| L. casei | 8.24 (0.05) | 8.26 (0.05) | 7.31 (0.09)* | 5.50 (0.08)* | 4.7 (0.1)* | ND |
| L. crispatus | 6.51 (0.03) | 6.35 (0.03) | 5.42 (0.04)* | 3.81 (0.09)* | 2.22 (0)* | ND |
| L. casei Shirota | 7.91 (0.07) | 6.16 (0.08)* | 5.26 (0.07)* | 4.1 (0.1)* | ND | ND |
| E. gallinarum | 7.9 (0.2) | 5.37 (0.05)* | 3.5 (0.3)* | ND | ND | ND |
| L. brevis NR1C1684 | 8.29 (0.07) | 6.2 (0.1)* | ND | ND | ND | ND |
| B. infantis ATCC 15697 | 8.61 (0.03) | 3.8 (0.2)* | ND | ND | ND | ND |
| S. bovis ATCC 43143 | 7.42 (0.01) | 2.9 (0.2)* | ND | ND | ND | ND |
| L. pentosus (B) | 8.4 (0.2) | ND | ND | ND | ND | ND |
| S. thermophilus DSM 20617 | 6.42 (0.01) | ND | ND | ND | ND | ND |
Cultures were incubated anaerobically at 37°C in MRS broth adjusted to pH 2.0 with HCl. ND, no CFU with zero dilution. Results are shown as means (standard deviations) (n = 3). *, mean significantly different from initial value (t = 0) with P < 0.005; **, mean significantly different from initial value (t = 0) with P < 0.05.
The data from in vitro studies show that some strains of L. acidophilus and Bifidobacterium longum are able to take up cholesterol into their cellular membrane (4, 7, 11, 13, 14). However, controversy still exists as to whether these and other strains of probiotic bacteria can exert any cholesterol-lowering action in vivo. In this study, we have examined the cholesterol assimilation abilities of nine strains of lactic acid bacteria and bifidobacteria of human origin together with 11 commercial probiotics. We assessed the survival abilities of these cultures after passage through conditions set to simulate the upper intestinal tract, i.e., by comparison of their acid and bile tolerance. Both of these traits are required for efficacious probiotics, and variability in survival rates may help to explain controversy about their beneficial aspects, including cholesterol reduction. Considering the purported mechanisms behind this phenomenon, it is rational to assume that probiotic survival in the gut is a prerequisite. The results presented here partly confirm the data of Gilliland et al. (7), Rašić et al. (14), and Tahri et al. (21) in that some strains of lactic acid bacteria and bifidobacteria are able to remove cholesterol from culture medium during anaerobic growth in the presence of bile acids. However, great variability was found between strains and even for the same strain during different experimental runs. On the other hand, small differences were observed in the growth and the specific cholesterol uptake rate of each culture among the three replicates of the same run, as indicated by small associated standard deviations (Table 2). This suggests that the cholesterol assimilation ability of the bacteria is highly dependent on their growth in each run, perhaps reflecting the growth stage of the inoculum used. Indeed, Tahri et al. (20, 21) demonstrated that the ability of some strains of bifidobacteria to take up cholesterol into their cellular membrane was growth associated, since resting cells did not interact with cholesterol. Although in the experiments carried out here the seed cultures were always grown till the end of the exponential phase, under the same conditions, and after at least three subculturings from the stock frozen cultures, differences were still observed in the growth of each strain between runs. This was particularly significant for L. brevis, L. pentosus, L. casei, Lactobacillus reuteri DSM 20016T, and S. bovis. Also, significant variation (P < 0.05) was found between strains in their growth in the test media used, and therefore, the direct comparison of their cholesterol reduction data may give misleading impressions. In order to take this into account, we determined the specific cholesterol uptake rate of each strain, which was standardized with the final dry weight of the culture. However, considerable variation with regard to the cholesterol assimilation in the different runs for the same strain was still observed in some cases. This could be partly explained by a coprecipitation of cholesterol with deconjugated bile acids, which is observed at pH values below 5.5 (9). Since the study was performed without pH control, it is probable that part of the cholesterol present in the growth medium precipitated when the pH had dropped below 5.5, due to bacterial fermentation and short-chain fatty acid formation. The amount of cholesterol coprecipitated with bile acids in each culture may differ between trials due to variations in bacterial growth and therefore the final pH of the medium. However, this phenomenon would be relevant for only those strains that were able to deconjugate bile acids. For the majority of strains tested, cholesterol reduction from the culture medium was higher (although not significantly so) in the presence of more bile acids. This is in agreement with the work of Tahri et al. (22), who reported higher cholesterol assimilation with increased oxgall concentrations in the growth medium. Cultures of L. pentosus (A), Lactobacillus plantarum NDV, L. fermentum F53, L. reuteri DSM 20016T, E. durans DSM 20633, and E. gallinarum all assimilated below 1 mg of cholesterol per g of cells with 0.2% (wt/vol) bile acids in the medium in at least two of the runs. Although the amount of cholesterol assimilated by these strains increased with 0.4% (wt/vol) oxgall, this concentration is unlikely to occur in vivo since the bile acid content of the small intestine is normally below this level (24). Alternatively, cultures of L. brevis NR1C1684, L. casei Shirota, and E. faecalis all assimilated more than 5 mg/g with 0.2% (wt/vol) oxgall in two of the runs. However, the variability found does not allow the conclusion that such strains were better cholesterol assimilators. Previous studies have shown that, in order to assimilate cholesterol, the organisms must be able to grow in the presence of bile (4, 7, 20, 21, 25). With the exception of L. crispatus ATCC 33820, all the cultures tested exhibited some degree of bile tolerance, being able to grow in medium containing up to a concentration of 4,000 mg/liter, four times the considered standard for bile tolerance (24). However, the strains with high bile tolerance did not necessarily assimilate more cholesterol than those with a lower tolerance. For example, L. acidophilus johnsonii was significantly more bile tolerant than L. casei Shirota but did not assimilate more cholesterol. In this study, intrinsic resistance to gastric acid was observed to be a rare property among the commercial probiotic cultures examined. With the exception of L. acidophilus johnsonii and Lactobacillus delbrueckii subsp. bulgaricus JCM 1002, the latter previously reported as poorly tolerant (3), all other commercial strains lost viability after 2 h in a medium at pH 2.0. From all the strains tested, only L. fermentum KC5b, L. delbrueckii JCM 1002, and L. acidophilus johnsonii retained good viability for up to 2 h at pH 2.0 and were therefore considered intrinsically tolerant to acid. In order to exert a beneficial effect in the gut, probiotic cultures must survive passage through the stomach and be tolerant to the bile concentrations in the small intestine (16). Survival at pH 3.0 for 2 h and growth in medium containing 1,000 mg of bile acids per liter are considered standards for acid and bile tolerance of probiotic cultures (24).
Results from the present study showed that the human strain L. fermentum KC5b isolated from the steady state of chemostats with media enriched with bile acids and cholesterol was comparable to the commercial probiotics L. delbrueckii subsp. bulgaricus JCM 1002 and L. acidophilus johnsonii in its ability to tolerate acid and bile. Furthermore, this strain is of human origin, which could be of advantage in its ability to compete with the indigenous microflora. Additional in vitro studies are needed, focused on determining the cholesterol reduction ability of this strain in a variety of media and growth conditions in pH-controlled experiments, as well as the mechanism(s) behind the purported assimilation process. Also, the influence of this strain on the indigenous microflora and overall metabolic activity of the gut should be further assessed in vitro in mixed-culture and mixed-substrate environments prior to the design of any clinical intervention trials.
Acknowledgments
D. Pereira gratefully acknowledges the Portuguese Ministry of Science and Technology (FCT) for a Ph.D. scholarship (PRAXISXXI/BD/19520/99) during the course of this work.
REFERENCES
- 1.Agerbaek, M., L. U. Gerdes, and B. Richelsen. 1995. Hypocholesterolemic effect of a new fermented milk product in healthy middle-aged men. Eur. J. Clin. Nutr. 49:346-352. [PubMed] [Google Scholar]
- 2.Bertolami, M. C., A. A. Faludi, and M. Batlouni. 1999. Evaluation of the effects of a new fermented milk product (Gaio) on primary hypercholesterolemia. Eur. J. Clin. Nutr. 53:97-101. [DOI] [PubMed] [Google Scholar]
- 3.Conway, P. L., S. L. Gorbach, and B. R. Goldin. 1987. Survival of lactic acid bacteria in the human stomach by adhesion to intestinal cells. J. Dairy Sci. 70:1-12. [DOI] [PubMed] [Google Scholar]
- 4.Dambekodi, P. C., and S. E. Gilliland. 1998. Incorporation of cholesterol into the cellular membrane of Bifidobacterium longum. J. Dairy Sci. 81:1818-1824. [DOI] [PubMed] [Google Scholar]
- 5.Danielson, A. D., E. R. Peo, Jr., K. M. Shahani, A. J. Lewis, P. J. Whalen, and M. A. Amer. 1989. Anticholesterolemic property of Lactobacillus acidophilus yogurt fed to mature boars. J. Anim. Sci. 67:966-974. [DOI] [PubMed] [Google Scholar]
- 6.De Roos, N. M., and M. B. Katan. 2000. Effects of probiotic bacteria on diarrhea, lipid metabolism, and carcinogenesis: a review of papers published between 1988 and 1998. Am. J. Clin. Nutr. 71:405-411. [DOI] [PubMed] [Google Scholar]
- 7.Gilliland, S. E., C. R. Nelson, and C. Maxwell. 1985. Assimilation of cholesterol by Lactobacillus acidophilus. Appl. Environ. Microbiol. 49:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray, C. D., and P. R. Kinnear. 2000. SPSS for Windows made simple: release 10. Psychology Press Ltd., Hove, United Kingdom.
- 9.Klaver, F. A. M., and R. van der Meer. 1993. The assumed assimilation of cholesterol by lactobacilli and Bifidobacterium bifidum is due to their bile salt-deconjugating activity. Appl. Environ. Microbiol. 59:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen, L. A., A. Raben, N. Haulrik, A. S. Hansen, M. Manders, and A. Astrop. 2000. Effect of 8 week intake of probiotic milk products on risk factors for cardiovascular diseases. Eur. J. Clin. Nutr. 54:288-297. [DOI] [PubMed] [Google Scholar]
- 11.Lin, M.-Y., and T.-W. Chen. 2000. Reduction of cholesterol by Lactobacillus acidophilus in culture broth. J. Food Drug Anal. 8:97-102. [Google Scholar]
- 12.Mann, G. V. 1974. Studies of a surfactant and cholesteremia in the Maasai. Am. J. Clin. Nutr. 27:464-469. [DOI] [PubMed] [Google Scholar]
- 13.Noh, D. O., S. H. Kim, and S. E. Gilliland. 1997. Incorporation of cholesterol into the cellular membrane of Lactobacillus acidophilus ATCC 43121. J. Dairy Sci. 80:3107-3113. [DOI] [PubMed] [Google Scholar]
- 14.Rašić, J. L., I. F. Vujičić, M. Škrinjar, and M. Vulić. 1992. Assimilation of cholesterol by some cultures of lactic acid bacteria and bifidobacteria. Biotechnol. Lett. 14:39-44. [Google Scholar]
- 15.Salè, F. O., S. Marchesini, P. H. Fishman, and B. Berra. 1984. A sensitive enzymatic assay for determination of cholesterol in lipid extracts. Anal. Biochem. 142:347-350. [DOI] [PubMed] [Google Scholar]
- 16.Sanders, M. E. 2000. Considerations for use of probiotic bacteria to modulate human health. J. Nutr. 130:384S-390S. [DOI] [PubMed] [Google Scholar]
- 17.Schaafsma, G., W. J. A. Meuling, W. van Dokkum, and C. Bouley. 1998. Effects of a milk product, fermented by Lactobacillus acidophilus and with fructo-oligosaccharides added, on blood lipids in male volunteers. Eur. J. Clin. Nutr. 52:436-440. [DOI] [PubMed] [Google Scholar]
- 18.Shaper, A. G., K. W. Jones, M. Jones, and J. Kyobe. 1963. Serum lipids in three nomadic tribes of northern Kenya. Am. J. Clin. Nutr. 13:135-146. [DOI] [PubMed] [Google Scholar]
- 19.Sĵovall, J. 1959. On the concentration of bile acids in the human intestine during absorption. Acta Physiol. Scand. 46:339-345. [DOI] [PubMed] [Google Scholar]
- 20.Tahri, K., J. Crociani, J. Ballongue, and F. Schneider. 1995. Effects of three strains of bifidobacteria on cholesterol. Lett. Appl. Microbiol. 21:149-151. [DOI] [PubMed] [Google Scholar]
- 21.Tahri, K., J. P. Grill, and F. Schneider. 1996. Bifidobacteria strains' behaviour toward cholesterol: coprecipitation with bile salts and assimilation. Curr. Microbiol. 33:187-193. [DOI] [PubMed] [Google Scholar]
- 22.Tahri, K., J. P. Grill, and F. Schneider. 1997. Involvement of trihydroxyconjugated bile salts in cholesterol assimilation by bifidobacteria. Curr. Microbiol. 34:79-84. [DOI] [PubMed] [Google Scholar]
- 23.Taylor, G. R. J., and C. M. Williams. 1998. Effects of probiotics and prebiotics on blood lipids. Br. J. Nutr. 80(Suppl. 2):S225-S230. [PubMed] [Google Scholar]
- 24.Usman, H. A. 1999. Bile tolerance, taurocholate deconjugation, and binding of cholesterol by Lactobacillus gasseri strains. J. Dairy Sci. 82:243-248. [DOI] [PubMed] [Google Scholar]
- 25.Walker, D. K., and S. E. Gilliland. 1993. Relationships among bile tolerance, bile salt deconjugation and assimilation of cholesterol by Lactobacillus acidophilus. J. Dairy Sci. 76:956-961. [DOI] [PubMed] [Google Scholar]


