Abstract
We studied how the introduction of an additional ATP-consuming reaction affects the metabolic fluxes in Lactococcus lactis. Genes encoding the hydrolytic part of the F1 domain of the membrane-bound (F1F0) H+-ATPase were expressed from a range of synthetic constitutive promoters. Expression of the genes encoding F1-ATPase was found to decrease the intracellular energy level and resulted in a decrease in the growth rate. The yield of biomass also decreased, which showed that the incorporated F1-ATPase activity caused glycolysis to be uncoupled from biomass production. The increase in ATPase activity did not shift metabolism from homolactic to mixed-acid fermentation, which indicated that a low energy state is not the signal for such a change. The effect of uncoupled ATPase activity on the glycolytic flux depended on the growth conditions. The uncoupling stimulated the glycolytic flux threefold in nongrowing cells resuspended in buffer, but in steadily growing cells no increase in flux was observed. The latter result shows that glycolysis occurs close to its maximal capacity and indicates that control of the glycolytic flux under these conditions resides in the glycolytic reactions or in sugar transport.
Lactic acid bacteria are used extensively in the dairy industry, where the production of lactic acid is important for texture, flavor, and preservation purposes. In addition, lactic acid bacteria are also used for industrial lactate production, which has numerous applications, such as cosmetics, cleaning agents, and biodegradable polylactic acid polymers. From an industrial point of view there is great interest in improving the performance of these organisms with respect to both the rate and the yield of lactate production.
In spite of the importance of glycolysis for fermentation purposes, it is still not known what controls the glycolytic flux in microbial bioreactors. It has been suggested that the enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) has a high level of control (estimated to be 90% of the control) over the glycolytic flux in nonproliferating cells of Lactococcus lactis (33). However, it has recently been shown that GAPDH has no control over the glycolytic flux in steadily growing L. lactis cells (Solem, Koebmann, and Jensen, unpublished data). The control over the glycolytic flux exerted by lactate dehydrogenase was also reported to be close to zero (2).
According to metabolic control theory (16, 25), flux control can reside in any of the steps in a system; i.e., it can reside in the numerous processes that consume the ATP generated in glycolysis (8, 17). Indeed, we have recently shown that at least 75% of the control over glycolysis in aerobic Escherichia coli cultures occurs in the ATP-consuming reactions (26). This result was obtained by overexpression of genes encoding part of the F1 unit of the (F1F0) H+-ATPase, which resulted in uncoupling of glycolysis from biomass production and a 70% increase in the glycolytic flux.
In this paper we show that expression of genes encoding F1-ATPase can also induce uncoupling of glycolysis from biomass production in L. lactis. Interestingly, the glycolytic flux was not increased in cells steadily growing on glucose, and we concluded that control of the glycolytic flux under these conditions resides in glycolysis itself or perhaps in sugar transport.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Cloning and plasmid propagation were performed by using E. coli BOE270 (6), which was derived from MC1000 (7). Plasmid-free L. lactis subsp. cremoris strain MG1363 (15) was used for studying the effects of uncoupled ATPase activity on growth, biomass yield, and glycolytic flux.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or characteristicsa | Reference or source |
|---|---|---|
| L. lactis strains | ||
| MG1363 | Plasmid-free derivative of NCDO712 | 15 |
| BK1010 | MG1363 transformed with pAK80, Ermr | This study |
| BK1094 | MG1363 transformed with pCP34::atpAGD, Ermr | This study |
| BK1502 | MG1363 transformed with pCPC3::atpAGD, Ermr | This study |
| BK1503 | MG1363 transformed with pCPC4::atpAGD, Ermr | This study |
| BK1506 | MG1363 transformed with pCPC7::atpAGD, Ermr | This study |
| BK1511 | MG1363 transformed with pCPC21::atpAGD, Ermr | This study |
| BK1517 | MG1363 transformed with pCPC33::atpAGD, Ermr | This study |
| BK1525 | MG1363 transformed with pCPC46::atpAGD, Ermr | This study |
| BK1536 | MG1363 transformed with pCPC59::atpAGD, Ermr | This study |
| BK1540 | MG1363 transformed with pCPC63::atpAGD, Ermr | This study |
| BK1542 | MG1363 transformed with pCPC65::atpAGD, Ermr | This study |
| BK1546 | MG1363 transformed with pCPC69::atpAGD, Ermr | This study |
| BK1552 | MG1363 transformed with pCPC75::atpAGD, Ermr | This study |
| BK1557 | MG1363 transformed with pCPC80::atpAGD, Ermr | This study |
| E. coli BOE270 | Cloning host derived from strain MT102, which is an hsdR derivate of MC1000 [araD139 Δ(ara-leu)7679 galU galK lac174 rpsL thi-1] | 6 |
| Plasmids | ||
| pMOSBlue | E. coli cloning vector, pUC18 ori, MCS in lacZ′, Ampr | Pharmacia |
| pAK80 | Promoter probe vector, promoterless lacLM encoding the reporter enzyme β-galactosidase, shuttle vector between E. coli and L. lactis, Ermr | 20 |
| pCP34 | pAK80 derivative carrying constitutive promoter CP34-lacLM, Ermr | 23 |
| pCPC library | Library of pAK80 derivative carrying constitutive promoters with different strengths upstream of lacLM, Ermr | 23 |
| pMOS::atpAGD | pMOSBlue HincII-SpeI::atpAGD SpeI amplified with primer 5704F and primer 6243R from chromosomal DNA of MG1363, 4 kb (positions 3216 to 7240), Ampr | This study |
| pCPCx::AGDb | pCPC library BamHI-BamHI::atpAGD BamHI-BamHI fragment from pMOS::atpAGD, 4 kb (positions 3216 to 7240), Ermr | This study |
| pCP34::atpAGD | pCP32 BamHI-BamHI::atpAGD BamHI-BamHI fragment from pMOS::atpAGD, 4 kb (positions 3216 to 7240), Ermr | This study |
The feature of a plasmid is indicated by the vector ligated to the insert. The restriction endonuclease used for digestion is shown. The kilobase values indicate the sizes of inserts. The coordinates in parentheses are the sequence positions in the atp operon deposited in the National Center for Biotechnology Information under accession no. AF059739. Ampr, ampicillin resistance gene; Ermr, erythromycin resistance gene.
A library of 98 plasmids with different promoters was obtained in this study.
Media and growth conditions.
E. coli was routinely grown with agitation at 30°C in Luria-Bertani (LB) broth (36). L. lactis was routinely cultivated at 30°C without aeration in M17 broth (40) or in chemically defined SA medium (21) supplemented with 5 to 10 g of glucose per liter and appropriate antibiotics.
Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml (for selection of a pMOSBlue derivative in E. coli); and erythromycin, 5 μg/ml (for selection of pAK80 derivatives in L. lactis) and 200 μg/ml (for selection of pAK80 derivatives in E. coli).
Growth experiments with L. lactis were carried out at 30°C by using batch cultures in flasks containing 100 ml of SA medium (21) supplemented with 1.0 g of glucose per liter and 5 μg of erythromycin per ml. The strains were inoculated by using growing overnight cultures at low densities 6 to 10 h before optical densities were first measured in order to obtain exponentially growing cells. A strain containing promoter cloning vector pAK80 was used as a reference. Slowly rotating magnets were used to keep the cultures homogeneous. Regular measurements of optical density at 450 nm (OD450) were obtained, and samples were withdrawn and used for determination of ATP and ADP concentrations and for high-performance liquid chromatography (HPLC) to measure glucose and by-product contents. The cell density was correlated with the cell mass of L. lactis as follows: 0.19 g (dry weight)/liter of SA medium was equivalent to an OD450 of 1 (31). The biomass yield was determined from the cell density divided by the glucose concentration by using a molar weight of glucose of 198 g/mol. The glycolytic flux was routinely calculated from the specific growth rate and the biomass yield (specific growth rate/biomass yield), assuming steady-state conditions, and was validated by HPLC measurements. The fluxes measured by HPLC matched the fluxes deduced from specific growth rate/biomass yield with an error of less than 3%.
Growth of resuspended cells.
L. lactis was grown in 100 ml of SA medium supplemented with 2 g of glucose per liter to an OD450 of 0.9. The cultures were put on ice. After cooling, the cells were centrifuged (7,000 × g for 10 min) and washed once with SA medium supplemented with 2 g of glucose per liter but without amino acids or vitamins. The cells were resuspended in the latter medium to an OD450 of 0.9. Samples were withdrawn for measuring the ATP and ADP concentrations, and samples were also used for HPLC to measure glucose and product concentrations at 10, 30, 70, 150, and 310 min after resuspension.
DNA techniques and DNA isolation.
Extraction of chromosomal DNA from L. lactis was carried out as previously described (24), with the following modifications: a final concentration of 15.5 mg of lysozyme per ml for 30 min was used for lysis, and the sample was incubated with sodium dodecyl sulfate for 10 min at 37°C and then for 10 min at 65°C. PCR amplification with Pfu DNA polymerase (Stratagene, La Jolla, Calif.) was performed as recommended by the manufacturer. Plasmid DNA from E. coli for analytic purposes was isolated by using an alkaline lysis method, and plasmid DNA from E. coli for preparative purposes was isolated by using Qiagen columns (Qiagen, Hilden, Germany). Plasmid DNA from L. lactis was isolated as described by Birnboim and Doly (5), with the following modification: a final concentration of 15.5 mg of lysozyme per ml for 20 min at 37°C was used for lysis of the cell wall. Digestion with restriction enzymes (Gibco BRL, Pharmacia, New England Biolabs) and treatment with T4 DNA ligase (Gibco BRL) and calf intestine alkaline phosphatase (Pharmacia) were carried out by using standard recombinant DNA techniques as described by Sambrook et al. (36) and as recommended by the manufacturers. DNA fragments were purified from agarose gels by using GFX columns (Pharmacia) or a High Pure PCR product purification kit (Boehringer Mannheim). Linearized cloning vectors were treated with calf intestine alkaline phosphatase to avoid religation of the vector.
Transformation.
E. coli strains were made competent by using a CaCl2 treatment (36). After transformation, the cells were regenerated in LB medium (36) and subsequently transferred to LB agar plates supplemented with the appropriate antibiotic. Cells of L. lactis were made competent by growing them in M17 medium supplemented with 5 g of glucose per liter and 10 g of glycine per liter and resuspending them in a solution containing 100 g of glycerol per liter and 0.5 M sucrose as described by Holo and Nes (18). Plasmid DNA was used to transform the cells by electroporation (18), and the cells were allowed to regenerate in Schmidt-Ruppin medium (30) for 2 h and then plated on Schmidt-Ruppin agar plates supplemented with the appropriate antibiotics. Histochemical screening for lacLM was carried out with 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) (Sigma) at a final concentration of 100 μg/ml.
PCR amplification and cloning of atpAGD from L. lactis MG1363.
A 4.0-kb DNA fragment encoding atpAGD (positions 3205 to 7240) from the atp operon of L. lactis MG1363 (sequence positions 3205 to 7240 of the atp operon deposited in the National Center for Biotechnology Information data bank under accession no. AF059739) was amplified by PCR with primer 5704F (5′-CGGGATCCAGCTAAATAGCCTTGAACTAG-3′) and primer 6243R (5′-CTGGATCCTTTCATAAGAAATCGAATTAATAACCC-3′) purchased from Hobolth DNA Synthese, Hillerød, Denmark (Fig. 1A). The resulting PCR product was digested with SpeI, positioned immediately upstream of the ribosome binding site of atpA, and cloned in plasmid cloning vector pMOSBlue (Pharmacia) digested with SpeI and HincII (2.7 kb), resulting in pMOS::atpAGD.
FIG. 1.
Cloning of PCR fragment of atpAGD from L. lactis MG1363. (A) PCR amplification of the atpAGD genes from L. lactis MG1363. A 4.0-kb PCR fragment encoding the α, γ, and β subunits was amplified with primers 5704F and 6243R (sequence positions 3205 to 7240 of the atp operon deposited in the National Center for Biotechnology Information data bank under accession no. AF059739) and cloned in pMOSBlue. (B) Plasmids used for introduction of F1-ATPase in L. lactis. A 4.1-kb fragment encoding atpAGD was cloned downstream of constitutive promoters in a transcriptional fusion with the reporter gene lacLM coding for β-galactosidase. The open reading frames are indicated by boxes, and the designations of the genes and their products are shown above the boxes. The arrows indicate the promoters and show the direction of transcription. The plasmid designation is shown to the left of the linear plasmid. The plasmid is not drawn to scale. In this study a library of new synthetic constitutive promoters was employed. CPC, individual constitutive promoters (23); ori, origin of replication; erm, erythromycin resistance; lacLM, β-galactosidase.
Cloning of atpAGD under control of synthetic constitutive promoters.
The relevant features of the plasmid constructs are illustrated in Fig. 1B and listed in Table 1. A 4.1-kb fragment containing atpAGD was obtained from pMOS::atpAGD by digestion with BamHI. The atpAGD fragment was cloned after synthetic promoters from a pCPC library (23) digested with BamHI, resulting in 98 clones (pCPCx::atpAGD) which expressed the atpAGD genes to different extents.
Measurement of β-galactosidase activity.
Sampling was carried out by pipetting 1-ml portions of culture into ice-cooled Eppendorf tubes containing 12.5 μl of 0.1% sodium dodecyl sulfate and 25 μl of chloroform to permeabilize the cells. After vortexing for 10 s, the samples were placed on ice until the enzyme activities were measured. The extracts were diluted with suitable volumes of Z-buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4; pH 7.0) to obtain a total volume of 1 ml in each case. Each diluted sample was preheated at 30°C for 5 min, and the enzyme activity was determined by a standard procedure described by Miller (28) and modified by Israelsen and coworkers (20). β-Galactosidase activities greater than 400 Miller units were subject to larger errors due to instability resulting from high levels of expression of the genes encoding F1-ATPase.
Measurement of the intracellular ATP and ADP concentrations.
Samples (0.5 ml) were quenched in 0.5 ml of 80°C buffer-saturated phenol (equilibrated with 10 mM Tris-1 mM EDTA, pH 8.0) containing 0.6 g of glass beads (diameter, 106 μm; Sigma) as previously described (26). Treatment and measurement of the intracellular concentrations of ATP and ADP were carried out as previously described (26). Determinations of ratios of ATP to ADP greater than 15 were more uncertain due to relatively low levels of ADP.
Measurement of G6P and FBP concentrations.
Extracts were prepared from batch cultures at OD450 of 0.6 to 1.8 by quenching in 80°C buffer-saturated phenol (equilibrated with 10 mM Tris-1 mM EDTA, pH 8.0) supplemented with 0.6 g of glass beads (diameter, 106 μm; Sigma) as previously described for measurement of ATP and ADP concentrations (26). After chloroform treatment, the concentrations of glucose 6-phosphate (G6P) and fructose 1,6-bisphosphate (FBP) were measured by recording the increase in NADH fluorescence as described previously (1, 44), with the following modification: the buffer contained 50 mM triethanolamine (pH 7.5) instead of imidazole hydrochloride. Glucose-6-phosphate dehydrogenase, aldolase, and glycerol-3-phosphate dehydrogenase were obtained from Boehringer GmbH, Mannheim, Germany. Intracellular concentrations of sugar phosphates (42) were calculated by assuming that 1 g (dry weight) corresponded to 1.67 ml of intracellular volume.
Quantification of glucose and end products by HPLC.
Glucose and product concentrations were determined by HPLC as previously described (1). The carbon balance was calculated by determining the recovery of substrates converted into products in terms of C-moles.
RESULTS
Expression of genes encoding F1-ATPase activity in L. lactis.
To analyze how increased ATP consumption affected L. lactis physiology, genes encoding the hydrolytic part of F1-ATPase were overexpressed in L. lactis MG1363. In reconstitution experiments the combination of gene products α, γ, and β has been found to be most active with respect to ATP hydrolysis in vitro (10), and in vivo experiments with E. coli have confirmed this observation (26). To study the effect of a gradual increase in uncoupled ATPase activity in L. lactis, the atpA, atpG, and atpD genes encoding the α, γ, and β subunits, respectively, were cloned into a library of synthetic constitutive promoters having different strengths (23). Clones were obtained in which the atpAGD genes were inserted into an operon structure together with the lacLM genes encoding a β-galactosidase (Fig. 1B). The specific β-galactosidase activities in these cells should therefore reflect the expression of the F1-encoding genes.
We found a good correlation between the measured β-galactosidase activities and the biomass yields; greater expression of the lacLM reporter gene resulted in a gradually decreasing growth yield. This indicates that there was increased uncoupling of glycolysis from biomass production as a result of increased ATPase activity (Fig. 2). At the highest β-galactosidase activities the biomass yield was reduced to one-half of the wild-type biomass yield.
FIG. 2.
Correlation between specific β-galactosidase activities and biomass yield for the F1-ATPase library. The specific β-galactosidase activities and biomass yields were measured for overnight cultures of L. lactis strains grown in SA medium supplemented with 1.5 g of glucose per liter and 5 μg of erythromycin per ml. (A) Primary data for L. lactis strains containing ATPase plasmids from the pCPCx::atpAGD plasmid library. The bars and lines indicate the specific β-galactosidase activities and biomass yields, respectively. (B) Correlation between specific β-galactosidase activity and biomass yield. The symbols represent clones with a range of expression of genes coding for F1-ATPase.
Effect of F1-ATPase activity on the intracellular energy state.
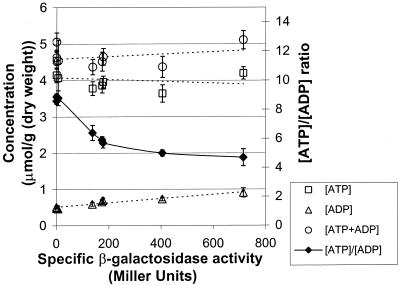
We then measured how expression of atpAGD affected the intracellular energy state. Nine selected strains were grown in SA medium supplemented with glucose, and samples were withdrawn and used to determine the intracellular ATP and ADP concentrations (Fig. 3). As the F1-ATPase activity increased, as estimated indirectly by the β-galactosidase activity, the intracellular ATP concentration decreased gradually and the ADP concentration increased, which resulted in a gradual decrease in the [ATP]/[ADP] ratio from 9 to 4.7 (Fig. 3). The total concentrations of ATP plus ADP remained comparable.
FIG. 3.
Effect of uncoupled F1-ATPase on the intracellular energy level: correlation of specific β-galactosidase activity with the ATP, ADP, and ATP plus ADP pools and the [ATP]/[ADP] ratio. The experimental data for the ATP, ADP, and ATP plus ADP pools are fitted to linear curves indicated by dotted lines, and the [ATP]/[ADP] ratio is fitted to a curve indicated by a solid line. The error bars indicate the standard deviations of the measurements.
Effect of ATPase activity on growth and glycolytic flux of L. lactis.
We subsequently characterized 10 strains in batch fermentations with respect to specific growth rate, biomass yield, and glycolytic flux in chemically defined SA medium (21) supplemented with 1 g of glucose per liter and 5 μg of erythromycin per ml (Table 2 and Fig. 4). Expression of genes encoding F1-ATPase had a negative effect on the growth rate; as the expression level increased, the growth rate decreased gradually to 69% of the value for the reference strain. At the highest level of expression of F1-ATPase the growth yield dropped to 69%. The uncoupling of glycolysis from biomass production is illustrated by the glucose consumption expressed as a function of biomass production (Fig. 5). The uncoupling had very little effect on the glycolytic flux, regardless of the expression of the F1 genes employed (Table 2).
TABLE 2.
Growth and sugar consumption of derivatives of strain L. lactis MG1363 with uncoupled F1-ATPase activity (α, γ, and β subunits)
| Strain | Plasmid | β-Galactosidase sp act (Miller units)a | Biomass yield (g [dry wt]/mol of glucose) | Growth rate (h−1) | Glucose flux (mmol of glucose/ h/g [dry wt])b | Biomass yield (%)c | Growth rate (%)c | Glucose flux (%)c |
|---|---|---|---|---|---|---|---|---|
| BK1010 | pAK80 | 0 | 30.5 | 0.74 | 24.2 | 100 | 100 | 100 |
| BK1546 | pCPC69::atpAGD | 0.2 | 30.2 | 0.73 | 24.1 | 99 | 99 | 100 |
| BK1540 | pCPC63::atpAGD | 5.9 | 30.1 | 0.73 | 24.4 | 99 | 100 | 101 |
| BK1094 | pCP34::atpAGD | 95 | 28.0 | 0.71 | 25.3 | 92 | 96 | 105 |
| BK1506 | pCPC7::atpAGD | 137 | 27.8 | 0.67 | 24.2 | 91 | 92 | 100 |
| BK1542 | pCPC65::atpAGD | 173 | 26.6 | 0.66 | 24.9 | 87 | 90 | 103 |
| BK1552 | pCPC75::atpAGD | 177 | 26.4 | 0.65 | 24.6 | 87 | 88 | 102 |
| BK1503 | pCPC4::atpAGD | 307 | 23.4 | 0.58 | 25.0 | 77 | 79 | 103 |
| BK1557 | pCPC80::atpAGD | 405 | 23.9 | 0.56 | 23.5 | 78 | 76 | 97 |
| BK1502 | pCPC3::atpAGD | 714 | 21.0 | 0.51 | 24.2 | 69 | 69 | 100 |
Expression of the lacLM genes downstream of atpAGD is indicated by the β-galactosidase specific activity (28) and reflects expression of the atpAGD genes.
The glycolytic flux was calculated from the specific growth rate and the biomass yield as follows: specific growth rate/biomass yield.
Values are relative to wild-type levels in BK1010.
FIG. 4.
Growth curves of derivatives of L. lactis MG1363 with uncoupled F1-ATPase. Cell density (OD450) is plotted as a function of time for the cultures. The cultures were grown in batches without aeration at 30°C in SA medium (21) supplemented with 1 g of glucose per liter and 5 μg of erythromycin per ml. The strains and their doubling times (td) (in minutes) are indicated on the right. The growth curves are offset for clarity. The standard deviations of the calculated doubling times were <3%.
FIG. 5.
Steady-state consumption of glucose in L. lactis strains with uncoupled F1-ATPase during batch fermentation. The consumption of glucose in a time course experiment as measured by HPLC is plotted as a function of cell density (OD450). The plasmids of the individual strains are indicated at the bottom. pAK80 is a vector without atp genes. The samples were taken from an independent experiment.
L. lactis remained almost homolactic in spite of a reduced energy state.
End product formation in the growth experiment described above was measured by HPLC. Formate was used to evaluate the activity of the mixed-acid pathway. Interestingly, the strains with expression of F1-ATPase remained essentially homolactic during growth with concentrations of lactate ranging from 8.8 to 8.6 mM (Fig. 6). Only a small amount of glucose was converted to the mixed-acid products with concentrations of formate below 1.1 mM even at a high level of expression of atpAGD. The levels of carbon recovery were 85 to 95% (based on C-moles), which is in agreement with results obtained previously with defined medium (1, 2, 32).
FIG. 6.
Lactate and formate production by strains with incorporated F1-ATPase in the batch experiment described in the legend to Fig. 4. Samples for HPLC measurements were taken when the cultures entered the stationary phase. The level of recovery of C-moles ranged between 85 and 95%.
Metabolite measurements.
We then measured how the decrease in the [ATP]/[ADP] ratio affected the levels of the intracellular metabolites by measuring concentrations of G6P and FBP in the wild-type strain MG1363 and in BK1502, which has a high level of expression of atpAGD. G6P and FBP are upstream and downstream metabolites, respectively, of the ATP-driven glycolytic enzyme phosphofructokinase (PFK). Interestingly, the reduction in the energy level resulted in a twofold increase in the G6P concentration, from 16 to 32 mM, whereas the FBP concentration decreased by approximately 20%, from 26.4 to 21.5 mM (Table 3).
TABLE 3.
Pools of G6P and FBPa
| Strain | G6P concn (mM) | FBP concn (mM) |
|---|---|---|
| MG1363 | 16.4 ± 0.9 | 26.4 ± 1.2 |
| BK1502 | 31.5 ± 2.2 | 21.5 ± 0.82 |
The intracellular metabolite concentrations were obtained from three determinations. The values are means ± standard deviations.
Glycolytic flux in nongrowing cells increased when F1-ATPase was incorporated.
Since no increase in the glycolytic flux was observed in the exponentially growing cells, it was tempting to conclude that glycolysis was already running close to its maximal capacity under these conditions and that the activity of the ATP-demanding processes was already so high that the cells were unable to compensate for additional removal of ATP by increasing the glycolytic flux. If this scenario is correct, then it should be possible to increase the glycolytic flux in cells which have a reduced glycolytic flux. To test this hypothesis, we performed an experiment with cells that exhibited different levels of expression of genes encoding F1-ATPase and were resuspended in buffer (SA medium without vitamins and amino acids). Under such nongrowth conditions the ATP-demanding anabolic reactions decreased as a result of the lack of essential nutrients. In the wild-type cells the glycolytic flux was 33% of the glycolytic flux in steadily growing cells (Fig. 7). Interestingly, expression of genes encoding F1-ATPase increased the glycolytic flux until the flux observed in growing cells was approached, but not beyond this point. The results indicated that glycolysis was running close to its maximal capacity in cells growing exponentially in SA medium supplemented with glucose. These observations also show that under conditions where the activity of the ATP-demanding processes is reduced, it is possible to increase the flux through glycolysis by introducing an additional ATP-consuming reaction.
FIG. 7.
Effect of uncoupled F1-ATPase in nongrowing cells. The glycolytic fluxes in nongrowing cells are plotted as a function of the β-galactosidase activities of strains measured with steadily growing cells. The glycolytic fluxes were estimated from measurements of glucose consumption by HPLC. The strains used were MG1363, BK1511, BK1503, BK1525, BK1517, and BK1536, which exhibit gradually increased expression of genes coding for F1-ATPase. The glycolytic flux in steadily growing L. lactis MG1363 in the experiment is indicated by a solid line.
We also measured how the introduced F1-ATPase affected the [ATP]/[ADP] ratio in these cells. In nongrowing wild-type cells the ratio was more than 25, whereas introduction of F1-ATPase resulted in a gradual decrease in the [ATP]/[ADP] ratio to 10 (Table 4).
TABLE 4.
Intracellular [ATP]/[ADP] ratios in resuspended cells
| Strain | Plasmid | β-Galactosidase sp act (Miller units)a | [ATP]/[ADP] ratiob |
|---|---|---|---|
| MG1363 | None | 0 | >25 |
| BK1525 | pCPC46::atpAGD | 451 ± 23 | 20 ± 4 |
| BK1517 | pCPC33::atpAGD | 982 ± 70 | 10 ± 1 |
| BK1536 | pCPC59::atpAGD | 1,082 ± 8 | 10 ± 1 |
Expression of the lacLM genes downstream of atpAGD is indicated by the β-galactosidase specific activity (28) and reflects expression of the atpAGD genes. The values are means ± standard deviations for four measurements.
Intracellular [ATP]/[ADP] ratios were obtained from three or four determinations. The values are means ± standard deviations.
Control of glycolysis exerted by the ATP-consuming processes in L. lactis.
It was clear from the primary data that virtually no increase in the glycolytic flux occurred when an additional ATP-consuming process was introduced into steadily growing cells. This demonstrated that the ATP-consuming processes have no control over the glycolytic flux. This is in sharp contrast to our observations with E. coli, in which the ATP-consuming reactions had at least 75% of the control (26). If we assume that ATP consumption is strictly coupled to growth (17, 45), the energy metabolism of L. lactis can be divided into a catabolic module (ATP supply) and an anabolic module (ATP demand), which are linked by the [ATP]/[ADP] ratio: e1e2 Substrate ΔGp Growth where ΔGp is the cellular energy state and e1 and e2 represent the anabolic and catabolic modules, respectively. The flux control of this simple metabolic scheme can be expressed in terms of the elasticities (the metabolic control analysis term for sensitivities) (J. A. Burns, A. Cornish-Bowden, A. K. Groen, R. Heinrich, H. Kacser, J. W. Porteous, S. M. Rapoport, T. A. Rapoport, J. W. Stucki, J. M. Tager, R. J. A. Wanders, and H. V. Westerhoff, Letter, Trends Biochem. Sci. 10:16, 1985) of the two blocks:
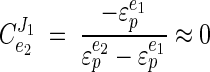 |
This equation states that the control by the ATP-consuming reactions of the glycolytic flux (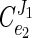 ) can be calculated if we know how elastic the catabolic reactions (
) can be calculated if we know how elastic the catabolic reactions (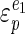 ) and the anabolic reactions (
) and the anabolic reactions (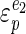 ) are towards changes in the cellular energy state. A high elasticity coefficient means that a block is very sensitive, while a value close to zero means that a block is completely insensitive. The absence of control of the glycolytic flux by ATP-consuming reactions can then be explained either by a low elasticity of catabolism (
) are towards changes in the cellular energy state. A high elasticity coefficient means that a block is very sensitive, while a value close to zero means that a block is completely insensitive. The absence of control of the glycolytic flux by ATP-consuming reactions can then be explained either by a low elasticity of catabolism (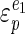 ) towards the cellular energy state or by a relatively higher elasticity of anabolism (
) towards the cellular energy state or by a relatively higher elasticity of anabolism ( ) towards the cellular energy state.
) towards the cellular energy state.
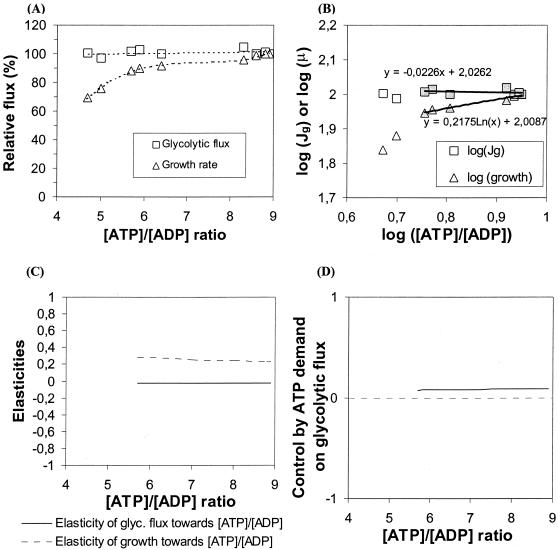
The relationship among the growth rate, the glycolytic flux, and the cellular energy state ([ATP]/[ADP]) is plotted in Fig. 8. The glycolytic flux does not change much over a twofold change in the [ATP]/[ADP] ratio, while the growth rate changes even when a small change in the [ATP]/[ADP] ratio is induced. Figure 8B shows a log-log plot of the glycolytic flux and the growth rate as a function of the intracellular [ATP]/[ADP] ratio in strains with modulated expression of genes encoding F1-ATPase. Different curves (logarithmic, linear, exponential, power) were fitted to the data points with low to moderate changes in the [ATP]/[ADP] ratio. We then determined the elasticities of the catabolic and anabolic blocks towards the [ATP]/[ADP] ratio (Fig. 8C), which are simply the slopes of the two curves in Fig. 8B. The elasticity of the catabolic block was estimated to be close to zero (−0.02), which confirms that glycolysis is very insensitive to changes in the energy level. The elasticity of the anabolic block (growth rate) ranged between 0.22 and 0.26 in the absence of ATPase, depending on the applied curve fit, and increased at the highest level of ATPase.
FIG. 8.
Dependence of glycolytic flux and growth rate on the [ATP]/[ADP] ratio and calculation of elasticity and flux control coefficients. Cultures of L. lactis MG1363 derivatives were grown as described in the legend to Fig. 4. (A) Relative glycolytic fluxes and growth rates plotted as a function of the [ATP]/[ADP] ratio for derivatives of L. lactis MG1363 in which genes encoding F1-ATPase are expressed from synthetic promoters with different strengths. (B) Logarithmic (scaled) relative glycolytic fluxes (Jg) and growth rates (μ) plotted as functions of logarithmic [ATP]/[ADP] ratios for derivatives of L. lactis MG1363 in which the atpAGD genes are expressed at five different strengths. The experimental data points indicated by shaded symbols are fitted to curves shown by solid lines. The equations are indicated above and below the plots. (C) Elasticities of the glycolytic flux (glyc. flux) and the growth rate towards the intracellular [ATP]/[ADP] ratio. The elasticities are the slopes of the scaled fluxes towards the [ATP]/[ADP] ratios shown in panel B, calculated from the fitted equations. (D) Flux control by the demand for ATP on the glycolytic flux as a function of the [ATP]/[ADP] ratio, calculated by using the equation in the text.
The control exerted by the ATP-consuming processes was then calculated from the equation given above (Fig. 8D). The calculations showed that the ATP-consuming reactions have less than 10% of the flux control over glycolysis (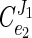 ≈ 0.09). According to metabolic control analysis, the flux control values should add up to 1, which means that the catabolic module (i.e., glycolysis itself and/or the sugar uptake system) might have a flux control value close to 1.
≈ 0.09). According to metabolic control analysis, the flux control values should add up to 1, which means that the catabolic module (i.e., glycolysis itself and/or the sugar uptake system) might have a flux control value close to 1.
DISCUSSION
Introduction of uncoupled ATPase activity by expression of genes encoding F1-ATPase.
We showed that overexpressing genes coding for the F1-part of the H+-ATPase in L. lactis introduces ATPase activity in vivo. The uncoupled F1-ATPase significantly reduced the biomass yield and the intracellular energy level. The significant reduction in biomass yield was not a result of non-growth-related energy consumption (maintenance), since maintenance has been estimated to account for less than 3% of the reduction in biomass yield under energy-limiting conditions in defined medium (4, 39). The fact that the biomass yield decreased in combination with the reduction in the intracellular energy level is therefore a strong indication that an uncoupled ATPase is active in vivo, as recently found for E. coli (26). Our results are also in agreement with early reconstitution experiments, in which complexes of α, γ, and β subunits reconstituted from isolated individual subunits of E. coli (9, 10, 11), Bacillus strain PS3 (46), and Salmonella enterica serovar Typhimurium (19) were found to have high ATPase activities.
The reduction in the intracellular energy level with an increased level of uncoupled F1-ATPase did not result in a shift of metabolism from homolactic fermentation to a mixed-acid pattern that would otherwise have provided the cell with extra ATP (14, 43). This is in agreement with the results of previous chemostat experiments, which showed that cells remained homolactic at a dilution rate of 0.5 h−1 (27, 43).
Glycolytic flux in growing L. lactis is close to the maximal capacity.
The glycolytic flux was virtually unaffected by the expression of genes encoding F1-ATPase in growing cells. This means that the flux control by the ATP-demanding processes was close to zero, indicating that glycolysis in L. lactis is close to the maximal capacity under these conditions. The lack of control by demand for ATP on the glycolytic flux is in sharp contrast to previous results obtained with E. coli, in which the glycolytic flux was observed to increase gradually in response to increased expression of genes encoding F1-ATPase (26). The difference may reflect differences in energy generation. The homofermentative metabolism of L. lactis yields only 2 mol of ATP per mol of glucose, whereas aerobic organisms that possess oxidative phosphorylation can obtain 8 to 10 mol of ATP per mol of hexose (22). L. lactis therefore requires a considerably higher flux through glycolysis for energy production (∼24 mmol of glucose/h/g [dry weight] at 30°C in defined medium) than E. coli requires (∼7 mmol of glucose/h/g [dry weight] at 30°C in minimal medium) (26) in order to obtain sufficient energy for rapid growth.
To test this hypothesis, experiments were performed with nongrowing cells. In nongrowing cells the glycolytic flux of the wild type was approximately threefold lower than that in growing cells, since no ATP was being used for biomass production. The glycolytic flux increased gradually with increased expression of F1 genes until the level found in exponentially grown cultures was approached. Thus, it appears that when there is excess glycolytic capacity, the flux through glycolysis can indeed be increased by increasing the ATP turnover in the cells until the full capacity is being employed. The finding that there was control by ATP demand under conditions under which anabolism was limited is in agreement with a mechanistic model for glycolysis in L. lactis that describes the sugar metabolism in nongrowing cells (29).
A decrease in the intracellular energy state of the cell may affect the kinetics of enzymatic steps involved in metabolism. Such a kinetic effect has, for instance, been demonstrated for PFK by simulations with erythrocytes (34). The reaction catalyzed by PFK is an ATP-requiring step, after which further metabolism yields surplus ATP, also referred to as a turbo design (41). Lowering the [ATP]/[ADP] ratio may therefore ultimately reduce the activity of PFK. The excess capacity of PFK in L. lactis was recently found to be small, and a slight reduction in PFK activity results in an increased pool of upstream metabolites (1). Indeed, in growing cells the level of the upstream metabolite G6P increased twofold in the presence of F1-ATPase compared to the level in the reference strain, while the concentration of FBP was found to be slightly decreased. The increased concentration of G6P and the decreased concentration of FBP observed here fit well with the potential risk of a turbo design, as suggested by Teusink and coworkers (41), resulting in decreased glycolytic flux at low [ATP]/[ADP] ratios.
According to metabolic control theory flux control is usually distributed over several steps. Our results indicate that control resides either in glycolysis itself or perhaps in glucose uptake. Interestingly, the glycolytic enzymes in yeast and also sugar transport in E. coli have been found to exert no significant control over the glycolytic flux (35, 37). It has been suggested that in L. lactis GAPDH has a high level of flux control in nongrowing cells (33). However, it was recently found that GAPDH has no control over the glycolytic flux in growing cells of L. lactis (Solem et al., unpublished). Lactate dehydrogenase had no control over the glycolytic flux either (2). A recent study of sugar catabolism in L. lactis IL-1403 also suggested a catabolic flux limitation (12), and due to concomitant high in vivo glycolytic enzyme capacities it was suggested that the control was probably at the level of sugar transport (12).
Based on the existing data we suggest that simultaneous modulation of several metabolic steps might be necessary to increase the glycolytic flux. Thus, by increasing the glycolytic capacity in L. lactis, the control of glycolysis is likely to shift to ATP demand, which can then be increased by introduction of uncoupled F1-ATPase, as shown previously for E. coli (26) and here for nongrowing L. lactis. This would be in agreement with a recent study of glycolysis in Trypanosoma brucei, in which experiments and computer simulations indicated that control of the glycolytic flux is shared by several steps in the pathway (3).
It may also be important to achieve appropriate levels of each of the individual enzymes involved in order to maintain proper metabolite concentrations (1, 13). At first, the task of simultaneous and accurate modulation of many genes in a cell may seem impracticable. However, the so-called SPL technology developed recently in our laboratory (23, 38) involving synthetic promoter libraries has greatly facilitated such an undertaking.
Acknowledgments
This work was supported by The Danish Academy of Technical Sciences (ATV), The Danish Research Agency, and Chr. Hansen A/S.
We thank Regina Shürmann for excellent technical assistance.
REFERENCES
- 1.Andersen, H. W., C. Solem, K. Hammer, and P. R. Jensen. 2001. Twofold reduction of phosphofructokinase activity in Lactococcus lactis results in strong decreases in growth rate and in glycolytic flux. J. Bacteriol. 183:3458-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, H. W., M. B. Pedersen, K. Hammer, and P. R. Jensen. 2001. Lactate dehydrogenase has no control on lactate production but has a strong negative control on formate production in Lactococcus lactis. Eur. J. Biochem. 268:6379-6389. [DOI] [PubMed] [Google Scholar]
- 3.Bakker, B. M., H. V. Westerhoff, F. R. Opperdoes, and P. A. Michels. 2000. Metabolic control analysis of glycolysis in trypanosomes as an approach to improve selectivity and effectiveness of drugs. Mol. Biochem. Parasitol. 106:1-10. [DOI] [PubMed] [Google Scholar]
- 4.Benthin, S. 1992. Growth and product formation of Lactococcus cremoris. Ph.D. thesis. Technical University of Denmark, Lyngby, Denmark.
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boogerd, F. C., L. Boe, O. Michelsen, and P. R. Jensen. 1998. atp mutants of Escherichia coli fail to grow on succinate due to a transport deficiency. J. Bacteriol. 180:5855-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casabadan, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 8.Cornish-Bowden, A., J.-H. S. Hofmeyr, and M. L. Cardenas. 1995. Strategies for manipulating metabolic fluxes in biotechnology. Bioorg. Chem. 23:439-449. [Google Scholar]
- 9.Dunn, S. D., and M. Futai. 1980. Reconstitution of a functional coupling factor from the isolated subunits of Escherichia coli F1 ATPase. J. Biol. Chem. 255:113-118. [PubMed] [Google Scholar]
- 10.Dunn, S. D., and L. A. Heppel. 1981. Properties and functions of the subunits of the Escherichia coli coupling factor ATPase. Arch. Biochem. Biophys. 210:421-436. [DOI] [PubMed] [Google Scholar]
- 11.Ekuni, A., H. Watanabe, N. Kuroda, K. Sawada, H. Murakami, and H. Kanazawa. 1998. Reconstitution of F1-ATPase activity from Escherichia coli subunits α, β and subunit γ tagged with six histidine residues at the C-terminus. FEBS Lett. 427:64-68. [DOI] [PubMed] [Google Scholar]
- 12.Even, S., N. D. Lindley, and M. Cocaign-Bousquet. 2001. Molecular physiology of sugar catabolism in Lactococcus lactis IL1403. J. Bacteriol. 183:3817-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fell, D. A., and S. Thomas. 1995. Physiological control of metabolic flux: the requirement for multisite modulation. Biochem. J. 311:35-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fordyce, A. M., V. L. Crow, and T. D. Thomas. 1984. Regulation of product formation during glucose or lactose limitation in nongrowing cells of Streptococcus lactis. Appl. Environ. Microbiol. 48:332-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinrich, R., and T. A. Rapoport. 1974. A linear steady-state treatment of enzymatic chains: general properties, control and effector-strength. Eur. J. Biochem. 42:89-95. [DOI] [PubMed] [Google Scholar]
- 17.Hofmeyr, J.-H. S., and A. Cornish-Bowden. 2000. Regulating the cellular economy of supply and demand. FEBS Lett. 467:47-51. [DOI] [PubMed] [Google Scholar]
- 18.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu, S.-Y., M. Senda, H. Kanazawa, T. Tsuchiya, and M. Futai. 1984. Comparison of F1's of oxidative phosphorylation from Escherichia coli and Salmonella typhimurium and demonstration of interchangeability of their subunits. Biochemistry 23:988-993. [DOI] [PubMed] [Google Scholar]
- 20.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, P. R., and K. Hammer. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, P. R., O. Michelsen, and H. V. Westerhoff. 1995. Experimental determination of control by the H(+)-ATPase in Escherichia coli. J. Bioenerg. Biomembr. 27:543-554. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, P. R., and K. Hammer. 1998. The sequence between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 64:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen, E., and A. Kibenich. 1992. Characterization of Leuconostoc isolates from commercial mixed strain mesophilic starter cultures. J. Dairy Sci. 75:1186-1191. [Google Scholar]
- 25.Kacser, H., and J. A. Burns. 1973. Rate control of biological processes. Symp. Soc. Exp. Biol. 27:65-104. [PubMed] [Google Scholar]
- 26.Koebmann, B. J., H. V. Westerhoff, J. L. Snoep, D. Nilsson, and P. R. Jensen. 2002. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 184:3909-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melchiorsen, C. R., N. B. Jensen, B. Christensen, K. Vaever Jokumsen, and J. Villadsen. 2001. Dynamics of pyruvate metabolism in Lactococcus lactis. Biotechnol. Bioeng. 74:271-279. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Neves, A. R., A. Ramos, M. C. Nunes, M. Kleerebezem, J. Hugenholtz, W. M. de Vos, J. Almeida, and H. Santos. 1999. In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 64:200-212. [DOI] [PubMed] [Google Scholar]
- 30.Okamato, T., Y. Fujita, and R. Irie. 1983. Protoplast formation and regeneration of Streptococcus lactis cells. Agric. Biol. Chem. 47:259-263. [Google Scholar]
- 31.Pedersen, M. B. 1997. Metabolic control analysis on glycolysis in Lactococcus lactis. M.S. thesis. Technical University of Denmark, Lyngby, Denmark.
- 32.Platteeuw, C., J. Hugenholtz, M. Starrenburg, I. Alen-Boerrigter, and W. M. de Vos. 1995. Metabolic engineering of Lactococcus lactis: influence of the overproduction of alpha-acetolactate synthase in strains deficient in lactate dehydrogenase as a function of culture conditions. Appl. Environ. Microbiol. 61:3967-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poolman, B., B. Bosman, J. Kiers, and W. N. Konings. 1987. Control of glycolysis by glyceraldehyde-3-phosphate dehydrogenase in Streptococcus cremoris and Streptococcus lactis. J. Bacteriol. 169:5887-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rapoport, T. A., R. Heinrich, and S. M. Rapoport. 1976. The regulatory principles of glycolysis in erythrocytes in vivo and in vitro. A minimal comprehensive model describing steady states, quasi-steady states and time-dependent processes. Biochem. J. 154:449-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruyter, G. J. G., P. W. Postma., and K. van Dam. 1991. Control of glucose metabolism by enzyme IIGlc of the phosphoenolpyruvate-dependent phosphotransferase system in Escherichia coli. J. Bacteriol. 173:6184-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schaaff, I., J. Heinisch, and F. K. Zimmermann. 1989. Overproduction of glycolytic enzymes in yeast. Yeast 5:285-290. [DOI] [PubMed] [Google Scholar]
- 38.Solem, C., and P. R. Jensen. 2002. Modulation of gene expression made easy. Appl. Environ. Microbiol. 68:2397-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stouthamer, A. H., and C. Bettenhaussen. 1973. Utilization of energy for growth and maintenance in continuous and batch cultures of microorganisms. Biochim. Biophys. Acta 301:53-70. [DOI] [PubMed] [Google Scholar]
- 40.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teusink, B., M. C. Walsh, K. van Dam, and H. V. Westerhoff. 1998. The danger of metabolic pathways with turbo design. Trends Biochem. Sci. 23:162-169. [DOI] [PubMed] [Google Scholar]
- 42.Thomas, J. 1976. Characteristics and energy requirements of an α-aminoisobutyric acid transport system in Streptococcus lactis. J. Bacteriol. 127:719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, T. D., D. C. Ellwood, and V. M. Longyear. 1979. Change from homo- to heterolactic fermentation by Streptococcus lactis resulting from glucose limitation in anaerobic chemostat cultures. J. Bacteriol. 138:465-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J., and T. D. Thomas. 1977. Phosphoenolpyruvate and 2-phosphoglycerate: endogenous energy source(s) for sugar accumulation by starved cells of Streptococcus lactis. J. Bacteriol. 130:583-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westerhoff, H. V., and K. van Dam. 1987. Thermodynamics and control of biological free energy transduction. Elsevier, Amsterdam, The Netherlands.
- 46.Yoshida, M., H. Okamoto, N. Sone, H. Hirata, and Y. Kagawa. 1977. Reconstitution of thermostable ATPase capable of energy coupling from its purified subunits. Proc. Natl. Acad. Sci. USA 74:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]