Abstract
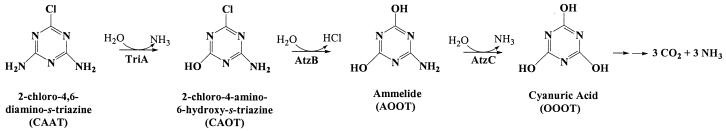
2-Chloro-4,6-diamino-s-triazine (CAAT) is a metabolite of atrazine biodegradation in soils. Atrazine chlorohydrolase (AtzA) catalyzes the dechlorination of atrazine but is unreactive with CAAT. In this study, melamine deaminase (TriA), which is 98% identical to AtzA, catalyzed deamination of CAAT to produce 2-chloro-4-amino-6-hydroxy-s-triazine (CAOT). CAOT underwent dechlorination via hydroxyatrazine ethylaminohydrolase (AtzB) to yield ammelide. This represents a newly discovered dechlorination reaction for AtzB. Ammelide was subsequently hydrolyzed by N-isopropylammelide isopropylaminohydrolase to produce cyanuric acid, a compound metabolized by a variety of soil bacteria.
s-Triazine herbicides, such as atrazine and simazine, commonly undergo oxidative dealkylation reactions in soils. Soil fungi (14, 15, 17) and Rhodococcus strains (3, 19) have been shown to catalyze dealkylation reactions with s-triazine substrates. In the Rhodococcus strains, a cytochrome P450 monooxygenase enzyme (ThcB) catalyzes a single dealkylation reaction (3, 19). However, double dealkylated metabolites, such as 2-chloro-4,6-diamino-s-triazine (CAAT), have been detected in soils and with pure cultures of fungi (13, 17). Klebsiella pneumoniae strain AZ was isolated for its ability to grow with CAAT as a nitrogen source, but the enzymatic basis for this metabolism was not defined in that or subsequent studies (9).
In contrast, the bacterial metabolism of atrazine to carbon dioxide and ammonia is well defined and known to occur via a series of hydrolytic reactions. For example, atrazine chlorohydrolase (AtzA) from Pseudomonas sp. strain ADP catalyzes the hydrolytic dechlorination of N-alkylated s-triazines, such as atrazine and simazine (Fig. 1A) (6). Subsequent hydrolytic deamination reactions are catalyzed by hydroxyatrazine ethylaminohydrolase (AtzB) (4) and N-isopropylammelide isopropylaminohydrolase (AtzC) (21). Atrazine chlorohydrolase, however, does not transform the double dealkylated CAAT (23). Aminotriazines such as melamine (Fig. 1B) are hydrolyzed by melamine deaminase (TriA) from Acidovorax avenae subsp. citrulli strain NRRL B-12227 (22). TriA has an amino acid sequence that is 98% identical to that of AtzA, yet it has a very different substrate specificity (22, 23).
FIG. 1.
Chemical structures of atrazine (A) and melamine (B).
In the present study, TriA, AtzB, and AtzC were found to hydrolytically displace the one chloride and two amino substituents of CAAT. While all of these enzymes were previously observed to catalyze only deamination reactions, here we show that AtzB catalyzes a dechlorination reaction. The final product of these three reactions is cyanuric acid, which is readily mineralized by soil bacteria.
Methods.
The substrate CAAT was obtained from Aldrich (Milwaukee, Wis.). 2,4-Dichloro-6-amino-s-triazine (CCAT) was prepared from 2,4,6-trichloro-s-triazine (Aldrich) as previously described (26). CCAT was hydrolyzed to 2-chloro-4-amino-6-hydroxy-s-triazine (CAOT) by adding a 2.5 M equivalent of aqueous potassium hydroxide. The mixture was chilled in an ice bath and acidified with acetic acid to produce the precipitated CAOT. Ammelide was produced directly from CCAT as described by Smolin and Rapoport (25). Ammeline was prepared by acidifying CAAT in 20% (vol/vol) acetic acid in the presence of a potassium bromide catalyst, followed by buffering with sodium acetate. Identity of the standard compounds was confirmed by silination of the hydroxyl groups with 1,1,1,3,3,3-hexamethyldisilazane (Aldrich)/N,O-bis-trimethylsilyl-acetamide (Aldrich) and analysis of the resulting product by gas chromatography (GC)-mass spectrometry (MS). Synthetic standards and enzyme reaction products were also analyzed by high-pressure liquid chromatography (HPLC) using a Hewlett-Packard 1100 Series system equipped with a photodiode array detector interfaced to a Hewlett-Packard ChemStation. A Waters IC-PAK anion high capacity (HC) column (4.6 by 150 mm) was used with a 100 mM phosphate buffer, pH 7, mobile phase and a 1-ml min−1 flow rate for detection of CAAT and CAOT. The chlorinated triazines were detected at 255 and 200 nm, respectively. A 10 mM phosphate buffer, pH 5, mobile phase with a 0.25-ml min−1 flow rate was used to analyze ammeline and ammelide at 200 nm. HPLC-MS was done in cooperation with Tom Krick at the Mass Spectrometry Consortium for Life Sciences at the University of Minnesota. Ammonium acetate buffer was substituted as a mobile phase for MS detection. Ammonia release was detected as previously described (22). The triA gene was isolated as previously described (22), except that it was subsequently cloned into the EcoRI/HindIII sites of a pTrc99A vector. The resulting vector, pJS5, was transformed into Escherichia coli DH5α. Basal transcription levels produced optimal soluble protein levels. Other recombinant strains used in these studies were described elsewhere: AtzB (E. coli[pATZB2]) (4) and AtzC (E. coli[pATZC]) (21) (N. Shapir, unpublished data).
Enzymatic reactions and formation of products were confirmed in both resting cell and cell extract experiments. Resting cell assays were conducted in 50 ml of 25 mM phosphate buffer, pH 7, containing the given substrate. Cells were removed by centrifugation for 2 min at 13,000 × g in a tabletop microcentrifuge, and the supernatants were analyzed by HPLC and HPLC-MS. To prepare cell extracts, cells were harvested by centrifugation as described above, resuspended in ice-cold, 25 mM phosphate buffer, pH 7, and lysed by sonication (3- to 20-s bursts at 80% intensity on a Biosonik sonicator; Bronwill Scientific, Rochester, N.Y.). The resulting solution was centrifuged at 4°C to remove cell debris (13,000 × g for 5 min in a tabletop microcentrifuge). The cell extract was incubated with substrate overnight and was analyzed by HPLC and HPLC-MS. The identity of products was confirmed by comparison to standards. Bacteria for growth curve studies were grown in R medium (24), with equimolar concentrations of the various s-triazines as sole nitrogen sources. Samples were removed at specified time points and were analyzed spectophotometrically at 600 nm. Due to the low water solubility of CAAT, maximum growth occurred at approximately an optical density at 600 nm of 0.15. Similar growth was observed with equimolar ammonia concentrations (data not shown). Substrate stability was tested over a period of 1 month. None of the compounds used throughout this study showed significant degradation in phosphate buffer at the pH values used over this time period. E. coli DH5α cells, lacking the recombinant genes, served as a control, and no degradation of the substrates was observed with this strain.
CAAT hydrolysis by TriA
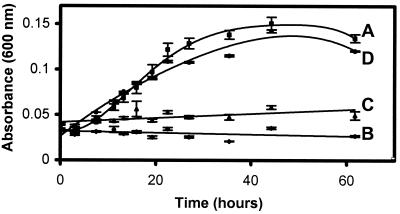
. Resting cell suspensions of E. coli(pJS5) containing the triA gene were incubated with CAAT, and the culture filtrate was analyzed by HPLC and HPLC-MS. Over time, the CAAT peak disappeared with the concomitant increase of another peak. The product was identified as CAOT by coelution with an authentic standard on HPLC, UV spectroscopy, and mass spectrometry. There was no further metabolism of CAOT by E. coli(pJS5). Additional experiments showed that ammonia release paralleled CAOT formation and that E. coli(pJS5) grew on CAAT as a sole source of nitrogen for growth (Fig. 2).
FIG. 2.
Growth of recombinant E. coli on several potential nitrogen sources. Shown are results for E. coli(pJS5) (A) with CAAT as the sole nitrogen source, E. coli DH5α (B) with CAAT as the sole nitrogen source, E. coli(pATZB) (C) with CAOT as the sole nitrogen source, and E. coli(pATZC) (D) with ammelide as the sole nitrogen source (n = 3). Standard error bars are shown by vertical lines at each time point.
Grossenbacher et al. previously showed that the parent strain of triA, A. avenae subsp. citrulli strain NRRL B-12227 (formerly Pseudomonas sp. strain NRRLB-12227 and Pseudomonas sp. strain A) was capable of CAAT degradation, but the enzyme responsible for the activity was not identified (10). In the present study, we showed that TriA is the responsible enzyme. s-Triazine hydrolase (TrzA) from Rhodococcus corallinus NRRL B-15444R is a homolog to TriA and also catalyzes turnover of CAAT (18). Data from ammonia release assays and UV spectrophotometry were used to hypothesize that the product of the TrzA reaction was CAOT; however, no standard was available in that study to confirm the identity of the product. TrzA and TriA are homologous but share only 45% sequence identity. TriA is much more highly related to AtzA, sharing 98% sequence identity. However, AtzA shows no detectable reactivity with CAAT (23).
It was previously reported that CAAT was degraded by TriA to yield ammelide (22). However, in the present work, we showed that the acidic conditions used for the previous MS analysis caused hydrolysis of the labile chloride substituent of CAOT to produce ammelide. The stability of CAOT at pH 7 was confirmed by experiments with standards, and MS at neutral pH identified that CAOT was the only product produced by TriA. These observations further support the conclusion that the distinct specificities of TriA, which catalyzes deamination reactions, and AtzA, which catalyzes dechlorination reactions, are determined by one or more of the nine amino acids that differ between these two proteins.
CAOT hydrolysis by AtzB.
The enzymatic fate of CAOT has not been reported previously. In this study, CAOT was shown to be hydrolyzed by cell extracts prepared from an E. coli strain containing the enzyme AtzB. A parallel extract from a control E. coli strain lacking AtzB did not hydrolyze CAOT. E. coli(pATZB2) transformed CAOT stoichiometrically to ammelide as determined by UV spectroscopy and HPLC, with comparison to standards. This was the only product detected. Consistent with the observation that only a chloride substituent was removed, E. coli(pATZB2) did not grow on CAOT in the absence of additional nitrogen sources (Fig. 2).
AtzB was previously identified for its ability to catalyze the deamination of hydroxyatrazine to produce N-isopropylammelide (4). Results from these studies indicate that AtzB can hydrolyze N-alkyl or chloride substituents and is less strict in its specificity than AtzA or TriA. All three of these enzymes belong to the same amidohydrolase superfamily, which includes such enzymes as adenosine deaminase and dihydroorotase (11, 21, 22). As the superfamily name suggests, most of these enzymes catalyze the hydrolysis of amino and amido groups. However, there is some precedence for enzymes that physiologically catalyze deamination reactions to also catalyze fortuitous dechlorination reactions (1, 2). It is remarkable in this respect that TriA shows a high degree of specificity for deamination without any significant dechlorination activity.
Ammelide hydrolysis by AtzC.
Ammelide is known to be metabolized by bacteria, but the enzymes responsible for its transformation have not been purified and characterized previously (7, 8). Here we show that purified AtzC from Pseudomonas sp. strain ADP catalyzes the hydrolytic deamination of ammelide to produce cyanuric acid. Product identity was confirmed with UV spectroscopy and HPLC, with comparison to standards. The kcat for the reaction was 2 s−1, compared to 14 s−1 for the hydrolysis of N-isopropylammelide. Consistent with these data, E. coli(pATZC) was observed to grow on ammelide as a sole nitrogen source (Fig. 2). A detailed purification and characterization of AtzC will be published elsewhere.
Ammelide metabolism has been observed with numerous bacteria. The ammelide amidohydrolase gene (trzC) from A. avenae subsp. citrulli strain NRRL B-12227 has been cloned and sequenced (7) (GenBank protein accession number AAK00493), but the enzyme was not purified or characterized. It is shown in the present study that AtzC has substantial activity with ammelide and will support bacterial growth on ammelide as the sole nitrogen source. The product of the AtzC reaction, cyanuric acid, is metabolized by many soil bacteria. Genes and enzymes involved in the metabolism of cyanuric acid have been reported by several authors (5, 7, 8, 12, 16).
Taken together, the data obtained in this study allow us to depict a complete hypothetical pathway for the metabolism of CAAT (Fig. 3). This constellation of genes has not yet been detected in any single bacterium. However, atzABC genes, known to reside on a broad-host-range plasmid in Pseudomonas sp. strain ADP (16), differ by only 9 nucleotides out of 4,081 from a hypothetical clustering of triA, atzB, and atzC, rendering it plausible that such a clustering exists in nature. Also TriA and TrzC are found in the same bacterium, A. avenae subsp. citrulli strain NRRL B-12227. The addition of atzB to this bacterium would also result in a functional pathway for CAAT.
FIG. 3.
Proposed hypothetical pathway for the degradation of CAAT.
A previous study showed that changes in a small number of amino acids caused significant alterations in s-triazine substrate specificity for AtzA and TriA (20). The present study shows that changes in the clustering of specific s-triazine-degrading enzymes can broaden the specificity of a metabolic pathway. The new pathway described here has potential environmental relevance, since CAAT is an important intermediate of s-triazine herbicide metabolism in soil. Knowledge of enzyme sequence and substrate plasticity may increasingly be used to predict the course of metabolism in the environment.
Acknowledgments
We thank Tom Krick of the Mass Spectrometry Consortium for Life Sciences at the University of Minnesota for his assistance in the HPLC-MS analysis.
This research was supported in part by a grant from Syngenta Crop Protection, National Institutes of Health training grant GM08347, and a postdoctoral fellowship from the United States-Israel Binational Agriculture Research and Development (BARD) fund grant FI-295-99 (to N.S.).
REFERENCES
- 1.Baer, H. P., G. I. Drummond, and E. L. Duncan. 1966. Formation and deamination of adenosine by cardiac muscle enzymes. Mol. Pharmacol. 2:67-76. [PubMed] [Google Scholar]
- 2.Bar, H. P., and G. I. Drummond. 1966. On the mechanism of adenosine deaminase action. Biochem. Biophys. Res. Commun. 24:584-587. [DOI] [PubMed] [Google Scholar]
- 3.Behki, R., E. Topp, W. Dick, and P. Germon. 1993. Metabolism of the herbicide atrazine by Rhodococcus strains. Appl. Environ. Microbiol. 59:1955-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boundy-Mills, K., M. L. de Souza, R. M. Mandelbaum, L. P. Wackett, and M. J. Sadowsky. 1997. The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl. Environ. Microbiol. 63:916-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook, A. M., P. Beilstein, H. Grossenbacher, and R. Huetter. 1985. Ring cleavage and degradative pathway of cyanuric acid in bacteria. Biochem. J. 231:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Souza, M. L., M. J. Sadowsky, and L. P. Wackett. 1996. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization. J. Bacteriol. 178:4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton, R. W., and J. S. Karns. 1991. Cloning and analysis of s-triazine catabolic genes from Pseudomonas sp. strain NRRLB-12227. J. Bacteriol. 173:1215-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton, R. W., and J. S. Karns. 1991. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s-triazine-degrading bacterial strains. J. Bacteriol. 173:1363-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst, C., and H. J. Rehm. 1995. Utilization of chlorinated s-triazines by a new strain of Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 42:763-768. [DOI] [PubMed] [Google Scholar]
- 10.Grossenbacher, H., C. Horn, A. M. Cook, and R. Hütter. 1984. 2-Chloro-4-amino-1,3,5-triazine-6(5H)-one: a new intermediate in the biodegradation of chlorinated s-triazines. Appl. Environ. Microbiol. 48:451-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holm, L., and C. Sander. 1997. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins 28:72-82. [PubMed] [Google Scholar]
- 12.Karns, J. S. 1999. Gene sequence and properties of an s-triazine ring-cleavage enzyme from Pseudomonas sp. strain NRRLB-12227. Appl. Environ. Microbiol. 65:3512-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufman, D. D., and J. Blake. 1970. Degradation of atrazine by soil fungi. Soil Biol. Biochem. 2:73-80. [Google Scholar]
- 14.Kaufman, D. D., P. C. Kearney, and T. J. Sheets. 1965. Microbial degradation of simazine. J. Agric. Food Chem. 13:238-242. [Google Scholar]
- 15.Kearney, P. C., and D. D. Kaufman. 1965. Enzyme from soil bacterium hydrolyzes phenylcarbamate herbicides. Science 147:740-741. [DOI] [PubMed] [Google Scholar]
- 16.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masaphy, S., D. Levanon, J. Vaya, and Y. Henis. 1993. Isolation and characterization of a novel atrazine metabolite produced by the fungus Pleurotus pulmonarius, 2-chloro-4-ethylamino-6-(1-hydroxyisopropyl)amino-1,3,5-triazine. Appl. Environ. Microbiol. 59:4342-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulbry, W. W. 1994. Purification and characterization of an inducible s-triazine hydrolase from Rhodococcus corallinus NRRL B-15444R. Appl. Environ. Microbiol. 60:613-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy, I., F. Compernolle, K. Ghys, J. Vanderleyden, and R. De Mot. 1995. A single cytochrome P-450 system is involved in degradation of the herbicides EPTC (s-ethyl dipropylthiocarbamate) and atrazine by Rhodococcus sp. strain NI86/21. Appl. Environ. Microbiol. 61:2056-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raillard, S. A., A. Krebber, Y. Chen, J. E. Ness, E. Bermudez, R. Trinidad, R. Fullem, C. Davis, M. Welch, J. L. Seffernick, L. P. Wackett, W. P. C. Stemmer, and J. Minshull. 2001. Novel enzyme activities and functional plasticity revealed by recombining highly homologous enzymes. Chem. Biol. 8:891-898. [DOI] [PubMed] [Google Scholar]
- 21.Sadowsky, M. J., Z. Tong, M. L. de Souza, and L. P. Wackett. 1998. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J. Bacteriol. 180:152-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seffernick, J. L., M. L. de Souza, M. J. Sadowsky, and L. P. Wackett. 2001. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J. Bacteriol. 183:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seffernick, J. L., G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2000. Substrate specificity of atrazine chlorohydrolase and atrazine-catabolizing bacteria. Appl. Environ. Microbiol. 66:4247-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selinofova, O., and T. Barkay. 1994. Role of Na+ in transport of Hg+ and induction of the Tn21 mer operon. Appl. Environ. Microbiol. 60:3503-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smolin, E. M., and L. Rapoport. 1959. s-Triazines and derivatives, p. 272. Interscience Publishers Inc., New York, N.Y.
- 26.Thurston, J. T., J. R. Dudley, D. W. Kaiser, I. Hechenbleikner, F. C. Schaefer, and D. Holm-Hansen. 1951. Cyanuric chloride derivatives: aminochloro-s-triazines. J. Am. Chem. Soc. 73:2981-2983. [Google Scholar]