Abstract
As part of a study to elucidate the environmental parameters that control microbial perchlorate respiration, we investigated the reduction of perchlorate by the dissimilatory perchlorate reducer Dechlorosoma suillum under a diverse set of environmental conditions. Our results demonstrated that perchlorate reduction by D. suillum only occurred under anaerobic conditions in the presence of perchlorate and was dependent on the presence of molybdenum. Perchlorate reduction was dependent on the presence of the enzyme chlorite dismutase, which was induced during metabolism of perchlorate. Anaerobic conditions alone were not enough to induce expression of this enzyme. Dissolved oxygen concentrations less than 2 mg liter−1 were enough to inhibit perchlorate reduction by D. suillum. Similarly to oxygen, nitrate also regulated chlorite dismutase expression and repressed perchlorate reduction by D. suillum. Perchlorate-grown cultures of D. suillum preferentially reduced nitrate in media with equimolar amounts of perchlorate and nitrate. In contrast, an extended (40 h) lag phase was observed if a similar nitrate-perchlorate medium was inoculated with a nitrate-grown culture. Perchlorate reduction commenced only when nitrate was completely removed in either of these experiments. In contrast to D. suillum, nitrate had no inhibitory effects on perchlorate reduction by the perchlorate reducer Dechloromonas agitata strain CKB. Nitrate was reduced to nitrite concomitant with perchlorate reduction to chloride. These studies demonstrate that microbial respiration of perchlorate is significantly affected by environmental conditions and perchlorate reduction is directly dependent on bioavailable molybdenum and the presence or absence of competing electron acceptors. A microbial treatment strategy can achieve and maintain perchlorate concentrations below the recommended regulatory level, but only in environments in which the variables described above can be controlled.
Environmental contamination with oxyanions of chlorine, especially perchlorate (ClO4−), has recently been recognized as posing a significant health threat (39; Environmental Protection Agency [EPA], wysiwyg://2/http://www.epa.gov/ncea/perch.htm). In general, chlorine oxyanions in the environment result from anthropogenic sources, including disinfectants, bleaching agents, and herbicides (2, 15, 35), as well as munitions (38). Although all chlorine oxyanions are toxic to various extents, recent attention has focused on environmental contamination with perchlorate, which is extensively used by the munitions industry and the defense department as a major component of explosives and rocket fuels (38, 39). Prior to 1997, perchlorate was an unregulated compound; however, the recent discovery of perchlorate contamination in drinking water resources throughout the United States, especially in the southwestern states of Nevada, Utah, and California (31), prompted the California Department of Health Services (DHS) together with the California EPA to establish a provisional action level for perchlorate of 18 μg liter−1 in 1997. This recommended action level was subsequently increased by the U.S. EPA to 32 μg liter−1, with a final action level to be determined based on the findings of ongoing toxicological studies (30). In January 2002, as a result of the publication of the first draft of the EPA review on toxicological and risk characterization data associated with perchlorate contamination, the California DHS revised and lowered its original provisional action level to 4 μg liter−1 (http://www.dhs.ca.gov/ps/ddwem/chemicals/perchl/actionlevel.htm), which is at the limits of detection according to current technologies (42).
Because of its unique chemical stability under environmental conditions and its high solubility (39), microbial reduction of perchlorate was identified as the most feasible method of remediation of contaminated environments (39). However, until recently, very little was known about the diversity or ubiquity of microorganisms that can grow by dissimilatory chlorate or perchlorate respiration (39). Although microbial reduction of chlorate has been known for more than 70 years (4), this metabolism was associated with nitrate-respiring organisms, and chlorate was assumed to be a coincidental substrate for the nitrate reductase (13, 18, 19). However, this assumption could not explain the presence of specialized enzymes, such as the chlorate reductase C purified from Proteus mirabilis, which could only use chlorate as a substrate (29). Now it is known that specialized microorganisms have evolved that can couple growth to the anaerobic reduction of chlorate or perchlorate and completely reduce these compounds to chloride (1, 6, 10, 24, 26, 27, 32, 34, 37, 41, 43). These organisms are phylogenetically diverse (10, 27, 41) with members in the α-, β-, γ-, and ɛ-subclasses of the Proteobacteria (1, 10, 27, 41); however, the majority are in the β-subclass of the Proteobacteria and are members of the genus Dechloromonas or Dechlorosoma (1, 10). Phenotypic characterization studies have demonstrated that the known perchlorate-reducing bacteria (ClRB) exhibit a broad range of metabolic capabilities, including the oxidation of hydrogen (41), simple organic acids and alcohols (6, 10, 26, 27, 32), aromatic hydrocarbons (8), hexoses (26), reduced humic substances (6, 8, 9), both soluble and insoluble ferrous iron (6, 7, 10, 22, 23, 27), and hydrogen sulfide (6, 10). All of the known ClRB are facultatively anaerobic or microaerophilic (6, 10, 27, 32, 41) and some, but not all, alternatively respired nitrate, supporting the suggestion that perchlorate reduction is unrelated to nitrate reduction (6, 10). A central step in the reductive pathway of perchlorate or chlorate that is common to all ClRB is the dismutation of chlorite, which is mediated by the enzyme chlorite dismutase (CD) (6, 10, 36, 40). Studies with a recently developed immunoprobe specific for purified CD from Dechloromonas agitata strain CKB indicated that the CD is highly conserved among the ClRB, regardless of their phylogenetic affiliation (28). In addition, these studies also demonstrated that the CD enzyme was present on the outer membrane of all ClRB and was only expressed when the organisms were grown anaerobically in the presence of perchlorate or chlorate (28).
Although recent studies have significantly increased our knowledge of the microorganisms involved in microbial perchlorate reduction, there is still relatively little information available on the variables that control this metabolism in the natural environment. Here we report on a further investigation into the environmental factors that control microbial reduction of perchlorate and determine the conditions required for perchlorate respiration to occur.
MATERIALS AND METHODS
Source of organisms.
The facultative (per)chlorate-reducing bacteria Dechlorosoma suillum strain PS (1, 27) and Dechloromonas agitata strain CKB (1, 6) were taken from our laboratory culture collection of ClRB, where they have been maintained as frozen stocks at −70°C.
Growth medium and culture conditions.
Unless otherwise stated, the medium used was a modification of the basal medium (BM) previously described for ClRB (6). The BM was modified by replacing the carbonate buffer with a 10 mM phosphate buffer (pH 7.0). Acetate was amended as the electron donor from a concentrated stock solution (1.0 M) of the sodium salt to give the desired final concentration. Sodium salts of perchlorate and nitrate were also added from stock solutions (1 M) as suitable electron acceptors when required. For anaerobic serum bottle cultures, the prepared medium was made anoxic by purging with oxygen-free nitrogen, and the bottles were crimp sealed with thick butyl rubber stoppers under a nitrogen headspace before being autoclaved at 121°C for 20 min.
Batch fermentation.
Experiments were carried out in a 6-liter automatically controlled (pH, temperature, and dissolved oxygen [DO]) batch fermentor (model Bioflow 2000; New Brunswick Scientific Co., Inc., Edison, N.J.) with an operating volume of 5 liters unless otherwise stated. The vessel was filled with fresh medium containing the appropriate electron donor and acceptor, respectively; sealed with the stainless steel lid equipped with probes for monitoring pH, DO, and temperature; and autoclaved at 121°C for 35 min. The pH meter was calibrated before autoclaving. Immediately after autoclaving, the fermentor was connected to the control systems, and all of the parameters were set to their desired value. The DO content of each culture was maintained by balancing the mixed ratio of air to oxygen-free N2 gas. The premixed gas was then filtered through a sterile 0.2-μm-pore-diameter filter and bubbled through the media. DO content was monitored with a previously calibrated DO sensor. Anoxic sterile solutions of 0.5 M H2SO4 and 1.0 M NaOH were pumped through automatic acid and alkali pumps, respectively, of the pH controller to maintain the pH. After reaching steady state relative to the set parameters, 500 ml of a 20-h growth culture of the pertinent perchlorate-reducing organism was used to inoculate the vessel. The speed of agitation in anaerobic run was fixed at 400 rpm, whereas in the aerobic run, the lower and upper limits of the speed of agitation were 400 and 650 rpm, respectively, to control the DO concentration at the desired level. All cultures were incubated at 37°C. Samples were collected anaerobically when required in sterile containers at regular time intervals for analysis.
CD activity determination.
CD activity in collected culture samples was determined in triplicate by mixing 0.8 ml of the culture sample with 0.2 ml of a freshly prepared aqueous sodium chlorite stock solution (20 mM) in a 1.5-ml Eppendorf tube. At appropriate time points (0 to 1.5 min), 100-μl subsamples were collected and transferred immediately into 1.5-ml Eppendorf tubes in an 80°C water bath to stop the reaction. The residual chlorite content in the heat-killed subsamples was determined by colorimetric microassay based on horseradish peroxidase and ortho-dianisidine dye as previously described (10; J. D. Coates and S. M. O'Connor, submitted for publication).
Analyses.
The concentration of perchlorate in samples was determined by ion chromatography coupled to suppressed conductivity with a Dionex IonPac AS 11 (4 by 250 mm) column (Dionex Corp., Sunnyvale, Calif.) with a 100 mM NaOH mobile phase at a flow rate of 1ml min−1. The eluting perchlorate was then detected by a conductivity detector (Shimadzu model CDD-6A) that was suppressed with a Dionex ASRS (anion self-regenerating suppressor)-Ultra apparatus operating in the external water mode. The suppressor controller was set at 300 mA for the analysis. With an injection volume of 200 μl, and the ASRS apparatus operating with an external water source, the detection limit for perchlorate was 6.0 μg liter−1. Chlorate, chloride, nitrate, and nitrite in the culture medium were determined with a Dionex DX500 ion chromatograph (Dionex Corporation, Sunnyvale, Calif.) equipped with a GP50 gradient pump, CD20 conductivity detector, ASRS-Ultra apparatus for suppressed conductivity, and PeakNet 6 controlling software. An IonPac AS9-SC (4 by 250 mm) column was used for analysis with bicarbonate buffer containing 2 mM sodium carbonate and 0.75 mM sodium bicarbonate at a flow rate of 2 ml min−1 as the eluent. The SRS current was set at 100 mA for all the analysis. Acetate concentrations were determined by high-performance liquid chromatography (Shimadzu, model SPD-10A), using a UV-VIS detector, at a wavelength of 210 nm by using an HL-75H+ cation-exchange column (Hamilton, model no. 79476) and a mobile phase of 0.016 N H2SO4 at a flow rate of 0.4 ml min−1. Culture growth was monitored both by optical density at 600 nm (OD600) with a spectrophotometer (Spectronic Genesis 5; Spectronic Instruments, Inc., Rochester, N.Y.) and by total cell count with a phase-contrast microscope as previously described (10).
RESULTS AND DISCUSSION
Effect of oxygen on CD activity and perchlorate reduction by D. suillum.
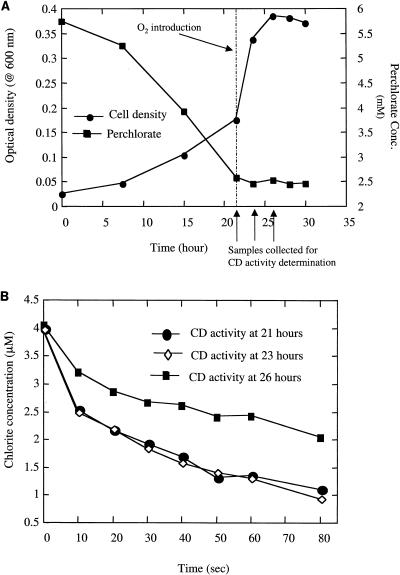
When D. suillum was grown anaerobically under an N2 gas phase with perchlorate as the sole electron acceptor, perchlorate removal was rapid and CD activity was observed (Fig. 1A and B). However, if air was introduced into the headspace of the anaerobic culture after 21 h, perchlorate reduction was immediately inhibited, despite the continued increase in cell density. Measurement of CD activity indicated that chlorite was readily dismutated by the cells at the 21-h time point (Fig. 1B) when O2 was introduced to the culture. Subsequent analyses 2 and 5 h after introduction of air indicated that CD was still active, although the dismutation rate was significantly lower 5 h after O2 introduction. Previous studies have suggested that the presence of oxygen similarly inhibited microbial reduction of chlorate (25). Furthermore, thermodynamic consideration of the respective reductive pathways suggested that the aerobic metabolism of acetate is energetically more favorable than acetate oxidation coupled to perchlorate reduction (11). However, this may be an oversimplification, because genetic regulation may also play an important role. In support of this, studies with the ClRB strain GR-1 demonstrated that the CD activity was significantly higher when the cells were grown on perchlorate relative to aerobic cultures, suggesting that the CD was induced under anaerobic conditions (32). In addition, immunoprobe studies of the ClRB D. agitata indicated that the CD enzyme was only expressed when the cells were grown on perchlorate (28). Similar to these findings, CD activity in D. suillum was only observed in cells collected from an active culture growing anaerobically with perchlorate as the electron acceptor (data not shown). No CD activity was detectable if the culture was grown aerobically.
FIG. 1.
Effects of the introduction of oxygen after 21 h of incubation on growth and perchlorate reduction (A) and CD activity (B) by an active perchlorate-respiring culture of D. suillum.
Growth and CD expression under anaerobic conditions.
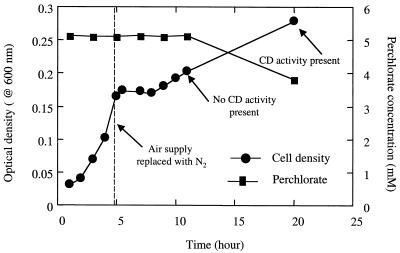
To determine if anoxic conditions alone would induce the production of CD by D. suillum, the O2 content of an actively metabolizing aerobic culture was replaced with N2 after 4.5 h of incubation during log-phase growth. Growth was immediately arrested relative to that of an unamended control culture (data not shown). CD activity was not observed in samples collected at 3.5, 5, and 7 h of incubation from either the aerobic control culture or the culture switched to anaerobic conditions (data not shown), suggesting that anaerobic conditions are not enough to initiate expression of an active CD. In addition, if D. suillum was grown aerobically in the presence of 5 mM perchlorate in batch culture at a 100% DO saturation (∼40 mg of DO liter−1), CD activity was not observed in cells throughout culture growth and perchlorate reduction did not occur (data not shown). Similar results were observed if the culture was grown at lower DO concentrations (70, 40, and 5% of saturation) in the presence of 5 mM perchlorate, indicating that the presence of perchlorate even at low DO concentrations was not enough to induce perchlorate reduction by D. suillum. If, however, the O2 content was replaced with N2 during log-phase growth, both perchlorate reduction and CD activity were measurable after an extended period (15 h) (Fig. 2). As observed above, growth was initially arrested on removal of the O2, but was recovered after a lag phase of 4 h (Fig. 2). Interestingly, renewed growth was observed prior to detectable perchlorate reduction or CD activity (Fig. 2), although this may be a function of the sensitivity of the various measurement techniques utilized.
FIG. 2.
Effect on growth and CD activity of switching an aerobic culture of D. suillum amended with perchlorate to anaerobic conditions during log-phase growth.
Effect of nitrate on perchlorate reduction and CD expression by D. suillum.
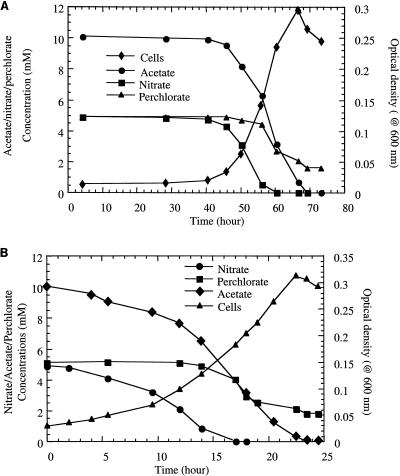
Previous studies have demonstrated that all but one of the known ClRB can alternatively utilize nitrate as a suitable electron acceptor. D. agitata strain CKB is the only ClRB described to date that cannot support growth by nitrate respiration (6). If a nitrate-grown culture of D. suillum was transferred into fresh anoxic medium, nitrate reduction and growth occurred immediately, with no apparent lag phase, and CD activity was not observed throughout the incubation (data not shown). In contrast, if a nitrate-grown culture of D. suillum was used to inoculate medium containing equimolar amounts of perchlorate and nitrate, all growth and respiration were inhibited for an extended lag period (40 h), after which nitrate reduction occurred prior to perchlorate reduction (Fig. 3A). When a similar experiment was performed with a perchlorate-grown culture inoculated into fresh medium containing equimolar amounts of both nitrate and perchlorate, no lag phase was apparent, and again, nitrate reduction occurred prior to perchlorate reduction (Fig. 3B). Regardless of the electron acceptor utilized in the inoculum culture, neither CD activity (data not shown) nor perchlorate reduction was observed until nitrate was completely consumed (Fig. 3A and B).
FIG. 3.
Growth, nitrate, and perchlorate reduction by nitrate-grown (A) and perchlorate-grown (B) cultures of D. suillum.
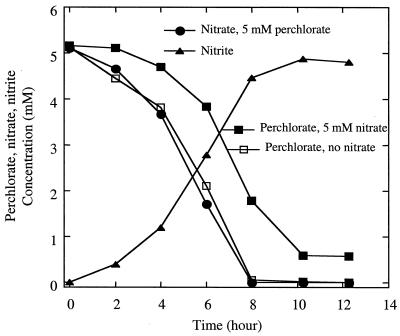
In contrast to D. suillum, the presence of nitrate had relatively little effect on perchlorate reduction by the ClRB D. agitata strain CKB. As outlined above, D. agitata cannot use nitrate as an alternative electron acceptor for growth (6). When an active perchlorate-respiring culture of D. agitata was transferred into freshly prepared anoxic medium containing both nitrate and perchlorate, the perchlorate was rapidly reduced (Fig. 4). Both the rate and extent of perchlorate utilization were lower than those observed in the absence of nitrate, suggesting that the nitrate was competitively inhibiting perchlorate reduction (Fig. 4). Interestingly, although D. agitata does not grow by nitrate reduction, the nitrate in the culture medium was concomitantly reduced to nitrite during perchlorate reduction, which accumulated in the culture broth (Fig. 4), suggesting that the nitrate was being coreduced by the (per)chlorate reductase. Previous studies performed with the ClRB strain perc1ace also indicated that nitrate was concomitantly reduced with perchlorate by this ClRB (20). However, in contrast to D. agitata, strain perc1ace could grow by nitrate reduction, and nitrite did not accumulate in the culture broth (20).
FIG. 4.
Growth, nitrate and perchlorate reduction, and nitrite production by a perchlorate-grown culture of D. agitata.
Effect of molybdenum on perchlorate reduction.
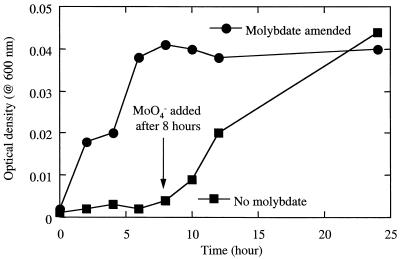
Recent molecular studies of the genetic systems associated with perchlorate reduction indicated the presence of a molybdenum-dependent chaperone gene in association with the genes encoding CD and perchlorate reductase in Dechloromonas aromatica strain RCB and Pseudomonas sp. strain PK (5). Furthermore, the perchlorate reductase enzyme recently purified from the ClRB strain GR-1 contained 1 mol of molybdenum per mol of the heterodimeric molecule (21), suggesting that molybdenum may play a functional role in the reduction of perchlorate. In support of this, growth and perchlorate reduction were completely inhibited when an active perchlorate-respiring culture of D. aromatica was transferred into medium from which molybdenum was omitted (Fig. 5). If the culture was amended with molybdenum after 8 h of incubation, growth was immediately recovered (Fig. 5), demonstrating a nutritional requirement for molybdenum. Omitting molybdenum from oxic medium had no effect on the aerobic growth of D. aromatica (data not shown). Similar inhibition of anaerobic growth and perchlorate reduction was observed with molybdenum-depleted cultures of D. agitata (data not shown). These results, in combination with the previous genetic and biochemical observations, indicate that molybdenum may be required by all perchlorate-respiring bacteria for the reduction of perchlorate. This may have important implications regarding bioremediative strategies for perchlorate in contaminated environments, because bioavailable molybdenum is quite often a limiting nutrient in many soils (12, 14). This is especially true in acidic soils, where adsorption reduces the availability of molybdenum salts at lower pHs (17).
FIG. 5.
Anaerobic growth of D. aromatica on perchlorate in the presence and absence of molybdate.
Significance.
The results presented here indicate that microbial perchlorate reduction is regulated by the presence of oxygen, molybdate, and, to various extents, nitrate. Although, respiration of oxyanions of chlorine was originally associated with the nitrate reduction pathway (13, 18, 19), it is now known that perchlorate reduction is a distinct respiratory pathway with its own unique enzymes (6, 10, 21, 28, 32, 36). The results of the present study indicate that perchlorate reduction is tightly regulated at a genetic level and that the expression of the enzymes associated with the reduction of perchlorate is controlled by the presence of both oxygen and nitrate. Nitrate, chlorate, and perchlorate [(per)chlorate] are structurally analogous to each other and may potentially be incorporated into the same enzyme active site, as is evidenced by the fact that chlorate can be used as a substrate by various nitrate reductases (3, 16, 33, 44). Thermodynamic considerations of perchlorate reduction and denitrification coupled to acetate oxidation suggest that the energy yield for the reduction of perchlorate (ΔGo = −801 kJ mol of acetate−1) is very similar to that of denitrification (ΔGo = −792 kJ mol of acetate−1) (11), and the fact that D. agitata can simultaneously reduce both perchlorate and nitrate would suggest that nitrate can alternatively be utilized as an analogous substrate by the (per)chlorate reductase enzymes. In support of this, the perchlorate reductase purified from the ClRB strain GR-1 could alternatively reduce nitrate as a substrate (21).
Although these compounds are analogous substrates to some reductase enzymes, the results presented here also indicate that the genetic regulatory mechanism of D. suillum, and presumably other ClRB can distinguish between them. The fact that the enzymes associated with perchlorate reduction are genetically regulated by different environmental parameters is interesting, especially in light of recent studies in which it was demonstrated that the CD enzyme is highly conserved among the tested ClRB regardless of their phylogenetic affiliation (28).
The results of the current studies have important implications with regards to the treatment of perchlorate-contaminated environments. Previously it was suggested that the inhibitory effects of oxygen and nitrate on in situ perchlorate reduction were the result of competitive thermodynamics (11). Here we demonstrate that this may have been an oversimplification and did not account for genetic regulation of the respective pathways. The results of the present studies demonstrate that the enzymes associated with perchlorate reduction are negatively regulated by DO content, in some cases nitrate, and require molybdenum as a cofactor for activity. These factors may explain the long-term stability of perchlorate in contaminated environments where perchlorate-reducing bacteria are known to be present.
Acknowledgments
This work was supported by grant no. DACA72-00-C-0016 from the U.S. Department of Defense to J.D.C. and L.A.A.
REFERENCES
- 1.Achenbach, L. A., R. A. Bruce, U. Michaelidou, and J. D. Coates. 2001. Dechloromonas agitata N.N. gen., sp. nov. and Dechlorosoma suillum N.N. gen., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. E vol. Microbiol. 51:527-533. [DOI] [PubMed] [Google Scholar]
- 2.Agaev, R., V. Danilov, V. Khachaturov, B. Kasymov, and B. Tishabaev. 1986. The toxicity to warm-blooded animals and fish of new defoliants based on sodium and magnesium chlorates. Uzb. Biol. Zh. 1:40-43. [Google Scholar]
- 3.Amy, N. K. 1981. Identification of the molybdenum cofactor in chlorate-resistant mutants of E. coli. J. Bacteriol. 148:274-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aslander, A. 1928. Experiments on the eradication of Canada thistle Cirsium arvense with chlorates and other herbicides. J. Agric. Res. 36:915-935. [Google Scholar]
- 5.Bender, K. S., S. M. O'Connor, R. Chakraborty, J. D. Coates, and L. A. Achenbach. Sequencing and transcriptional analysis of the chlorite dismutase gene of Dechloromonas agitata and its use as a metabolic probe. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 6.Bruce, R. A., L. A. Achenbach, and J. D. Coates. 1999. Reduction of (per)chlorate by a novel organism isolated from a paper mill waste. Environ. Microbiol. 1:319-331. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri, S. K., J. G. Lack, and J. D. Coates. 2001. Biogenic magnetite formation through anaerobic biooxidation of Fe(II). Appl. Environ. Microbiol. 67:2844-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coates, J. D., R. Chakraborty, J. G. Lack, S. M. O'Connor, K. A. Cole, K. S. Bender, and L. A. Achenbach. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039-1043. [DOI] [PubMed] [Google Scholar]
- 9.Coates, J. D., K. A. Cole, R. Chakraborty, S. M. O'Connor, and L. A. Achenbach. 2002. The diversity and ubiquity of bacteria utilizing humic substances as an electron donor for anaerobic respiration. Appl. Environ. Microbiol. 68:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. The ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coates, J. D., U. Michaelidou, S. M. O'Connor, R. A. Bruce, and L. A. Achenbach. 2000. The diverse microbiology of (per)chlorate reduction, p. 257-270. In E. D. Urbansky (ed.), Perchlorate in the environment. Kluwer Academic/Plenum, New York, N.Y.
- 12.Coventry, D. R., J. R. Hirth, and K. K. H. Fung. 1987. Nutritional restraints on subterranean clover grown on acid soils used for crop-pasture rotation. Aust. J. Agric. Res. 38:163-176. [Google Scholar]
- 13.de Groot, G. N., and A. H. Stouthamer. 1969. Regulation of reductase formation in Proteus mirabilis. I. Formation of reductases and enzymes of the formic hydrogen lyase complex in the wild type and in chlorate resistant mutants. Arch. Microbiol. 66:220-233. [PubMed] [Google Scholar]
- 14.Doerge, T. A., P. J. Bottomley, and E. H. Gardner. 1985. Molybdenum limitations to alfalfa growth and nitrogen content on a moderately acid, high-phosphorous soil. Agron. J. 77:895-901. [Google Scholar]
- 15.Germgard, U., A. Teder, and D. Tormund. 1981. Chlorate formation during chlorine dioxide bleaching of softwood kraft pulp. Paperi Puu 3:127-133. [Google Scholar]
- 16.Giordano, G., L. Saracino, and L. Grillet. 1985. Identification in various chlorate-resistant mutants of a protein involved in the activation of nitrate reductase in the soluble fraction of a chlA mutant of E. coli K-12. Biochim. Biophys. Acta 839:181-190. [DOI] [PubMed] [Google Scholar]
- 17.Graham, P. H. 1998. Biological dinitrogen fixation: symbiotic, p. 322-345. In D. M. Sylvia, J. J. Fuhrmann, P. G. Hartel, and D. A. Zuberer (ed.), Principles and applications of soil microbiology. Prentice-Hall, Inc., Englewood Cliffs, N.J.
- 18.Hackenthal, E. 1965. Die Reduktion von Perchlorat durch Bacterien. II. Die Identitat der Nitratreduktase und des Perchlorat Reduzierenden Enzyms aus B. cereus. Biochem. Pharmacol. 14:1313-1324. [DOI] [PubMed] [Google Scholar]
- 19.Hackenthal, E., W. Mannheim, R. Hackenthal, and R. Becher. 1964. Die Reduktion von Perchlorat durch Bakterien. I. Untersuchungen an Intaken Zellen. Biochem. Pharmacol. 13:195-206. [DOI] [PubMed] [Google Scholar]
- 20.Herman, D. C., and W. T. Frankenberger, Jr. 1999. Bacterial reduction of perchlorate and nitrate in water. J. Environ. Qual. 28:1018-1024. [Google Scholar]
- 21.Kengen, S. W. M., G. B. Rikken, W. R. Hagen, C. G. van Ginkel, and A. J. M. Stams. 1999. Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J. Bacteriol. 181:6706-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lack, J. G., S. K. Chaudhuri, R. Chakraborty, L. A. Achenbach, and J. D. Coates. 2002. Anaerobic biooxidation of Fe(II) by Dechlorosoma suillum. Microb. Ecol. 43:424-431. [DOI] [PubMed]
- 23.Lack, J. G., S. K. Chaudhuri, S. D. Kelly, K. M. Kemner, S. M. O'Connor, and J. D. Coates. 2002. Immobilization of radionuclides and heavy metals through anaerobic biooxidation of Fe(II). Appl. Environ. Microbiol. 68:2704-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Logan, B. E., H. Zhang, P. Mulvaney, M. G. Milner, I. M. Head, and R. F. Unz. 2001. Kinetics of perchlorate- and chlorate-respiring bacteria. Appl. Environ. Microbiol. 67:2499-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malmqvist, A., T. Welander, and L. Gunnarsson. 1991. Anaerobic growth of microorganisms with chlorate as an electron acceptor. Appl. Environ. Microbiol. 57:2229-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malmqvist, A., T. Welander, E. Moore, A. Ternstrom, G. Molin, and I.-M. Stenstrom. 1994. Ideonella dechloratans gen. nov., sp. nov., a new bacterium capable of growing anaerobically with chlorate as an electron acceptor. Syst. Appl. Microbiol. 17:58-64. [Google Scholar]
- 27.Michaelidou, U., L. A. Achenbach, and J. D. Coates. 2000. Isolation and characterization of two novel (per)chlorate-reducing bacteria from swine waste lagoons, p. 271-283. In E. D. Urbansky (ed.), Perchlorate in the environment. Kluwer Academic/Plenum, New York, N.Y.
- 28.O'Connor, S. M., and J. D. Coates. 2002. A universal immunoprobe for (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 68:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oltmann, L. F., W. N. M. Reijnders, and A. H. Stouthamer. 1976. Characterization of purified nitrate reductase A and chlorate reductase C from Proteus mirabilis. Arch. Microbiol. 111:25-35. [DOI] [PubMed] [Google Scholar]
- 30.Renner, R. 1999. EPA draft almost doubles safe dose of perchlorate in water. Environ. Sci. Technol. 33:110A-111A. [DOI] [PubMed] [Google Scholar]
- 31.Renner, R. 1998. Perchlorate-tainted wells spur government action. Environ. Sci. Technol. 32:210.A. [DOI] [PubMed] [Google Scholar]
- 32.Rikken, G., A. Kroon, and C. van Ginkel. 1996. Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45:420-426. [Google Scholar]
- 33.Roldan, M. D., F. Reyes, C. Moreno-Vivian, and F. Castillo. 1994. Chlorate and nitrate reduction in the phototrophic bacteria Rhodobacter capsulatus and Rhodobacter sphaeroides. Curr. Microbiol. 29:241-245. [Google Scholar]
- 34.Romanenko, V. I., V. N. Korenkov, and S. I. Kuznetsov. 1976. Bacterial decomposition of ammonium perchlorate. Mikrobiologiya 45:204-209. [PubMed] [Google Scholar]
- 35.Rosemarin, A., K. Lehtinen, and M. Notini. 1990. Effects of treated and untreated softwood pulp mill effluents on Baltic sea algae and invertebrates in model ecosystems. Nord. Pulp Paper Res. J. 2:83-87. [Google Scholar]
- 36.Stenklo, K., H. D. Thorell, H. Bergius, R. Aasa, and T. Nilsson. 2001. Chlorite dismutase from Ideonella dechloratans. J. Biol. Inorg. Chem. 6:601-607. [DOI] [PubMed] [Google Scholar]
- 37.Stepanyuk, V., G. Smirnova, T. Klyushnikova, N. Kanyuk, L. Panchenko, T. Nogina, and V. Prima. 1992. New species of the Acinetobacter genus Acinetobacter thermotoleranticus sp. nov. Mikrobiologiya 61:347-356. [Google Scholar]
- 38.Urbanski, T. 1988. Chemistry and technology of explosives, vol. 4, p. 602-620. Pergamon Press, Oxford, United Kingdom.
- 39.Urbansky, E. T. 1998. Perchlorate chemistry: implications for analysis and remediation. Biorem. J. 2:81-95. [Google Scholar]
- 40.van Ginkel, C., G. Rikken, A. Kroon, and S. Kengen. 1996. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch. Microbiol. 166:321-326. [DOI] [PubMed] [Google Scholar]
- 41.Wallace, W., T. Ward, A. Breen, and H. Attaway. 1996. Identification of an anaerobic bacterium which reduces perchlorate and chlorate as Wolinella succinogenes. J. Ind. Microbiol. 16:68-72. [Google Scholar]
- 42.Wirt, K., M. Laikhlman, J. Rohrer, and P. Jackson. 1998. Low-level perchlorate analysis in drinking water and groundwater by ion chromatography. Am. Environ. Lab. 10:1, 5. [Google Scholar]
- 43.Wu, J., R. F. Unz, H. Zhang, and B. E. Logan. 2001. Persistence of perchlorate and the relative numbers of perchlorate- and chlorate-respiring microorganisms in natural waters, soils, and wastewater. Biorem. J. 5:119-130. [Google Scholar]
- 44.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]