Abstract
Thirty-eight strains of Bacillus sporothermodurans isolated from ultra-high-temperature (UHT)-treated milk or sterilized milk (UHT isolates) and from animal feed or raw milk (farm isolates) were characterized by automated ribotyping and by repetitive extragenic palindromic (REP)-PCR fingerprinting. By investigating the genetic relationships among isolates from these various sources, the relative importance of different contamination sources could be evaluated. The results of the separate clustering analyses of the PvuII and EcoRI ribopatterns and the REP-PCR patterns were largely consistent with each other and revealed the existence of two main clusters; there was one homogeneous group containing all (REP-PCR) or most (ribotyping) of the UHT isolates, and there was a second more diverse group comprising the farm isolates. A combined three-dimensional analysis of all data showed that three German UHT isolates did not belong to the compact group containing the majority of the UHT isolates. These results demonstrate that B. sporothermodurans is more heterogeneous than previously assumed and that most of the UHT isolates form a genetically distinct subgroup and are capable of producing highly heat-resistant spores. The close genetic relationship of these UHT isolates suggests a clonal origin of a few predominant strains of B. sporothermodurans that can be found in UHT-treated or sterilized milk products.
Mesophilic aerobic sporeformers with extremely high heat resistance were first detected in ultra-high-temperature (UHT)-treated milk from southern Europe in 1985; later they were also detected in other countries in and outside Europe (6, 8, 17). These sporeformers belonged to the genus Bacillus and were recently classified as a new species, Bacillus sporothermodurans (20).
According to its original description (20), an important characteristic of B. sporothermodurans is its ability to produce highly heat-resistant spores (HRS) that may survive sterilization (115 to 120°C for 15 to 20 min) or UHT treatment (135 to 142°C for a few seconds). Huemer et al. (16) found D values at 140°C that varied between 3.4 and 7.9 s and z values that varied between 13.1 and 14.2°C for spore preparations from the original stock culture, but they also observed a significant decrease in the heat resistance after multiple laboratory culture passages. Surviving spores of B. sporothermodurans can germinate and multiply in products to a maximal concentration of ca. 105 cells/ml. Although the vegetative cells are not pathogenic (9, 10) and do not cause significant visual or taste deviations, their presence in sterilized and UHT-treated products is considered undesirable, as such products do not meet the legal requirements established by the European Union (1).
For detection and identification of B. sporothermodurans in raw and consumer milk, a PCR-based method, now called HRS-PCR (22), was developed by Herman et al. (13). Several molecular methods have been used successfully to differentiate and characterize B. sporothermodurans strains, including PCR methods like random amplified polymorphic DNA analysis, repetitive extragenic palindromic (REP)-PCR, and 16S ribosomal DNA (rDNA) sequence analysis (12, 18, 20). All these techniques showed that the different B. sporothermodurans strains isolated so far from European UHT-treated and sterilized milk are phylogenetically very closely related, forming the so-called HRS clone, named after the initial description of this highly heat-resistant spore-forming organism (8). Recently, B. sporothermodurans strains have also been isolated from raw milk and from animal feed (4, 22, 24). The majority of these farm isolates reacted negatively in the HRS-PCR of Herman et al. (13) but could be assigned to B. sporothermodurans by a polyphasic approach and/or a new 16S rDNA-based specific PCR identification test (22).
The purpose of the present study was to evaluate the genetic diversity of B. sporothermodurans isolates by using two molecular typing techniques (ribotyping and REP-PCR) targeted at different genomic sites as recommended by different authors (14, 25). REP-PCR has been recognized to be highly discriminatory for the differentiation of many bacterial species, including Escherichia coli and B. sporothermodurans (5, 11, 12, 18). In addition, ribotyping has proved to be useful for differentiation at the subspecies level, as demonstrated for species like Pseudomonas aeruginosa, Vibrio cholerae, Listeria monocytogenes, and Clostridium botulinum (3, 7, 19, 23). Both DNA fingerprinting techniques were applied to B. sporothermodurans strains isolated from a wide range of geographic areas, including Asia, South America, and Europe, and from different sources, as outlined above. The resulting genetic relationships found among the strains should allow workers to determine the relative contributions of different possible contamination sources.
MATERIALS AND METHODS
Bacterial strains used and isolation of new B. sporothermodurans strains.
The isolates from European UHT-treated and sterilized milk and the isolates from raw milk and animal feed (feed concentrate, silage, pulp, soy) have been described previously (12, 22, 24). A total of 2,113 UHT-treated or sterilized milk samples were collected worldwide (Pakistan, Ecuador, Dominican Republic, Mexico) in local supermarkets and factories during the second half of 1998. The products were incubated for 5 to 7 days at 37°C, and 0.1 ml of each product was streaked onto brain heart infusion broth (Oxoid) supplemented with vitamin B12 (1 mg/liter; Sigma) and bacteriological agar 1 (15 g/liter; Oxoid) (pH 6.8). Petri dishes were incubated at 37°C for 2 days. Typical colonies of B. sporothermodurans (20) and also spores were identified by phase-contrast microscopy and retained. All of the strains are listed in Table 1, including B. sporothermodurans type strain MB 581 (= DSMZ 10599) and a raw milk isolate of Bacillus oleronius, the closest phylogenetic neighbor of B. sporothermodurans (4, 10, 22).
TABLE 1.
B. sporothermodurans and B. oleronius strains used in this study and their originsa
| B. sporothermodurans strain(s) | Country of origin | Product | PCR (primers SH2-F1 and SH2-R)b | PCR (16S rDNA- based primers)c | PvuII ribogroup | EcoRI ribogroup | REP-PCR group |
|---|---|---|---|---|---|---|---|
| UHT isolates | |||||||
| MB 1320, MB 1325, MB 1512 | Dominican Republic | Sterilized milk | + | + | A1 | B3 | C2a |
| MB 1324 | Dominican Republic | Sterilized milk | + | + | A1 | B1 | C2a |
| MB 1511, MB 1517 | Dominican Republic | Sterilized milk | + | + | A1 | B1 | C2d |
| MB 1516 | Dominican Republic | Sterilized milk | + | + | A1 | B7 | C2d |
| MB 1327 | Ecuador | Sterilized milk | + | + | A1 | B3 | C2a |
| MB 1513 | Ecuador | Sterilized milk | + | + | A1 | B1 | C2d |
| MB 1326, MB 1514 | Ecuador | Sterilized milk | + | + | A1 | B1 | C2c |
| MB 1515 | Ecuador | UHT milk | + | + | A1 | B3 | C2b |
| MB 372 | Germany | UHT milk | + | + | A12 | B6 | C1 |
| MB 374 | Germany | UHT milk | + | + | A4 | B5 | C1 |
| MB 373 | Germany | UHT milk | + | + | A11 | B4 | C1 |
| MB 1321 | Pakistan | UHT milk | + | + | A1 | B3 | C2a |
| MB 1323 | Mexico | UHT milk | + | + | A1 | B3 | C2a |
| MB 582 (= TP12481)d | France | UHT milk | + | + | A1 | B19 | C2d |
| MB 338 | Italy | UHT milk | + | + | A1 | B1 | C2d |
| MB 581T (= DSMZ 10599T)e | Italy | UHT milk | + | + | A2 | B3 | C2d |
| MB 512 | Belgium | UHT milk | + | + | A1 | B2 | C2d |
| MB 404 | Belgium | UHT milk | + | + | A1 | B8 | C2a |
| MB 389, MB 390 | Belgium | UHT milk | + | + | A1 | B1 | C2d |
| MB 359 | Belgium | UHT milk | + | + | A1 | B3 | ND |
| Farm isolates | |||||||
| MB 1494 | Belgium | Soy | − | + | A8 | B9 | C5 |
| MB 1495 | Belgium | Soy | − | + | A15 | B10 | C13 |
| MB 1497 | Belgium | Feed concentrate | − | + | A5 | B20 | C4 |
| MB 1498 | Belgium | Feed concentrate | − | + | A7 | B14 | C12 |
| MB 1499 | Belgium | Feed concentrate | − | + | A3 | B12 | C13 |
| MB 1500 | Belgium | Feed concentrate | − | + | A9 | B13 | C9 |
| MB 1501 | Belgium | Feed concentrate | + | + | A5 | B17 | C3 |
| MB 1506 | Belgium | Feed concentrate | − | + | A13 | B16 | C6 |
| MB 1508 | Belgium | Feed concentrate | − | + | A6 | B15 | C10 |
| MB 1316 | Belgium | Feed concentrate | − | + | A14 | B11 | C14 |
| MB 1317 | Belgium | Feed concentrate | − | + | A10 | B9 | C7 |
| MB 1505 | Belgium | Silage | + | + | A13 | B17 | C2b |
| MB 385 | Belgium | Raw milk | − | + | A16 | B18 | C8 |
| B. oleronius MB 397 | France | Raw milk | − | − |
Identification and characterization were performed by using PCR, ribotyping, and REP-PCR as described in this paper.
PCR results obtained by using the method of Herman et al. (13).
PCR results obtained by using the method of Scheldeman et al. (22).
The designation in parentheses is the designation of Pettersson et al. (20).
DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany.
ND, not determined.
Identification of presumptive B. sporothermodurans isolates.
Presumptive B. sporothermodurans colonies were identified by two PCR methods. The first method, HRS-PCR (22) performed with sequences obtained by subtractive hybridization, was performed with the primers SH2-F1 (5′GATTCAGGCAGAATGTAGCA3′) and SH2-R (5′TTTCGCCACTTGATGGTACA3′) (Microsynth AG, Balgach, Switzerland) as described by Herman et al. (13). In addition, a newly developed primer pair, F2 (5′ACGGCTCAACCGTGGAG3′) and R2 (5′GTAACCTCGCGGTCTA3′), for amplification of a B. sporothermodurans-specific 16S rDNA fragment was used as described by Scheldeman et al. (22).
Automated ribotyping and data analysis.
All B. sporothermodurans isolates were processed at least two times by using the automated microbial characterization system (RiboPrinter; Qualicon, Inc., Wilmington, Del.) and restriction enzymes EcoRI and PvuII (Qualicon) as described elsewhere (2). All ribopatterns with similarity coefficients higher than 0.93 were considered identical by the RiboPrinter software and were grouped together based on the position and intensity of the bands to form a ribogroup (a set of isolates with indistinguishable ribotypes). Further refinement of the automated ribogrouping was performed by visual evaluation of closely related ribopatterns, which resulted in merger or separation of ribogroups.
REP-PCR.
Total genomic DNA from purified B. sporothermodurans strains was isolated by using the method of Pitcher et al. (21), as slightly modified by Heyndrickx et al. (15), and the DNA concentration was determined with a spectrophotometer. Total genomic DNA (25 ng) was used as the template in REP-PCR performed with primers REP1R-I (5′ IIIICGICGICATCIGGC3′) and REP2-I (5′ICGICTTATCIGGCCTAC3′) (Isogen Bioscience bv), and the REP fragments were separated by denaturing polyacrylamide gel electrophoresis and silver stained as described by Herman et al. (12). All strains were analyzed in the same REP-PCR experiment and on the same gel to minimize possible variations in patterns caused by experimentation (12). A REP-PCR pattern could not be obtained for strain MB 359.
Banding pattern data analyses.
Digitized images of the gel obtained with the RiboPrinter were converted with the GelConvert program (Qualicon) and were analyzed by the unweighted pair group arithmetic (UPGMA) clustering algorithm computed by the GelCompar software (version 4.1; Applied Maths, Sint-Martens-Latem, Belgium) by using the Pearson product moment correlation coefficient with optimization of 1%. REP-PCR patterns on the silver-stained gels were scanned with a flat-bed scanner (Agfa SnapScan1236S; Agfa-Gevaert N.V., Mortsel, Belgium), and images were analyzed by UPGMA clustering computed by BioNumerics software (version 2.0; Applied Maths) by using the Pearson product moment correlation coefficient with optimization of 1%. Normalized REP-patterns were also visually classified in REP groups.
Combined clustering of the two ribopatterns and the REP-PCR profiles was performed by using the BioNumerics 2.0 software. To do this, the converted ribopatterns obtained with EcoRI or PvuII were introduced into the BioNumerics 2.0 program, and each pattern was linked with the strain database. Combination of the three experiment types was implemented in such a way that the typing methods used (REP-PCR, EcoRI riboprinting, and PvuII riboprinting) were weighted 2:1:1. For UPGMA clustering for this combination of experiment types, the same cluster analysis settings that were used for each experiment type separately were used (i.e., Pearson product moment correlation coefficient). Finally, multidimensional scaling of the combined cluster analysis allowed visually interpretable grouping of the strains in a three-dimensional plot.
RESULTS
Identification.
Fourteen new isolates of B. sporothermodurans were obtained in this study from contaminated UHT-treated or sterilized consumer milk (designated UHT isolates) from the Dominican Republic, Ecuador, Pakistan, and Mexico. These strains reacted positively as determined by the two PCR-based methods used for identification of B. sporothermodurans (Table 1), the HRS-PCR developed by Herman et al. (13) and the recently developed PCR based on specific 16S rDNA sequence stretches described by Scheldeman et al. (22). The reaction pattern was similar to that for the 11 European isolates from UHT-treated and sterilized milk listed in Table 1 and reported by Scheldeman et al. (22). However, in the previous study, it was found that 11 of the 13 isolates from raw milk or animal feed (feed concentrate, soy, silage) (designated farm isolates) (Table 1) reacted negatively in the HRS-PCR and positively in the 16S rDNA-based PCR; only two farm strains included in the present study (MB 1501 and MB 1505) reacted positively in both PCR tests (Table 1).
Ribotyping.
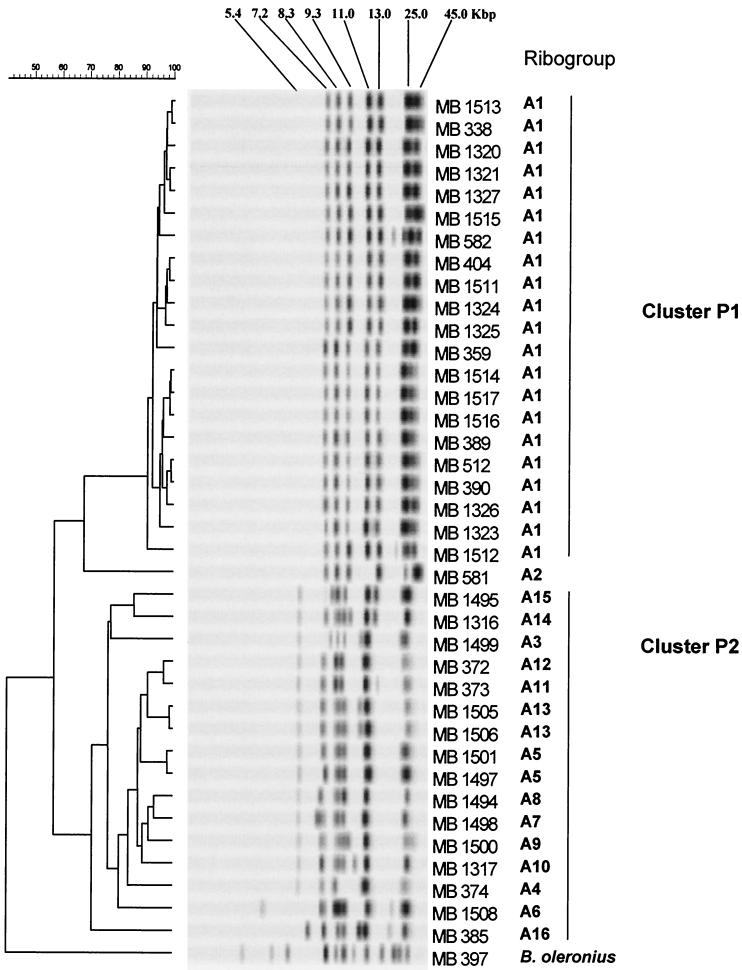
Molecular characterization of B. sporothermodurans strains and the B. oleronius reference strain was performed by automated ribotyping by using the restriction enzymes PvuII and EcoRI. Sixteen distinct major ribogroups were determined with PvuII (ribogroups A1 to A16) (Table 1 and Fig. 1). The three largest ribogroups, ribogroups A1, A5, and A13, contained 21, 2, and 2 strains, respectively (Tables 1 and 2), whereas the other 13 ribogroups were each represented by a single strain. Based on the cluster analysis performed with the ribopatterns of 39 strains (including B. sporothermodurans type strain MB 581 and the reference strain of B. oleronius), a dendrogram was constructed (Fig. 1). Two main clusters, clusters P1 and P2, were discerned visually and by cluster analysis at similarity levels of 91 and 71%, respectively. These two clusters exhibited 58% similarity to each other. The majority of the UHT isolates (21 of 25 isolates) were found in cluster P1, which included one ribogroup. These UHT isolates appeared to be closely related to each other, as reflected by the high similarity values (91 to 99%). All these isolates produced a typical pattern consisting of at least seven conserved bands at ca. 7.2, 8.3, 9.3, 11.0, 13.0, 25.0, and 45.0 kbp. In cluster P2, which was defined at a lower similarity level (71%), 14 different ribogroups were found; there were typical bands at ca. 5.4, 7.1, 7.5, 8.5, 9.0, 11.0, and 25.0 kbp in the patterns of most of the strains, while 13.0- and 45.0-kbp fragments were missing compared to the patterns of cluster P1 strains. Remarkably, UHT strains MB 372, MB 373, and MB 374, all of which originated from Germany, had clearly distinct patterns and were members of cluster P2 along with all 13 farm isolates. Type strain MB 581 was located separately between clusters P1 and P2.
FIG. 1.
Dendrogram of 38 strains of B. sporothermodurans and one strain of B. oleronius obtained after restriction with PvuII. The strain designations correspond to those shown in Table 1. The scale bar indicates the percentage of similarity.
TABLE 2.
Relationships between the different most important PvuII and EcoRI ribogroupsa
| PvuII ribogroup | EcoRI ribogroups |
|---|---|
| A1 (21) | B1 (9), B2 (1), B3 (8), B7 (1), B8 (1), B19 (1) |
| A5 (2) | B17 (1), B20 (1) |
| A13 (2) | B16 (1), B17 (1) |
The numbers in parentheses are the numbers of isolates in the ribogroups.
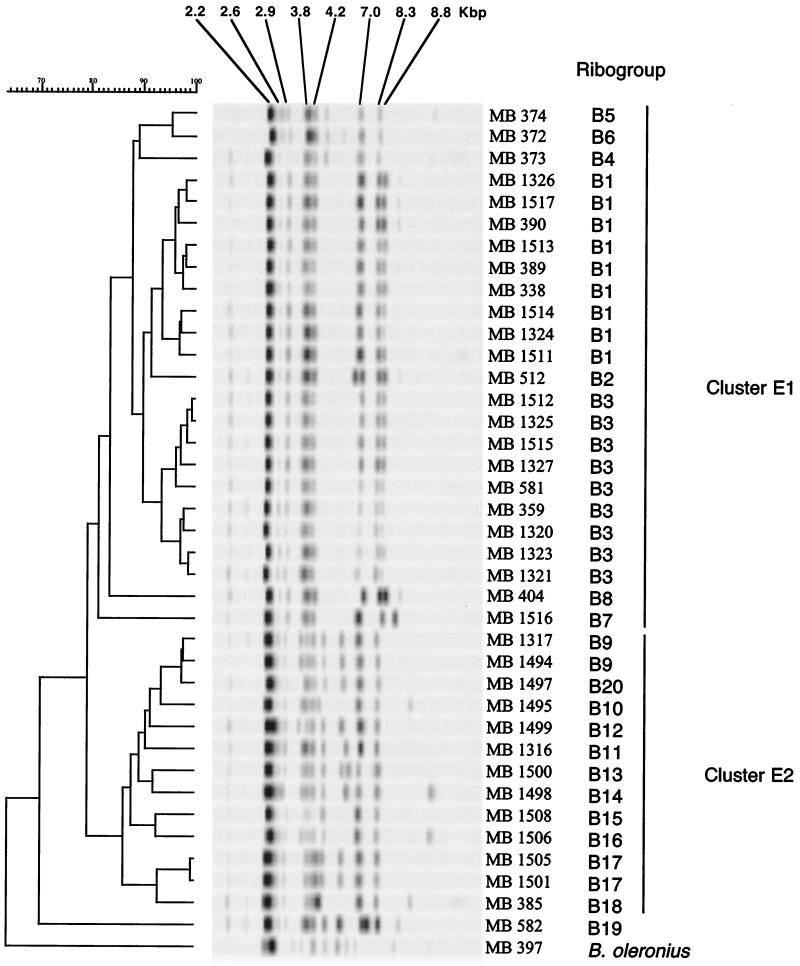
With EcoRI, 20 distinct ribogroups were found, and these ribogroups were designated ribogroups B1 to B20 (Table 1 and Fig. 2). Ribogroups B1 and B3 each contained nine strains, while two strains belonged to ribogroups B9 and B17. Sixteen ribogroups were represented by a single strain. All 38 strains of B. sporothermodurans produced at least five typical conserved bands, at ca. 2.2, 2.9, 3.8, 7.0, and 8.3 kbp. The dendrogram constructed with the EcoRI-generated patterns showed high degrees of similarity (ca. 80% without strain MB 582) among the B. sporothermodurans strains (Fig. 2). Despite the lower diversity, two main clusters, designated clusters E1 and E2, were discerned visually and by cluster analysis at similarity levels of 81 and 86%, respectively. Interestingly, cluster E1 encompassed 24 UHT isolates but did not contain UHT strain MB 582, whereas cluster E2 contained all 13 farm isolates. Compared to the ribopatterns of the UHT isolates, most of the ribopatterns in cluster E2 showed polymorphisms in the region between 4.0 and 6.0 kbp, and one or two extra bands were present. The farm isolates were scattered in 11 distinct ribogroups, confirming the genetic variability observed with the PvuII-generated patterns. UHT strain MB 582 (= TP1248) formed a separate ribogroup, ribogroup B19. This result is in agreement with the previous finding of Pettersson et al. (20), who described strain TP1248 as a strain that is slightly atypical based on ribotyping analysis.
FIG. 2.
Dendrogram of 38 strains of B. sporothermodurans and one strain of B. oleronius obtained after restriction with EcoRI. The strain designations correspond to those shown in Table 1. The scale bar indicates the percentage of similarity.
As illustrated in Table 2, the combination of restriction enzymes PvuII and EcoRI enabled a finer level of discrimination, as illustrated for strains MB 1497 and MB 1501. These strains, which constituted a single ribogroup (ribogroup A5) when PvuII was used, could be differentiated from each other with EcoRI (ribogroups B20 and B17). Conversely, strains MB 1317 and MB 1494, which were indistinguishable on the basis of typing with EcoRI (ribogroup B9), could be discriminated with PvuII (ribogroups A10 and A8). Also, strain TP1248 could be separated from all other strains only when EcoRI was used. All the PvuII ribogroups could be further differentiated with EcoRI and vice versa. However, the relationship between different PvuII and EcoRI ribogroups in some instances was rather complex.
B. oleronius MB 397 was ribotyped as an outlier. The PvuII- or EcoRI-generated ribopatterns of B. oleronius MB 397 were clearly different from those obtained with all the B. sporothermodurans strains (similarity levels, 40 and 57%, respectively).
REP-PCR.
Molecular characterization of B. sporothermodurans strains and the B. oleronius reference strain was performed by REP-PCR by using high-resolution separation of the bands by polyacrylamide gel electrophoresis and silver staining. Fourteen distinct major REP groups were visually determined (REP groups C1 to C14) (Table 1 and Fig. 3). The two largest REP groups, REP groups C1 and C2, contained 3 and 22 strains, respectively (Table 1), whereas the other 12 REP groups were each represented by a single strain. In comparison with B. oleronius, all B. sporothermodurans strains were characterized by a conserved major band at ca. 1,020 bp. Furthermore, strains belonging to REP groups C1 and C2, which contained all UHT isolates and only one farm isolate (MB 1505), were characterized by major conserved bands at ca. 875, 730, and 600 bp. Remarkably, the German UHT strains, MB 372 to MB 374, belonged to the same REP group, REP group C1, which could be differentiated from REP group C2, which contained the other UHT strains from different countries, by the absence of a major conserved band at ca. 850 bp in REP group C2. The largest REP group (REP group C2) could be further subdivided into four subgroups on the basis of minor polymorphisms (presence or absence of minor bands at various molecular weights). These four subgroups, subgroups C2a to C2d (Table 1 and Fig. 3), contained 8, 2, 2, and 10 strains, respectively.
FIG. 3.
Dendrogram of 37 strains of B. sporothermodurans and one strain of B. oleronius obtained by REP-PCR. The strain and REP group designations correspond to those shown in Table 1. The scale bar indicates the percentage of similarity as determined with the Pearson coefficient. The dark zones under the metric scale represent parts of the REP patterns omitted in the numerical analysis.
By performing a cluster analysis of the REP patterns, a dendrogram was constructed (Fig. 3). All of the B. sporothermodurans strains showed an overall similarity level of only 50%, indicating great genetic diversity. Only one major cluster, cluster R1, could be discerned visually and by cluster analysis at a meaningful similarity level (80%). All the UHT isolates were found in this major cluster, which contained REP groups C1 and C2. With the exception of two strains (MB 1494 and MB 1497) which exhibited 84% similarity to each other, the farm strains exhibited less than 80% similarity to each other and to the major cluster R1 strains. The closest relatives of cluster R1 of UHT isolates based on their REP patterns were farm strains MB 385 (isolated from raw milk) and MB 1317 (isolated from feed concentrate), at a similarity level of 76%. Surprisingly, farm strain MB 1505 belonging to REP group C2b (Table 1) clustered at only 69% similarity with cluster R1 containing the other REP group C2 strains. This can probably be explained by differences in band intensity, such as a less intense major band at ca. 1,020 bp in the pattern of MB 1505 (Fig. 3), which is known to influence clustering by the Pearson correlation coefficient (25).
The REP pattern of B. oleronius reference strain MB 397 was clearly different from the main B. sporothermodurans pattern.
Combined analysis of ribotyping and REP-PCR patterns.
The overall or consensus genetic relatedness among the B. sporothermodurans strains was inferred from a combined numerical analysis of the ribopatterns obtained with PvuII and EcoRI and the REP-PCR patterns by performing a UPGMA cluster analysis and by three-dimensional scaling. In the cluster analysis (data not shown), 21 of the 24 UHT strains included clustered together at a minimal similarity level of 81%. In this cluster, 19 UHT strains clustered together at 86% similarity, with strain MB 582 and type strain MB 581 joining at 84 and 81%, respectively. Conversely, 10 of the 13 farm strains and the three German UHT strains (MB 372 to MB 374) produced a cluster at a lower level of similarity (70%), exhibiting only 67% similarity to the major cluster of UHT strains. The remaining three farm strains (MB 1316, MB 1495, and MB 1499) clustered together at 76% similarity and exhibited a lower level of similarity (64%) to all other B. sporothermodurans strains.
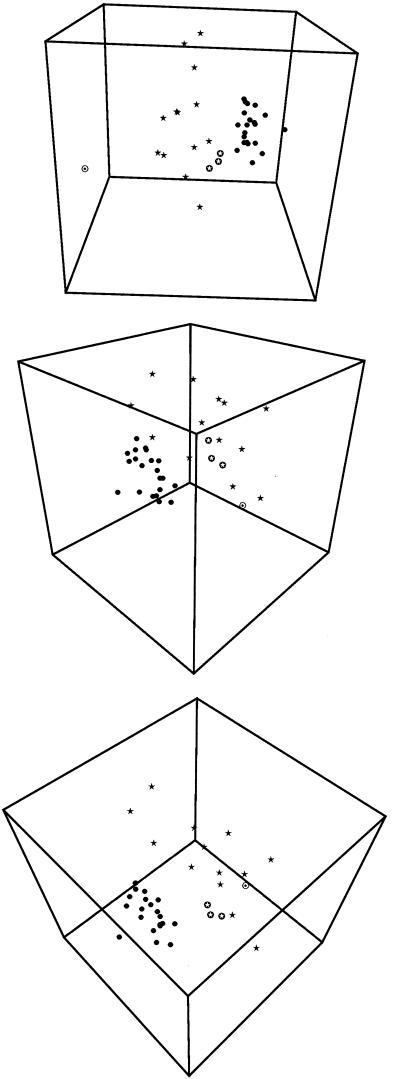
The results of the combined analysis of the data by three-dimensional scaling are shown in Fig. 4. In this nonhierarchical presentation of the relationships among the strains, essentially the same groups were obtained as in the hierarchical cluster analysis explained above. All UHT strains except the three German UHT strains formed a compact group, while all farm strains formed a very diffuse group clearly separated from the group of UHT strains. The only remarkable difference from the UPGMA cluster analysis was the grouping of the three German UHT isolates at the outer edge of the diffuse group of farm strains, showing a somewhat closer relationship to the other UHT isolates.
FIG. 4.
Visual three-dimensional representation of the combined clustering of the two ribotyping patterns (EcoRI and PvuII) and REP-PCR profiles of 37 strains of B. sporothermodurans and one strain of B. oleronius, obtained by multidimensional scaling of the cluster as explained in text. The strain designations correspond to those shown in Table1.
DISCUSSION
In this study, European isolates of B. sporothermodurans from UHT-treated and sterilized milk and isolates obtained from a large screening analysis of heat-resistant sporeformers from the farm environment (raw milk, feed concentrate, silage), as well as new non-European isolates from UHT-treated and sterilized milk, were characterized at different molecular levels. The new UHT isolates were obtained from different continents and countries (Mexico, Ecuador, Dominican Republic, Pakistan) and were found to be positive by both the HRS-PCR (13) and the general B. sporothermodurans-specific PCR (22). In contrast, the majority of the farm isolates reacted negatively in the HRS-PCR (22). This result confirms that the HRS-PCR is more discriminatory for the B. sporothermodurans strains which are relevant for UHT-treated products.
In a polyphasic typing approach, separate clustering analyses of PvuII and EcoRI ribopatterns and of REP-PCR patterns were largely consistent with each other and revealed the existence of two main clusters; one homogeneous group contained all (REP-PCR) or most (ribotyping) of the UHT isolates, and the second, more diverse group comprised the farm isolates (Fig. 1 to 3). The high level of genetic homology of most of the UHT isolates was further shown by a combined analysis of all molecular typing data in this study, both by cluster analysis and by three-dimensional scaling, which revealed a very compact cluster or group of isolates. The close genetic relationship of the UHT isolates suggests a clonal origin (HRS clone), which is particularly remarkable since B. sporothermodurans strains were isolated from UHT-treated and sterilized milk samples produced on three different continents.
The three German isolates were the only UHT strains whose genetic characteristics were quite different from those of the majority of the UHT strains. In a combined cluster analysis of all molecular typing data obtained in this study, the three German UHT strains clustered with the farm strains. A three-dimensional scaling analysis of all molecular typing data showed that these strains were at the border of the diffuse group of farm strains and directed to the compact group of UHT isolates. The latter observation and the REP-PCR and EcoRI ribotyping cluster analysis data suggest that the German UHT isolates have a remote genetic relationship with the HRS clone. This could also suggest that the extreme resistance of spores to sterilization temperatures is restricted to particular clones that have a possible common ancestor.
In contrast to the homogeneity found for the majority of the UHT isolates, the ribopatterns and REP patterns of the B. sporothermodurans strains isolated from animal feed (feed concentrate, silage, soy) and raw milk were much more diverse. Most of the ribogroups and all 12 REP types for the farm isolates were represented by a single strain (Table 1). Also, the two HRS-PCR-positive farm strains, which originated from feed concentrate and silage, produced patterns that were different from each other and from the patterns of the main group of UHT isolates (HRS clone) in the ribotyping analysis. Overall, it seems that there is no 100% concordance between a positive result in the HRS-PCR analysis and an HRS clone pattern determined by molecular typing.
Milk powder has been suggested to be a possible source of contamination of heat-treated dairy products (8). Since in some plants, UHT-treated or sterilized milk is prepared from imported milk powder, this practice could explain the spread of the same B. sporothermodurans HRS clone over different continents. Within one country, one can envisage a contamination route via raw milk that has been contaminated through animal feed at the farm level. Based on this assumption, one would expect to find B. sporothermodurans strains having similar ribopatterns or REP-PCR patterns in animal feed or raw milk, as well as in UHT-treated or sterilized milk. However, the combined analysis of all typing data definitely showed that none of the farm isolates genetically resembled any of the UHT isolates. Since all farm isolates in this study originated from Belgium, they probably represent only a limited part of the natural genetic diversity of B. sporothermodurans. The data presented here confirm the hypothesis that the regular occurrence of contaminated UHT-treated and sterilized milk in some European dairy plants in the mid-1990s, as well as the present sporadic occurrence of contamination, can also be caused by circulation of the HRS clone within and between UHT production units. Occasionally, contamination of UHT-treated milk by a new genetic type (e.g., a type originating at the farm level) occurs, as exemplified here by the German UHT isolates. At present, the data obtained in this study do not favor or eliminate any of the potential contamination routes mentioned above.
In conclusion, this molecular typing study showed that a few clones of B. sporothermodurans, including the so-called HRS clone, have been and are still responsible for the contamination of UHT-treated and sterilized milk and milk products due to the production of highly heat-resistant spores. In particular, the HRS clone has spread over several European countries and even between continents. The strains isolated from UHT-treated and sterilized milk show a close genetic relationship, suggesting a common ancestry for the production of highly heat-resistant spores. An intriguing question which emerges, is whether the capacity to produce highly heat-resistant spores that allow survival after certain heat treatments is restricted to the subgroup of UHT isolates or whether it is a property more widespread in B sporothermodurnans, including the farm isolates. Although heat resistance is not an absolute spore property and is influenced by several factors, such as repeated laboratory cultivation (16), preliminary studies indicate that some farm isolates produce spores with remarkably high heat resistance. However, it remains to be determined whether these spores can also survive UHT treatment (O. Guillaume-Gentil, P. Keijzer, P. Scheldeman, and M. Heyndrickx, unpublished data). The molecular typing techniques used in this study demonstrated the great genetic heterogeneity of B. sporothermodurans isolates from dairy farms, even though they had been isolated in only one country. Because of this observed heterogeneity, the original taxonomic description of B. sporothermodurans, which was based on only a few genetically homogeneous UHT isolates (20), may no longer be adequate.
Acknowledgments
We thank the Belgian Ministry of Small Enterprises, Traders and Agriculture, DG6—Division of Research, for financial support.
We thank E. Engels for performing REP-PCR, P. De Vos (University of Ghent, Ghent, Belgium) for the use of the BioNumerics software, and E. Bidlas for determination of ribopatterns.
REFERENCES
- 1.Anonymous. 1992. Directive 92/46/EEC. Council of the European Communities of 16 July. Health rules for the production and the trade of raw milk, heat-treated milk, and products based on milk. Off. J. Eur. Community L268:1-32.
- 2.Bruce, J. 1996. Automated system rapidly identifies and characterizes micro-organisms in food. Food Technol. 50:77-81. [Google Scholar]
- 3.Dalsgaard, A., P. Echeverria, J. L. Larsen, R. Siebeling, O. Serichantalergs, and H. H. Huss. 1995. Application of ribotyping for differentiating Vibrio cholerae non-O1 isolated from shrimp farms in Thailand. Appl. Environ. Microbiol. 61:245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Silva, S., B. Pettersson, M. De Muro, and F. Priest. 1998. A DNA probe for the detection and identification of Bacillus sporothermodurans using the 16S-23S rDNA spacer region and phylogenetic analysis of some field isolates of Bacillus which form highly heat resistant spores. Syst. Appl. Microbiol. 21:388-407. [DOI] [PubMed] [Google Scholar]
- 5.Dombek, P., L. Johnson, S. Zimmerley, and M. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foschino, R., A. Galli, and G. Ottogali. 1990. Research on the microflora of UHT milk. Ann. Microbiol. 40:47-59. [Google Scholar]
- 7.Gendel, S. M., and J. Ulaszek. 2000. Ribotype analysis of strain distribution in Listeria monocytogenes. J. Food Prot. 63:179-185. [DOI] [PubMed] [Google Scholar]
- 8.Hammer, P., F. Lembke, G. Suhren, and W. Heeschen. 1995. Characterization of a heat resistant mesophilic Bacillus species affecting quality of UHT-milk—a preliminary report. Kiel. Milchwirtsch. Forschungsber. 47:303-311. [Google Scholar]
- 9.Hammer, P., G. Suhren, and W. Heeschen. 1995. Pathogenicity testing of unknown mesophilic heat resistant bacilli from UHT-milk. Bull. Int. Dairy Fed. 302:56-57. [Google Scholar]
- 10.Herman, L., M. Heyndrickx, M. Vaerewijck, and N. Klijn. 2000. Bacillus sporothermodurans—a Bacillus forming highly heat-resistant spores. 3. Isolation and methods of detection. Bull. Int. Dairy Fed. 357:9-14. [Google Scholar]
- 11.Herman, L., and M. Heyndrickx. 2000. The presence of intragenically located REP-like elements in Bacillus sporothermodurans is sufficient for REP-PCR typing. Res. Microbiol. 151:255-261. [DOI] [PubMed] [Google Scholar]
- 12.Herman, L., M. Heyndrickx, and G. Waes. 1998. Typing of Bacillus sporothermodurans and other Bacillus species isolated from milk by repetitive element sequence based PCR. Lett. Appl. Microbiol. 26:183-188. [DOI] [PubMed] [Google Scholar]
- 13.Herman, L., M. Vaerewijck, R. Moermans, and G. Waes. 1997. Identification and detection of Bacillus sporothermodurans spores in 1, 10, and 100 milliliters of raw milk by PCR. Appl. Environ. Microbiol. 63:3138-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heyndrickx, M., N. Rijpens, and L. Herman. 2001. Molecular detection and typing of foodborne bacterial pathogens: a review, p. 193-238. In A. Durieux and J.-P. Simon (ed.), Trends in bio/technology. Applied microbiology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 15.Heyndrickx, M., L. Vauterin, P. Vandamme, K. Kersters, and P. De Vos. 1996. Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J. Microbiol. Methods 26:247-259. [Google Scholar]
- 16.Huemer, I., N. Klijn, H. Vogelsang, and L. Langeveld. 1998. Thermal death kinetics of spores of Bacillus sporothermodurans isolated from UHT-milk. Int. Dairy J. 8:851-855. [Google Scholar]
- 17.Kessler, H. G., J. Pfeifer, and C. Schwöppe. 1994. Untersuchungen über hitze resistente mesophile Bacillus-Sporen in UHT-Milch. Dtsch. Milchwirtsch. 13:588-592. [Google Scholar]
- 18.Klijn, N., L. Herman, L. Langeveld, M. Vaerewijck, A.Wagendorp, I. Huemer, and A. Weerkamp. 1997. Genotypical and phenotypical characterization of Bacillus sporothermodurans strains, surviving UHT sterilization. Int. Dairy J. 7:421-428. [Google Scholar]
- 19.Nociari, M. M., M. Catalano, M. Torrero, and D. O. Sordelli. 1996. Pseudomonas aeruginosa ribotyping: stability and interpretation of ribosomal operon restriction patterns. Diagn. Microbiol. Infect. Dis. 25:27-33. [DOI] [PubMed] [Google Scholar]
- 20.Pettersson, B., F. Lembke, P. Hammer, E. Stackebrandt, and F. Priest. 1996. Bacillus sporothermodurans, a new species producing highly heat-resistant endospores. Int. J. Syst. Bact. 46:759-764. [DOI] [PubMed] [Google Scholar]
- 21.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 22.Scheldeman, P., L. Herman, J. Goris, P. De Vos, and M. Heyndrickx. 2002. Polymerase chain reaction identification of Bacillus sporothermodurans from dairy sources. J. Appl. Microbiol. 92:983-991. [DOI] [PubMed]
- 23.Skinner, G. E., S. M. Gendel, G. A. Fingerhut, H. A. Solomon, and J. Ulaszek. 2000. Differentiation between types and strains of Clostridium botulinum by riboprinting. J. Food Prot. 63:1347-1352. [DOI] [PubMed] [Google Scholar]
- 24.Vaerewijck, M. J. M., P. De Vos, L. Lebbe, P. Scheldeman, B. Hoste, and M. Heyndrickx. 2001. Occurrence of Bacillus sporothermodurans and other aerobic sporeforming species in feed concentrate for dairy cattle. J. Appl. Microbiol. 91:1074-1084. [DOI] [PubMed] [Google Scholar]
- 25.Vandamme, P., B. Pot, M. Gillis, P. De Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]