Abstract
Background
The Amazon region is home to more than 30% of the sand flies species in Colombia, including vectors of Leishmania mainly in the genus Lutzomyia and Psychodopygus. Advances in morphological and molecular taxonomy of sand flies facilitate the development of updated and robust species inventories in understudied areas, such as the departments of Amazonas and Caquetá. Currently, integrating the detection of blood meal sources and Leishmania DNA represents a key approach under the “One Health” concept by providing insights into human and animal health and the dynamics of different ecosystems.
Methodology/principal findings
This study characterized the sand flies fauna in Amazonas and Caquetá using an integrative taxonomic approach that included DNA detection from blood meal and Leishmania sources. Sand flies were collected using CDC, Shannon, Prokopack traps and mouth aspirators. DNA was analyzed by conventional PCR targeting COI, Cytb, 12S rDNA and HSP-70N markers, respectively. A total of 1,104 specimens were collected, representing 12 genera and 30 species, 10 are recognized vectors of Leishmania, including Nyssomyia antunesi and Psychodopygus amazonensis. Our findings include new reports of regional distribution, particularly the first report of Sciopemyia fluviatilis in Colombia. Homo sapiens (28.8% Cytb; 18.6% 12S) and Sus scrofa (16.9% Cytb; 6.8% 12S) were the main food sources detected. While Nyssomyia fraihai (2.6%), Trichophoromyia cellulana (1.3%), Nyssomyia yuilli pajoti (1.3%) and Evandromyia (Aldamyia) walkeri (1.0%) grouped the highest detection rate of Leishmania DNA (9.0%).
Conclusions/significance
The integration of molecular tools for the confirmation of phlebotomine species allowed the resolution of taxonomic conflicts, especially in the genus Trichophoromyia. These findings provide key information on ecological interactions (vectors-ingesta-Leishmania) related to leishmaniasis in the Colombian Amazon, suggesting a high diversity of sand flies and a significant zoonotic potential.
Author summary
Knowledge of the ecoepidemiology of leishmaniasis, an endemic disease in Colombia, is essential, especially in areas of triple frontier such as the Colombian, Brazil, and Peru Amazon, where the circulation of vectors, hosts and pathogens may be favored. For this reason, we analyzed the ecological interactions of sand flies in areas of transmission of tegumentary leishmaniasis in the departments of Amazonas and Caquetá. Sand flies were identified using an integrative approach combining classical and molecular tools, that also allowed identifying vertebrates from blood remains and detecting Leishmania DNA in different environments. Our findings reflect the need to update taxonomic inventories to facilitate tracking of circulating species. We report new records at the departmental and national level and increase the diversity of haplotypes of the COI gene for sand flies. The diversity of vertebrates identified as blood food source of Amazonian sand flies was mainly represented by Homo sapiens, Sus scrofa, and even monkeys like Saimiri macrodon. Demonstrating the importance of understanding the level of dietary plasticity, especially in species vectors, a determining factor in the maintenance of the pathogen life cycle. Finally, the detection of Leishmania sp. DNA suggests an active circulation of the parasite in the areas studied.
1. Introduction
Tegumentary Leishmaniases are a group of zoonotic diseases caused by Leishmania of the subgenera Leishmania and Viannia, L. (L.) amazonensis, L. (L.) mexicana, L. (V.) braziliensis, L. (V.) guyanensis, and L. (V.) panamensis in the Colombian Amazon region [1,2]. These parasite species are transmitted by hematophagous Diptera belonging to the subfamily Phlebotominae. A significant number of leishmaniasis cases (4,403) mainly cutaneous (<95%) were confirmed in the Colombian Amazon region alone, of which 37.5% (1,651 cases) were recorded in Amazonas and Caquetá departments during the period of 2020–2024 [3,4].
The fauna and flora biodiversity, the variety of ecologically distinct landscape features, as well as the constant changes in land use in the Amazon region, especially in both departments [5,6], allow the optimal development of sand flies and the increase of pathogen transmission risk [6–8].
In this context, human mobility between forest and jungle areas, driven by tourism, forced displacement, and cross-border economic activities, has led to significant disturbances in natural habitats. Additionally, deforestation, illegal mining, and extensive cattle ranching aggravate the problem of vector-borne diseases (ETVs) such as leishmaniasis [9,10].
This has favored the expansion of the geographical ranges and ecological niches of sand flies [11], increasing their interaction with wild and domestic vertebrates, which play a role in the epidemiological triad as food sources, but also as potential reservoirs of microorganisms [10,12–14].
Recent studies conducted in urban forest remnants and preserved forest environments in the Brazilian Amazon evidenced how the eclectic feeding behavior of sand flies in disturbed habitats increased the risk of contact with vertebrate reservoirs like Psychodopygus ayrozai which, besides resenting blood traces of Homo sapiens, was also associated with other sources of ingestion such as Dasypus novemcinctus, a potential reservoir of L. (V.) naiffí [15].
Other studies have detected blood traces of mammals such as Caniculus paca (lowland paca), a potential reservoir of L. (V.) lainsoni [16] and Tamandua tetradactyla (collared anteater), proven host of L. (V.) guyanensis [17], in Nyssomyia antunesi [16]. Trypanosoma minasense DNA has also been detected in individuals of this species along with blood traces of T. tetradactyla and H. sapiens [15]. Feeding plasticity in these insects favors their survival and establishment in recently colonized habitats or areas where the primary food source is scarce [18,19] as demonstrated in Ny. umbratilis, Lutzomyia gomezi, and Ny. antunesi recorded in Amazonas and Caquetá [20]. Notably, these insects have also been found infected with different Leishmania species [21–23].
Therefore, it is essential to understand the extent of dietary plasticity of these insects, especially in areas of the Colombian Amazon, where the availability of vertebrates could influence their role as primary or secondary vectors mainly of cutaneous leishmaniasis (CL) [24,25]. Which contributes to a better understanding of the epidemiology of the disease, particularly in border areas [26].
Specifically, in Belén de los Andaquíes, Milán, Solano and Solita Caquetá, the circulation of 46 species of sand flies distributed in 13 genera has been recorded, with Psychodopygus and Psathyromyia being the most abundant [20,24,27]. Meanwhile, in Amazonas, studies carried out mainly in the kilometers surrounding Leticia, Amacayacu National Natural Park and Puerto Nariño, have recorded 58 species belonging to 14 genera, the most representative being Lutzomyia, Psathyromyia and Psychodopygus [20,28–30]. These investigations have contributed to the knowledge on the distribution and diversity of sand flies, however, the role of the species in the transmission dynamics of the disease has yet to be addressed.
Entomological studies in the Caquetá department are limited to the year 2000, where species identification was conducted by comparing morphological characters [24]. Although with this approach it is possible to assign species, there are limitations associated with the presence of cryptic species mainly located within the genera Psychodopygus, Nyssomyia and Trichophoromyia. Among the limitations are the collection of individuals of a single sex, isomorphisms in female spermathecae, species complexes, the slow integration of newly described species into taxonomic keys and the use of ambiguous taxonomic characters that limit the correct differentiation of species [29]. For this reason, it is necessary to implement an approach that integrates classical and molecular taxonomy, referred to as “integrative taxonomy”, through the use of DNA barcoding and specifically the analysis of partial sequences of the Cytochrome Oxidase Subunit I (COI) gene that serves as a tool for the rapid and accurate identification and delimitation of phlebotomine species at different taxonomic levels and with different degrees of complexity [31–34].
Given the low number of sand flies studies recorded in the Colombian Amazon region and the current relevance of this region, which is highly susceptible to climate change and experiences high circulation of pathogens due to constant human and animal migration, it is crucial to provide new information to better understand the dynamics and risk of leishmaniasis transmission. In this context, the present study characterized the sand fly fauna associated with rural localities in Leticia, Amazonas, and Florencia, Caquetá, using an integrative taxonomy approach (taxonomical keys and COI as barcode), including the detection of Leishmania DNA, and the identification of blood ingestion sources through different molecular markers.
2. Materials and methods
2.1 Collection permits
The collection in the Amazonas and Caquetá departments was conducted under the Framework Permit for Collecting Specimens of Wild Species of Biological Diversity for Non-commercial Scientific Research Purposes, granted by the Autoridad Nacional de Licencias Ambientales (ANLA) to the Universidad Nacional de Colombia by Resolution No. 0255 of March 14, 2014 (article 3), in accordance with the Decree No. 1376, 2013 of the Ministerio de Ambiente y Desarrollo Sostenible, considering the mobility certificate 59453 and with prior consent of the landowners and indigenous community.
2.2 Study area
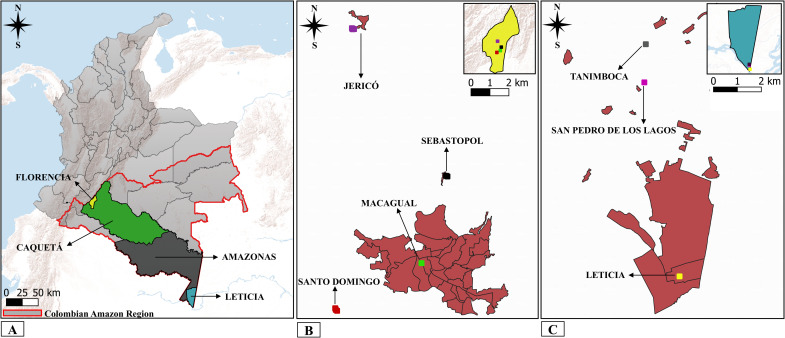
An entomological survey was conducted in San Pedro de los Lagos (S: -4.142304, W: -69.953815) and Tanimboca (S: -4.127948276, W: -69.95326736) localities of Leticia, Amazonas, and in Jericó (N:1.72855556, W: -75.64913), Sebastopol (N: 1.65894444, W: -75.605111), Santo Domingo (N: 1.59536111, W: -75.657417) localities and in the Amazonian Research Center-CIMAZ-Macagual (N: 1.61750000, W: -75.616750) of Florencia, Caquetá (Fig 1).
Fig 1. Geographic location of the study area and sand flies collection sites: (A) Departments of Amazonas and Caquetá, Colombia.
(B) Municipality of Florencia, locality Jericó, Macagual, Santo Domingo and Sebastopol. (C) Municipality of Leticia, locality Leticia, San Pedro de los Lagos and Tanimboca. Source of layers [39–42] and Qgis, v. 3.34.12 [43].
The study area is part of the Colombian Amazon region, characterized by the presence of tropical rainforest (Bh-T) [35]. The average annual temperature in the department of Amazonas ranges between 25.3°C and 25.7°C, the annual rainfall varies between 2,660 mm and 3,538 mm and the relative humidity is above 80% [36]. In the department of Caquetá, the average temperature ranges from 24.8°C to 25.9°C, the annual average precipitation varies between 4,385 mm and 2,483 mm and relative humidity is above 80% [36]. The most important productive activities in the region are livestock, agriculture, forestry, mining, ornamental fishing, and border trade [9,37].
Sand flies collection was conducted in secondary and primary forests. In the department of Amazonas, near the collection area, cassava and sugarcane crops, palm and fruit trees, domestic animals such as Canis lupus (dogs), Felis catus (cats) and Gallus gallus (hens) were observed. In the department of Caquetá, traps were placed in areas near vegetable gardens, arazá crops, cocoa, coffee, and banana plantations, all located close to natural water sources, and with presence of domestic animals such as Sus scrofa (domestic pig), C. lupus, F. catus, G. gallus, and Oryctolagus cuniculus (rabbits). However, in both departments, no evidence of animals associated with livestock was observed.
Sampling sites selection was made considering access conditions to the area, the history of leishmaniasis cases reported by health departments [38] and the existence of favorable microenvironments for sand flies development.
2.3 Sand flies collection and processing
During the year 2023, two collection campaigns were carried out, the first in August in Caquetá and the second in November in Amazonas. In each locality of both departments, 12 CDC (Centers for Disease Control and Prevention) light traps were installed in intra, peri and extradomiciliary areas, where they operated for 12 hours (18:00–6:00) each night, for three consecutive days. On the second night of collection, a Shannon trap was set up for one hour (18:00–19:00) at the point where the CDC traps recorded the highest abundance of sand flies on the first night. The collection was complemented with a search in potential resting sites, such as walls chicken coops, rabbit hutches and pig housing, using mouth and Prockopack [44] aspirators up for one hour (18:00–19:00). Each collection point was georeferenced with a GPSmap_60CSx, GARMIN. Specimens were stored and transported dry in 1.5 ml tubes, in racks with silica gel, and then to the laboratory of medical entomology of the Program for the Study and Control of Tropical Diseases (PECET), from the Universidad de Antioquia where all samples were stored at -20°C until processing.
Collected insects were fragmented for taxonomic identification. The head and the last three abdominal segments were mounted in Euparal medium (Carl Roth GmbH + Co. KG, Germany) for taxonomic allocation using the Galati classification [45], supported by the key of Young and Duncan [46] and bibliographical records of sand flies in the Colombian and Brazilian Amazon region. Some identified specimens were deposited in the Francisco Luis Gallego - MEFLG Entomological Museum of the Universidad Nacional de Colombia, at Medellín. Codes: 5089–65091, 65079–65081, 65082–65088, 65142–65146, 65147–65156, 65160–65162, 65157–65159, 65141, 65078. The use of generic and subgeneric abbreviations was conducted following Marcondes [47].
2.4 Molecular analyses: DNA extraction of sand flies
After sand flies identification by classical taxonomy, DNA extraction was performed from the thorax, legs, wings, and proximal abdominal segments of specimens using a high salt concentration protocol [48]. Individual abdomens from fed females were differentiated, separated and preserved in RNAlater until DNA extraction and blood source identification. Previous to extraction and in order to separate the tissue from the preservative salts, each specimen was transferred under sterile conditions to a new vial and washed with 70% ethanol for one minute at 2.000 revolutions per minute (rpm), then transferred to a new vial and washed with ultrapure water for one minute at 2.000 rpm, for finally transferred to the vial where DNA extraction would be performed.
The extraction process included mechanical lysis with pistil, thermal shock lysis, protein precipitation with proteinase K (Scientific Inc., Waltham, MA, USA), washing with alcohol at 70 and 96%, and final elution in TE Buffer (1X). DNA concentrations were determined using a NanoDrop spectrophotometer (IMPLEN, Germany) and stored at –20ºC for molecular identification, blood source and Leishmania detection.
2.4.1 PCR for phlebotomine barcoding.
The representation and total DNA of four females and four males of each phlebotomine species previously identified by classical taxonomy was considered, using partial amplification of the COI gene, using the primers LCO1490 5’-GGTCAACAAATCATAAAGATATTGG-3’ and HCO2198 5’-TAAACTTCA GGGTGACCAAAAAATC A-3’ [49], amplifying a product of approximately 710 base pairs (bps). PCR reaction contained 1X buffer, 0.25 mM MgCl2, 0.2 mM dNTPs, 0.4 μM of each oligonucleotide (forward and reverse), 1.5 U of Taq DNA polymerase (Excel taq, SMOBIO Technology, Hsinchu Science Park, Taiwan), 1mg/ml BSA (Scientific Inc., Waltham, MA, USA), 10–25 ng/μl of total DNA and ultrapure water until a final reaction volume of 25 μl was completed [31]. Ny. antunesi DNA was previously used in PCR standardization, whose identity was confirmed by sequencing, and which was negative for Leishmania infection and blood ingestion, was included positive control and ultrapure water as a negative control. The thermal profile consisted of an initial cycle of 1 min at 94°C, 1 min at 58°C and 2 min at 72°C followed by 35 cycles of 15 s at 94°C, 1 min at 58°C, 1.30 min at 72°C and finally a cycle of 15 s at 94°C, 1 min at 58°C, 7 min at 72°C [31].
2.4.2 Blood-meal source identification.
To increase the probability of detecting host DNA at different levels of preservation within the gut of females identified with blood traces, two molecular markers were used in independent PCR reactions. In the first reaction, the partial region of the Cytochrome B gene (Cytb) was amplified using the primers L14841 5’-AAAAAGCTTCCATCAACATC-3’ and H15148 5’-AAACTGCAGCCCCTCAGGATTTTGTTCCTC-3’ that amplify a fragment of approximately 305 bps [50]. In the second reaction, the 12S ribosomal gene was targeted with the primers L1085 5’-CCCAAACTGGGATTAGATACCC-3’ and H1259 5’-GTTTGCTGAAGATGGCGGTA-3’ amplifying a fragment of approximately 215 bps [51]. PCR reaction contained 1X buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.5 μM of each oligonucleotide (forward and reverse), 1.5 U of Taq DNA polymerase (Excel taq, SMOBIO Technology, Hsinchu Science Park, Taiwan) 1mg/ml BSA (Scientific Inc., Waltham, MA, USA), 10–25 ng/μl of total DNA and ultrapure water until a final reaction volume of 25 μl was completed. The thermal cycling conditions consisted of an initial cycle of 5 min at 95°C, 35 cycles of 30 s at 95°C, 15 s at 57°C, 30 s at 72°C and finally a 10 min cycle at 72°C [52]. DNA from Aedes aegypti fed with H. sapiens and C. lupus was used as a positive control and Ae. aegypti Rockefeller strain without blood meal ingestion, donated by the medical entomology laboratory of PECET, was also included as a negative control.
2.4.3 Molecular detection of Leishmania DNA.
The N-terminal region of the gene encoding the heat shock protein 70 (HSP-70) was amplified from the DNA of 37.9% of all females collected (with and without evidence of blood ingestion) to detect the presence of Leishmania DNA. These specimens were then organized into 168 samples, of which 41 contained 183 females grouped in pools of maximum six specimens, the remaining 127 samples belonged to individual females.
The protocol described by Montalvo et al. was used, with the primers F25 5’-GGACGCCGGCACGATTKCT-3’ and R617 5’-GAAGAAGTCCGATACGAGGGA-3’ amplifying a product of approximately 593 bps [53], L. (V.) braziliensis strain WA140 was donated by PECET and was used as a positive control, and Ny. antunesi DNA of wild type previously negative for parasite detection was used as a negative control.
PCR reaction contained 1X buffer, 1mM MgCl2, 0.2 mM dNTPs, 0.4 μM of each oligonucleotide (forward and reverse), 1 U of Taq DNA polymerase (Excel taq, SMOBIO Technology, Hsinchu Science Park, Taiwan), 1mg/ml BSA (Scientific Inc., Waltham, MA, USA), 10–40 ng/ μl of total DNA and ultrapure water until a final reaction volume of 25 μl was completed. The thermal cycling conditions consisted of an initial cycle of 1 min at 95°C, 35 cycles of 30 s at 95°C, 45 s at 61°C, 45 s at 72°C and finally a 10 min cycle at 72°C [53].
2.5 Sequence analysis PCR products
The PCR fragments were visualized with the EZ-Vision dye (Amresco, USA) on an agarose gel at 1.5%. 100 bp plus (Scientific Inc., Waltham, MA, USA) molecular weight marker was used, and the images were captured through the transilluminator Essential V6 (UVITEC-CAMBRIDGE). PCR products of the expected size were subjected to DNA sequencing in both directions using ABI PRISM 3700 DNA analyzer service of Applied Biosystem.
COI, 12S rRNA and Cytb sequences were edited and aligned using the Finch TV software V 4.0 (Geospiza, Inc.) and MEGA (Molecular Evolutionary Genetics Analysis) programs respectively [54] X v11. Sequence similarity analysis was performed by comparing sequences recorded in the GenBank database with the Basic Local Alignment Tool (BLASTN) [55].
The filtered reference sequences were downloaded, and multiple alignments were constructed with the sequences from this study using ClustalW with predetermined parameters. The phylogenetic analysis was performed by constructing dendrograms with the neighbor-joining (NJ) method with genetic distances per pair and a bootstrap value of 1000 replicates in the MEGA v11 software. Specifically in the COI marker sequences, the pair-based genetic distances for the maximum intra-specific and minimum inter-specific distances (nearest neighbor, NN) were generated using the Analysis tool with uncorrected distances (p) and the Kimura 2-parameter model (K2P) in the MEGA v11 software, while the number of haplotypes (h), polymorphic sites (s), and nucleotide diversity were determined using DnaSP 6.12.03 [56].
The graphs were made in the Linux environment. The sequence of Nemopalpus sp. (KM896679.1) was used as the external group for the COI dendrogram, while Bufo montfontamus (MK284968.1) was the external group selected for the blood intake dendrogram.
All sequences obtained in this study were submitted to GenBank with the following access codes: Cytochrome Oxidase Subunit I (COI): PQ783849 - PQ783902. Cytochrome B gene (Cytb): PQ963791 - PQ963793. Ribosomal 12S gene (12S): PQ895496 - PQ895500.
2.6 Analysis of interactions between blood-meal sources, Leishmania and sand flies species
The standardization of ranges in relation to the abundance of each species for each study site was calculated by the Standardized Index of Species Abundance (SISA), described by Roberts and Hsi [57]. The distribution and diversity of species was analyzed using the ANAFAU software, following the methodology described by Silveria et al. [58]. Through this analysis, species were classified into three categories: constant (species present in more than 50% of catches), incidental (present in 25–50% of catches) or accidental (present in less than 25% of catches). Shannon’s Diversity (H) and Equity (J) indices were used to estimate species richness and abundance uniformity. The alpha diversity indices were calculated in the PAST software (V 4.10). Diversity was calculated by the Simpson 1-D index, dominance by (Dominance_D) and equity was calculated by the Shannon_H index. The distribution of species was performed by a Principal Component Analysis (PCoA) with Bray-Curtis distances.
To estimate the infection rate in positive Leishmania samples, the minimum infection rate is the following formula: Minimal Infection Rate (MIR) = Number of infected sand flies/Total sand flies tested x 100. Finally, circus plot plots were used to show the interactions between sand flies species, blood intake source, Leishmania and their detection frequencies.
3. Results
3.1 Sand flies fauna recorded in the departments of Amazonas and Caquetá
A total of 1,104 sand flies were collected, with females comprising 68.4% (N = 755) and males 31.6% (N = 349). Of these, 191 specimens (17.3%) were collected in Amazonas and 913 (82.7%) in Caquetá. The specimens belonged to 30 species distributed across 12 genera. The most species-rich genera were Psychodopygus (7 species), Nyssomyia (4 species), Trichophoromyia (3 species) and Evandromyia (3 species) (Table 1).
Table 1. Taxonomic inventory of sand flies, sex and feeding status of the females collected in the departments of Amazonas (Tanimboca and San Pedro de los Lagos localities) and Caquetá (Sebastopol, Jericó, Macagual, and Santo Domingo localities).
| Department | Caquetá | Amazonas | N (%) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Municipality | Caraño | Santo Domingo | San Martín | Leticia | ||||||||||||||||||||
| Locality | Sebastopol | Jericó | Santo Domingo | Macagual | Tanimboca | San Pedro de los Lagos | ||||||||||||||||||
| Genus | Species | ♀ | ♂ | ♀ | ♀ˆ | ♂ | ♀ | ♀ˆ | ♂ | ♀ | ♀ˆ | ♂ | ♀ | ♀ˆ | ♀ | ♀ˆ | ♂ | |||||||
| Brumptomyia | 1 | mesai ** | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | 1 | (0.1) | ||||
| Evandromyia | 2 | (Eva.) georgii ** | - | - | - | 2 | - | - | - | - | - | - | - | - | - | - | - | - | 2 | (0.2) | ||||
| 3 | (Eva.) saulensis | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | - | - | 1 | (0.1) | |||||
| 4 | (Ald.) walkeri | - | - | 4 | 9 | 73 | - | - | 1 | 1 | - | - | - | - | 17 | 7 | 21 | 133 | (12.1) | |||||
| Nyssomyia | 5 | antunesi + | - | - | - | - | - | 4 | 1 | 13 | 101 | 1 | - | - | - | 1 | - | 1 | 122 | (11.1) | ||||
| 6 | fraihai ** | 8 | - | 28 | 4 | - | 40 | 1 | 7 | 17 | - | - | - | - | 14 | 1 | - | 120 | (10.9) | |||||
| 7 | umbratilis + | - | - | - | - | - | - | - | - | - | - | - | 1 | 1 | - | - | - | 2 | (0.2) | |||||
| 8 | yuilli pajoti ** | 1 | - | - | 2 | 1 | 17 | 8 | 8 | 166 | 1 | 1 | - | - | - | - | - | 205 | (18.6) | |||||
| Lutzomyia | 9 | (Tri.) sherlocki ** | - | - | 1 | - | - | - | - | 1 | - | - | - | - | - | - | - | 1 | 3 | (0.3) | ||||
| 10 | (Hel.) tortura ** + | - | - | - | - | - | 25 | - | 15 | - | - | - | - | - | - | - | - | 40 | (3.6) | |||||
| Micropygomyia | 11 | (Mic.) pilosa | - | - | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | 1 | (0.1) | ||||
| Psychodopygus | 12 | amazonensis + | - | 2 | - | - | - | - | - | - | - | - | - | 1 | - | 22 | - | 3 | 28 | (2.5) | ||||
| 13 | ayrozai + | - | - | - | - | - | 2 | 1 | 2 | - | - | - | - | 1 | 4 | - | 5 | 15 | (1.4) | |||||
| 14 | carrerai thula ** | - | - | - | - | - | 3 | - | 2 | - | - | - | - | - | - | - | - | 5 | (0.5) | |||||
| 15 | chagasi + | 11 | 13 | - | - | - | 2 | - | 5 | - | - | - | - | - | - | - | - | 31 | (2.8) | |||||
| 16 | davisi + | - | - | - | - | - | 5 | - | - | - | - | - | - | - | 12 | 1 | 6 | 24 | (2.2) | |||||
| 17 | paraensis + | - | - | - | - | 1 | 6 | - | 3 | - | - | - | - | - | 15 | 2 | 2 | 29 | (2.6) | |||||
| 18 | panamensis + | - | - | 1 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | (0.1) | |||||
| Psathyromyia | 19 | (For.) aragaoi | 4 | 6 | - | - | - | - | - | - | - | - | - | - | - | 3 | - | 3 | 16 | (1.5) | ||||
| 20 | (Psa.) dendrophyla | - | - | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | 1 | (0.1) | |||||
| Pintomyia | 21 | (Pif.) serrana ** | - | - | - | - | - | 1 | - | 1 | - | - | - | - | - | - | - | - | 2 | (0.2) | ||||
| 22 | (Pif.) nevesi | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - | - | 1 | (0.1) | |||||
| Sciopemyia | 23 | fluviatilis * ° | - | - | - | - | - | - | - | - | - | - | - | 1 | - | 2 | - | - | 3 | (0.3) | ||||
| 24 | sordellii | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 1 | - | 2 | (0.2) | |||||
| Trichophoromyia | 25 | cellulana | - | - | 112 | 8 | 116 | - | - | - | - | - | - | - | - | - | - | - | 236 | (21.4) | ||||
| 26 | howardi ** | - | - | - | - | 2 | - | - | - | - | - | - | - | - | - | 5 | 7 | 14 | (1.3) | |||||
| 27 | velezbernali | - | - | - | - | - | - | - | - | - | - | - | - | - | 2 | - | - | 2 | (0.2) | |||||
| Trichopygomyia | 28 | witoto | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 15 | 15 | (1.4) | ||||
| Viannamyia | 29 | caprina * | - | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | - | 1 | (0.1) | ||||
| 30 | tuberculata *+ | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | - | - | 1 | (0.1) | |||||
| Lutzomyia | 31 | sp. | 3 | - | 11 | 1 | 3 | 8 | - | 3 | 8 | 1 | - | 1 | - | 5 | - | 3 | 47 | (4.3) | ||||
| Total for sex | 27 | 21 | 157 | 26 | 197 | 114 | 11 | 63 | 293 | 3 | 1 | 5 | 2 | 100 | 17 | 67 | 1104 | |||||||
| Total for locality | 48 | 380 | 188 | 297 | 7 | 184 | ||||||||||||||||||
| Total for department | 913 | 191 | ||||||||||||||||||||||
*New record for the department of Amazonas, ** New record for the department of Caquetá, ° New record for Colombia, + Species of epidemiological relevance, N (%): Relative abundance of captured sand flies, ♀: Female, ♀ˆ : Fed females, ♂: Male.
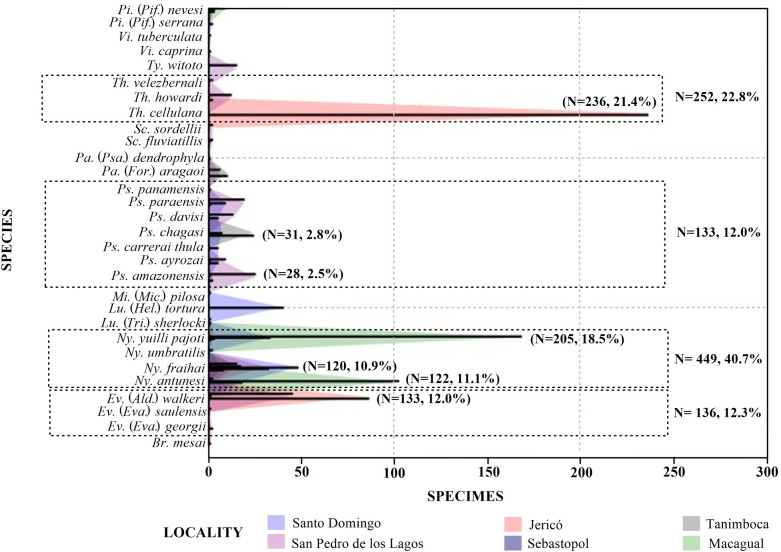
When considering the relative abundances between the two departments, Nyssomyia (n = 449, 40.7%), Trichophoromyia (n = 252, 22.8%), Evandromyia (n = 136, 12.3%) and Psychodophygus (n = 133, 12.0%) represented the most abundant genera (Fig 2), while at species level Th. cellulana (n = 236, 21.4%), Ny. yuilli pajoti (n = 205, 18.5%), Ev. (Ald.) walkeri (n = 133, 12.0%), Ny. antunesi (n = 122, 11.0%) and Ny. fraihai (n = 120, 10.8%) were the most abundant. Specifically in Amazon, Ev. (Ald.) walkeri (n = 45, 23.6%) and Ps. amazonensis (n = 26, 13.6%) were the most abundant species, while in Caquetá it was Th. cellulana (n = 236, 25.9%), Ny. yuilli pajoti (n = 205, 22.5%) and Ny. antunesi (n = 120, 13.3%).
Fig 2. Relative abundances of sand flies species collected in the departments of Amazonas (Tanimboca and San Pedro de los Lagos) and Caquetá (Sebastopol, Jericó, Macagual and Santo Domingo).
Dotted line: most abundant genera, N: Total specimens by species and by genus, %: Percentage abundance by species and by genus.
3.2 Ecological descriptors of sand flies captured
The analysis of the standardized index of species abundance (SISA) by sampling area shows that Ny. umbratilis (SISA = 1; 28.6%) was the most abundant species associated with the Tanimboca locality, while in San Pedro de los Lagos Ev. (Ald.) walkeri (SISA = 0.4; 24.4%) and Ps. amazonensis (SISA = 0.9; 13.5%) were the most abundant species. Conversely, Ev. (Eva.) saulensis, Lu. (Trl.) sherlocki, Ny. antunesi, Sciopemyia fluviatilis, Sc. sordellii, Viannamyia tuberculata, and Th. velezbernali were classified as accidental species (Chao-1 = 16.6) associated with the department of Amazonas (S1 Table). Regarding Caquetá, the most abundant species in Jericó was Th. cellulana (SISA = 1; 62.1%), followed by Ev. (Ald.) walkeri (SISA = 0.88; 22.6%). In Sebastopol, Ps. chagasi (SISA = 0.96; 55.3%) and Pa. (For.) aragaoi (SISA = 0.85; 20.8%) predominated.
Ny. fraihai (SISA = 0.7; 25.5%) was the most abundant in Santo Domingo, followed by Lu. (Hel.) tortura (SISA = 1; 21.3%) while Ny. yuilli pajoti (SISA = 0.98; 56.5%) and Ny. antunesi (SISA = 0.98; 34.3%) were predominated in San Martín; Ev. (Ald.) walkeri, Lu. (Trl.) sherlocki, Micropygomyia (Mic.) pilosa, Ps. davisi, Pa. (Psa.) dendrophyla and Ps. paraensis were classified as accidental and sporadic species (Chao-1 = 20) (S1 Table).
Alpha diversity indices evidenced that species richness varied considerably between locations. Santo Domingo, and San Pedro de los Lagos exhibited the highest richness with 15 and 16 species respectively, represent the localities with the greatest diversity (Simpson 1-D: 0.8235 and 0.8676), lowest dominance (Dominance_D: 0.1765 and 0.1324), and have an equitable distribution of species (Shannon_H: 2.014 and 2.268). Jericó and Macagual presented the highest dominance values (Dominance_D of 0.4814 and 0.4692), the lowest diversity indices (Simpson 1-D: 0.5186 and 0.5308) and the lowest evenness (Shannon_H: 0.9971 and 0.8676) (S1 Fig).
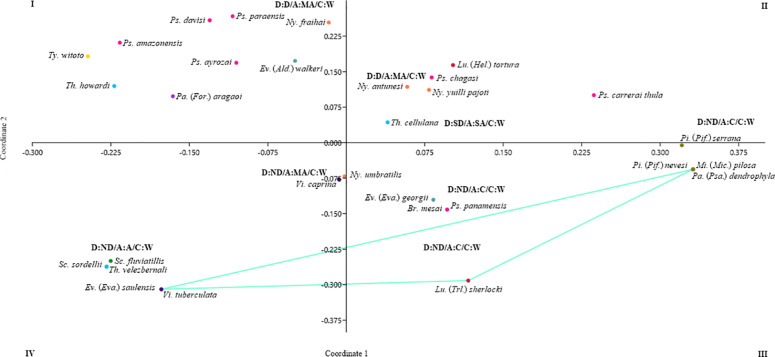
The PcoA analysis with Bray-Curtis distances exhibits clear differences in species composition between localities (Fig 3). In plane I species are grouped based on ecological indices of low dominance and abundance (D: ND-D; A:C-MA). These species are similar in terms of their distribution and abundance, indicating a balance in the community. Plan II mainly represents the most abundant and dominant species. In some sites, one or more species predominated within the community, such as Th. cellulana in Jericó Caquetá, which could influence community dynamics, affecting resource availability and interactions among species.
Fig 3. Principal component analysis (PCoA) indicates the distribution of sand flies species from Bray-Curtis similarity and standardized species abundance index (SISA).
SD: super dominant, D: dominant, ND: non-dominant; SA: super abundant, MA: Very abundant, C: common, A: Accidental, W: Constant.
Conversely, planes III and IV include non-dominant species, characterized by their rarity and low specimen counts. Their low frequency may be associated with unfavorable environmental conditions or limited interactions with dominant species, which could contribute to their displacement.
3.3 Sequence analysis, genetic diversity, and phylogenetic information
A total of 97 sand flies sequences were obtained, representing 12 genera and 30 species from the surveyed localities in the Colombian Amazon. For most species, between three and nine specimens were included in the analysis. However, in cases of low abundance, only one or two specimens were analyzed. Additionally, 16 new COI sequences were obtained for the species Lu. (Hel.) tortura (6), Th. cellulana (6), Trichopygomyia witoto (3) and Vi. caprina (1).
After manually editing, the consensus sequences fluctuated from 550 to 675 bps. The fragment was aligned between positions 17 and 669 of the 5’ segment of the mitochondrial COI gene of Ae. aegypti (OM214532.1) used as a reference genome, a region that coincides with that proposed by Hebert et al. as a barcode for species identification [59]. Visual inspection of alignment indicated the absence of termination codons in the medium of sequences suggesting the presence of pseudogenes or nuclear copies of mitochondrial origin NUMTs. The similarity percentage of between the sequences obtained when compared with the COI sequences of neotropical sand flies available at GenBank ranged from 93 to 100%.
During the nucleotide alignment including only the sand flies identified in our study, 442 preserved sites and 134 polymorphic sites were identified, of which 57 are sparingly informative. This is reflected by the nucleotide heterogeneity according to FINGERPRINT in Fig 4A. The A + T ratio (66.7%) was significantly higher than the G-C ratio (33.3%). We detected 64 haplotypes (S2 Table), from 97 sequences, indicating a high degree of total nucleotide diversity per site (π = 0.11596; Standard deviation of π = 0.00346).
Fig 4. Genetic divergence analysis from sand flies sequences collected in the departments of Amazonas and Caquetá.
(A) Nucleotide heterogeneity according to FINGERPRINT, (B) ABGD “barcode gap” K2P, (C) ABGD “barcode gap” uncorrected p distances. (D) Transitions/transversions rates versus genetic distance.
The number of haplotypes per species ranged from 1 to 9, the species with the greatest haplotype diversity (Hd) were Ps. davisi (N = 9; Hd: 0.9778), Ps. ayrozai (N = 5; Hd: 0.9333), Th. cellulana (N = 5; Hd: 0.593), Ps. chagasi (N = 4; Hd: 0.7143), and Lu. (Hel.) tortura (N = 4; Hd: 0.8000). In these species, the percentage of nucleotide diversity ranged from 0.1 to 2. 6% and 0.1 to 2.6% by the Pi and Pi (JC) models respectively (S2 Table). In contrast, Ev. (Eva.) georgii, Ps. paraensis and Ty. witoto presented unique haplotypes from different specimens (S2 Table).
The intraspecific p uncorrected and K2P genetic distances ranged from 0.0 to 2. 6% (average = 0.007; standard deviation = 0.002) and 0.0 to 2.6% (average = 0.007; standard deviation = 0.002), respectively (S2 Table). The species with the highest percentages of intraspecific distance were Pa. (For.) aragaoi (1. 2% p; 1. 2% K2P), Ps. ayrozai (1. 6% p; 1. 6% K2P), Ps. davisi (2.6% p; 2.6% K2P), Th. howardi (1.5% p; 1.5% K2P), and Ev. (Ald.) walkeri (1.3% p; 1.3% K2P). The minimum interspecific distance (NN) for each species ranged from 0.2% to 15.5% (p distances) and 0.2% to 16.6% (K2P distances) (S2 Table).
The genetic divergence analysis using the Automatic Barcode Gap Discovery ABGD method with the K2P model exhibited a marked “barcode gap” between the intra and interspecific distances of the sand flies collected in the Amazon, ranging from 0.25 to 0.75. This result suggests a clear separation between most of the species analyzed, as a delimiting criterion and indicator with an interspecific distance range from 0.075 to 0.175 (Fig 4B-4C). There is a slight overlap in the intraspecific distances (average 0.0042) associated with Th. cellulana, Th. howardi and Th. velezbernali which demonstrates limitations in the complete delimitation of these species and the need to use alternative markers with different replacement rate and inheritance. The transition/transversion rates versus genetic distance graph (Fig 4D) shows a grouping of transversions as genetic distance increases, and a pattern of substitutions that contributes to species differentiation, supporting interspecific separation.
3.4 Dendrogram constructed by the Neighbor-joining method (NJ), showing the phylogenetic relationship between species divided by genera
Intra-specific taxonomic association, that is clusters of sequences of the same species and redistribution and/or rearrangement of these clusters based on genetic divergence of different genera and/or other taxonomic levels of, in this case, Amazon sand flies, was obtained by constructing several dendrograms of NJ using 97 COI sequences from our study and 121 reference sequences available at GenBank. For a more detailed analysis, NJ was estimated by genera.
The NJ shows a correct specific clustering supported by high Bootstrap values, between 95 and 100%, for the genus Psychodopygus (Fig 5). The formation of two external clusters is evident. The first one formed by Ps. davisi that presented greater variability, along with the species Ps. amazonensis, Ps. panamensis, Ps. wellcomei, Ps. chagasi, and Ps. paraensis, while the second was formed by haplotypes of Ps. ayrozai (Fig 5).
Fig 5. Neighbor-joining gene tree and morphology of sand flies COI barcode sequences, with emphasis on Psychodopygus.
(A) NJ Dendrogram. (B-C) Male and female genitalia of Ps. davisi. (D-E) Male and female genitalia of Ps. chagasi. (F-G) Male and female genitalia of Ps. paraensis.
In Nyssomyia, lower Bootstrap values were also found, between 85 and 96%, and a main external cluster was formed including Ny. antunesi, Ny. yuilli pajoti and Ny. fraihai, while Ny. yuilli yuilli is shown in a different cluster (Fig 6).
Fig 6. Neighbor-joining gene tree and morphology of sand flies COI barcode sequences, with emphasis on Nyssomyia.
(A) NJ Dendrogram. (B-C) Male and female genitalia of Ny. antunesi. (D-E) Male and female genitalia of Ny. yuilli pajoti. (F-G) Male and female genitalia of Ny. fraihai.
The NJ dendrogram that integrated Evandromyia and Psathyromyia shows Bootstrap values of 100% for the grouping of all species (Fig 7), which demonstrates a clear consistency with the morphological allocation and low genetic variability of these species. Internally, a cluster was formed for Evandromyia (Bootstrap 81%) and two groups for Psathyromyia, one for Pa. (For.) aragaoi and another for Pa. (Psa.) dendrophyla, both with 100% support.
Fig 7. Neighbor-joining gene tree and morphology of sand flies COI barcode sequences, with emphasis on Evandromyia and Psathyromyia.
(A) NJ Dendrogram. (B) female genitalia of Ev. (Eva.) georgii. (C) female genitalia of Ev. (Eva.) saulensis. (D-E) male and female genitalia of Ev. (Ald.) walkeri. (F) male genitalia of Pa. (For.) aragaoi. (G) male genitalia of Pa. (Psa.) dendrophyla.
The dendrogram clustering species with morphologically indistinguishable females, shows the formation of two external clusters, the first of Trichophoromyia and the second Trichopygomyia represented only by Ty. witoto (Fig 8). Internally, the first cluster is divided into two groups. The first comprising Th. cellulana, Th. velezbernali sequences with high genetic variability (Bootstrap of 88%) while Th. howardi is grouped in the second with Bootstrap values of 87% (Fig 8).
Fig 8. Neighbor-joining gene tree and morphology of sand flies COI barcode sequences, with emphasis on Trichophoromyia and Trichopygomyia.
(A) NJ Dendrogram. (B-C) male and female genitalia of Th. cellulana. (D-E) male and female genitalia of Th. howardi. (F) male genitalia of Ty. witoto.
The dendrogram grouping the genera Lutzomyia and Sciopemyia (Fig 9) shows three outer clusters, all with Bootstrap support values of 100%. The first cluster comprises sequences of Lu. (Tri.) sherlocki, the second of Lu. (Hel.) tortura, and the third includes the Sciopemyia group, which is subdivided into three well-defined clusters: the first one corresponding to Sc. fluviatilis, the second to Sc. preclara, and the third to Sc. sordellii, each with Bootstrap supports of 100%.
Fig 9. Neighbor-joining gene tree and morphology of sand flies COI barcode sequences, with emphasis on Lutzomyia and Sciopemyia.
(A) NJ Dendrogram. (B-C) male and female genitalia of Lu. (Trl.) sherlocki. (D-E) male and female genitalia of Lu. (Hel.) tortura.
The NJ dendrogram with genera of low abundances (Fig 10) clusters well-defined species with Bootstrap values of 100%, with evident low genetic variability, demonstrating clear consistency with morphological allocation. Two external clusters were formed, the first comprising sequences of Pintomyia, Brumptomyia and Micropygomyia, while the second was formed by Vi. furcata and Vi. tuberculata.
Fig 10. Neighbor-joining gene tree and morphology of sand flies COI barcode sequences, with emphasis on genera of lesser abundance.
(B) male genitalia of Br. mesai. (C) male genitalia of Mi. (Mic.) pilosa. (D) male genitalia of Pi. (Pif.) serrana. (E) female genitalia of Vi. tuberculata. (F) female genitalia of Vi. caprina.
3.5 Identification of feeding sources
A total of 59 females out of the 755 collected presented visible traces of blood ingestion in the abdomen (7.8%). Ev. (Ald.) walkeri was the species with the highest number of females with evident blood intake (n = 17; 28.8%), followed by Ny. yuilli pajoti (n = 11; 18.6%), Th. cellulana (n = 8; 13.6%) and Ny. fraihai (n = 6; 10.2%). It was possible to detect the blood source in 12 of the 13 species analyzed, and the blood source was characterized in 39 females with the Cytb molecular marker (S3 Table) and 29 females (92.3%) with the marker rDNA 12S (S4 Table).
After manual editing, the size of the fragment sequenced with the 12S rDNA marker ranged from 150 to 155 bps, while the length of the Cytb sequences ranged from 280 to 345 bps. The identity percentage by using the two markers was greater than 98% with a coverage of 91–100%, except for the Bos taurus (cow) and Cheracebus lugens (white-chested Titi), with 85.9 and 82.0% of identity, respectively (S3-S4 Tables).
The dendrogram generated from the Cytb marker reveals four clearly defined clusters (Bootstrap support of 100%), which highlight the genetic relationships and associated blood sources. The first cluster groups sequences (n = 17, species = 7) corresponding to H. sapiens, confirming its role as a frequent source of blood. The second cluster, with a is related to Dasyprocta leporina (brazilian aguti) (n = 1, species = 1) and S. scrofa (n = 10, species = 3). Finally, the third cluster, composed of B. taurus (n = 1, species = 1) (Fig 11).
Fig 11. Neighbor-joining gene tree and blood food sources detected in sand flies collected in the Colombian Amazon region, using the molecular marker Cytochrome B (Cytb).
(A) Dendrogram NJ. (B) Vertebrate species identified.
The dendrogram grouping the vertebrates identified by the 12S marker reveals the formation of two main clusters, supported by Bootstrap values of 65% and 100%, respectively (Fig 12). The first cluster groups genetically close samples, such as S. scrofa and H. sapiens (n = 3, species = 2) with a Bootstrap value of 99%, evidencing genetic variability within each group. The second main cluster includes samples associated with H. sapiens (n = 11, species = 7) as well as C. lugens (n = 1, species = 1), Saimiri macrodon (ecuadorian squirrel monkey) (n = 1, species = 1) and F. catus (n = 1, species = 1).
Fig 12. Neighbor-joining gene tree and blood food sources detected in sand flies collected in the Colombian Amazon region, using the molecular marker 12S.
(A) Dendrogram NJ. (B) Vertebrate species identified.
H. sapiens was the most common blood source detected in 10 of the 12 species of sand flies analyzed, followed by S. scrofa detected in Ev. (Ald.) walkeri, Ny. fraihai, and Th. cellulana. DNA of B. taurus and F. catus was also detected in the latter two. In addition, mixed feeding (H. sapiens/D. leporina) was evidenced in a specimen of Pi. (Pif.) nevesi and S. scrofa/C. lugens in a specimen of Ev. (Ald.) walkeri, while S. macrodon was the only blood source detected in Ps. davisi (Fig 13).
Fig 13. Relationship between sources of blood ingestion and the detection of Leishmania DNA in sand flies from the department of Amazonas and Caquetá.
Ny.a: Ny. antunesi, Ny.u: Ny. umbratilis, Ny. yp: Ny. yuilli pajoti, Ev. w: Ev. (Ald.) walkeri, Ny. fr: Ny. fraihai, Th. ce: Th. cellulana, Pi. ne: Pi. (Pif.) nevesi, Sc. so: Sc. sordellii, Ps. da: Ps. davisi, Ps.pr: Ps. paraensis.
3.6 Detection of Leishmania
Leishmania DNA detection was performed in 37.9% (n = 310) (S2 Fig) of the total of females captured (n = 816), belonging to 28 species previously identified by integrative taxonomy. Using the HSP-70 N marker, Leishmania DNA was amplified in 28 samples from individual females with a minimum infection rate (MIR) of 9.0% (S5 Table).
The species with the highest Leishmania detection was Ny. fraihai (n = 8, 26%), collected in the peri and extra-domicile of all the localities sampled in Caquetá and in the extra-domiciliary of San Pedro de los Lagos (S5 Table). All Ny. fraihai specimens detected with DNA from S. scrofa and B. taurus collected in Caquetá were positive as well for Leishmania. Additionally, the DNA of the parasite was detected in a female sand flies of the same species classified as unfed and that was collected in the peridomicile areas of Sebastopol (Fig 13).
Th. cellulana was the second species with the highest detection rate (n = 4, 1.3%) (S5 Table). In these sand flies specimens collected in the extradomiciliar environment of Jericó, Leishmania DNA was detected in females with blood traces of H. sapiens as well as in those without evidence of blood feeding (Fig 13). Ny. yuilli pajoti presented the same rate as Th. cellulana (n = 4, 1.3%) (S5 Table). It was possible to detect Leishmania DNA in specimens collected from Santo Domingo that were also fed with H. sapiens (Fig 13). Ev. (Ald.) walkeri presented parasite detection rates lower than 0.9% in three specimens of this sand fly collected in the extradomicile of Jericó the presence of Leishmania was detected, one of them with blood meal associated with S. scrofa (S5 Table). Finally, Leishmania DNA was present in a single specimen of Pi. (Pif.) nevesi (n = 1, 0.3%) collected in the extra-domiciliary environment of Santo Domingo (S5 Table), which also exhibited a mixed feeding of D. leporina/H. sapiens (Fig 13).
4. Discussion
This study integrates morphological, molecular, and ecological approaches for the identification of sand flies in the Colombian Amazon, a region of high biodiversity and great vulnerability to the effects of climate change and human disturbances [60]. To the best of our knowledge, this work presents the first DNA barcoding (COI) sequences for sand flies from the department of Caquetá and the second study of this type in Amazonas [29]. Sixty-four new haplotypes were detected and sequences of Lu. (Hel.) tortura, Th. cellulana, Ty. witoto, and Vi. caprina not previously analyzed with this marker are provided. In addition, three new circulation records are presented for Amazonas, Vi. caprina, Vi. tuberculata and Sc. fluviatilis, nine Caquetá, Br. mesai, Ev. (Ald.) georgii, Ny. fraihai, Ny. yuilli pajoti, Lu. (Trl.) sherlocki, Lu. (Hel.) tortura, Ps. carrerai thula, Th. howardi and Pi. (Pif.) serrana, and one for Colombia, Sc. fluviatilis, as far as we know. Ten species of epidemiological relevance were identified, seven vertebrate species that act as a source of blood ingestion and the presence of Leishmania sp. was detected in areas where cases of tegumentary leishmaniasis have historically been recorded.
4.1 Sand flies and epidemiological relevance
In this study, Ev. (Ald.) walkeri and Ps. amazonensis were the most abundant species in the Amazonas department, representing 36.6% of the total number of specimens. Despite limited information on these species in the region, previous studies that use a similar collection approach evidenced that Ev. (Ald.) walkeri does not present high dominance [30]. This can be attributed to the ecotope selected for collection, the number of traps used, and the sampling effort time.
The department of Caquetá showed a greater richness of species, with 22 species found in the different localities, being a 18.2% higher when compared to the amount of species collected in the Amazonas department, which can be attributed to a greater sampling effort and a wider geographical range of collection in the first case. The results of species richness in this department are congruent with those recorded at Araracuara, Caquetá, during 1981 where Morales and Minter collected 35 species of sand flies, including six new records for the country and one new for science [27].
In Caquetá, Th. cellulana, Ny. yuilli pajoti and Ny. antunesi are the most abundant species, representing 61.4% of the total number of specimens caught. These results differ from previous studies in the same region, where collection during rainy seasons showed low abundance of these species [27,61,62].
Since the description of Th. cellulana in Caquetá in 1979 [62], it had not been recorded in this department until the present study. To date, its distribution is exclusively Amazonian, including regions of the Colombian Amazon (departments of Guaviare and Putumayo) [63,64], Peruvian and Ecuadorian [65,66], where the reported abundance does not exceed 3% of the total composition. Although the epidemiological importance of this species has not yet been explored, the growing interest in the ecoepidemiological aspects associated with the genus Trichophoromyia has led to consider several of its species as suspected vectors [67]. Such is the case of two of the five species recorded in Caquetá, Th. ubiquitalis, found with flagellates of L. (V.) lainsoni in Brazil [68], and detected with DNA from L. (V.) lainsoni, L. (L.) amazonensis and L. (V.) sp. in Brazil and Ecuador [69–71]. And Th. auraensis has been found infected with DNA from L. (V.) braziliensis, L. (V.) lainsoni and L. (V.) guyanensis in Peru and Brazil [72,73].
Ny. antunesi is another abundant species in Caquetá that exhibits highly anthropophilic behavior [74,75] and has been found during leishmaniasis outbreaks in the Meta department (Colombian eastern plains) with DNA of Leishmania not genotyped [76]. In Brazil, this species has been found infected with L. (V.) guyanensis [75], and is recognized as a vector of L. (L.) infantum and L. (V.) braziliensis [77]. Other sand fly like Ny. yuilli pajoti, are recognized to have the widest geographical distribution in Colombia with presence in Guaviare, Putumayo, Guainía, and Amazonas departments [20], has been registered as an anthropophilic species that is highly abundant in areas of intra, peri and extra-domiciliary in Putumayo [64], and has been collected frequently in rainy seasons and with low abundance in Amazonas and Guaviare department of Colombia [30,63].
During this study, a new geographical distribution of various sand flies species was found. Previous records of Vi. caprina have been associated with its distribution in the Colombian Amazon, specifically in the Guaviare department [63] but not in the Amazonas department as found in this study. This sand fly is not classified as a vector considering that it is not involved in the transmission cycle while its distribution in the country also includes the Andean and Pacific regions [20]; Vi. tuberculata previously documented in Colombia (Caquetá and Guaviare), as well as in the Andean and Pacific regions of Colombia, in addition to the Orinoco region [20], and in the Amazon region of Brazil where it was found to be naturally infected with L. (V.) utingensis [78], and Sc. fluviatilis, whose presence was unknown in the country until now, presents a jungle-like distribution behavior, with distributions recorded in French Guiana and several states of the Brazilian Amazon [45].
Although flagellate DNA has been detected in Sc. fluviatilis, it has not yet been considered as a probable vector of Leishmania [79]. Within Sciopemyia, blood traces of H. sapiens were also detected in specimens of Sc. sordellii. The anthropophilic habit had been previously documented [80], as well as the preference for other vertebrates, including C. lupus, G. gallus, S. scrofa, Bokermannohyla martinsi and Rattus rattus (black rat) [81–84] and the presence of DNA from L. (V.) guyanensis, L. (L.) infantum and L. (V.) braziliensis in specimens from different areas of Brazil [80,85,86]. These findings highlight the need to investigate the possible role of Sc. sordellii in the transmission of Leishmania in Amazonia, as well as to deepen the dimension of its feeding plasticity, key aspects for its survival and ecology.
Among the nine species reported for the first time in the department of Caquetá, two represent, to our knowledge, the first record for the Colombian Amazon region, Pi. (Pif.) serrana, an anthropophilic species previously recorded in the Andean, Caribbean, and Orinoco regions [20], is associated with an endemic zone of L. (V.) braziliensis in Norte de Santander [87], while other species like Ps. carrerai thula, previously recorded in the Pacific and Andean regions of the country, is not currently associated with Leishmania transmission [20,88].
In addition to the species previously recorded with epidemiological relevance due to their proven or suspected role in the transmission of Leishmania parasites, the presence of Ny. umbratilis, the main vector of L. (V.) guyanensis in Brazil [89], is noteworthy. In Colombia, no studies have examined its role as a vector, however, it is closely associated with L. (V.) guyanensis in the Amazonas [90]. Currently, there are no records in Colombia of Leishmania infection in Ny. fraihai, however, this species has been detected with trypanosomatids of the genus Endotrypanum in Porto Velho, Brazil [91]. In the case of Lu. (Tri.) sherlocki, captured in Caquetá and Amazonas in this study, there are records of this species in an endemic area for CL and visceral leishmaniasis (VL) in Pará, Brazil [92] and detected with Leishmania sp. DNA in a highly endemic area for leishmaniasis in the Peruvian Amazon [93]. Sand flies like Lu. (Hel.) tortura, have been reported in eight municipalities in the department of Putumayo and in Puerto Nariño locality, in the Amazonas department [30,64,94]. This species is highly anthropophilic, and similar to this study, it has been captured in the peri and extradomiciliary areas [64,95]. It has not been detected with Leishmania DNA in the Colombian territory, but in 2013 Kato and collaborators detected high rates of natural infection by L. (V.) naiffi suggesting its participation in the transmission cycle of this parasite in the Ecuadorian Amazon region [95].
The significance of finding sand flies like Ps. ayrozai, Ps. davisi, and Ps. amazonensis relies on the fact that they have been detected with L. (V.) naiffi in some regions of Brazil [96]. Specifically, Ps. ayrozai has also been detected with DNA from L. (L.) amazonensis, L. (V.) braziliensis, and L. (V.) guyanensis [80,97]. Ps. panamensis, associated with cutaneous leishmaniasis outbreaks, was recently classified as a level 4 vector [98], is highly anthropophilic, predominates in intra and peridomiciliary areas [99], and has been detected with DNA from L. (V.) panamensis [99] and L. (L.) amazonensis [100]. Ps. chagasi is of epidemiological relevance in Brazil and recently, DNA from L. (V.) braziliensis and L. (V.) lainsoni was detected in the municipality of Itapuã [97]. Although the role of Ev. (Ald.) walkeri in the transmission of leishmaniasis in Colombia has not been conclusively demonstrated, DNA from L. (V.) braziliensis [13] and L. (V.) guyanensis [75] has been detected in Acre state, Brazil.
4.2 Barcoding, a tool for species identification sand flies
Genetic analysis of the COI gene made it possible to differentiate species where the minimal macrometric differences of the males, associated with their parameres, require considerable expertise for their identification, as is the case with Ny. yuilli pajoti and Ny. fraihai. Improvements in the Barcode library allowed the consolidation of taxonomy within the genera Trichophoromyia and Trichopygomyia, facilitating the differentiation of species with isomorphic females, mainly Th. cellulana, Th. howardi, and Ty. witoto. The use of COI to differentiate species is justified by clear intraspecific (2.6%) and interspecific (16.6%) genetic distances. In some species with a considerable degree of morphological differentiation, as in the case of genera like Nyssomyia, Evandromyia and Psychodopygus may indicate the presence of a “barcode gap” within species evidenced by the clear separation into two groups in the NJ.
However, when species are closely related, the analysis of genetic distances may imply an overlapping of data, preventing the recognition of a clear gap, as occurred in this study with Th. cellulana and Th. velezbernali. This is consistent with the evidence reported for species of the genus Trichophoromyia collected in the department of Amazonas [29]. The limitation of barcoding to discriminate sand flies species has been documented in several cases, especially when genetic distances overlap due to a possible relatively recent divergence as reported in Ev. (Ald.) carmelinoi, Ev. (Ald.) evandroi and Ev. (Ald.) lenti [101] or Ps. complexus, and Ps. wellcomei [102]. This type of integrative taxonomy studies can be complemented with the use of multilocus tools that include nuclear markers such as the Second Inward Transcribed Spacer (ITS2) or Elongation Factor alpha 1 (Ef1-α) [103], in addition to other markers such as cytochrome b (Cytb) and the 28S ribosomal RNA gene [104] that can be essential for the complete delimitation of taxa, especially for those closely related [105,106].
4.3 Hosts and Leishmania in the Amazon region
Despite the challenges posed by the disturbance of the Colombian Amazon rainforest, such as population growth, deforestation, and environmental changes due to climate change [60], rural areas still favor the conservation of biodiversity and the richness of phlebotomines, whose wild behavior continues to shape their distribution and feeding habits. Among the vertebrate species that are considered as sand flies blood meal sources found in the Colombian Amazon localities analyzed, H. sapiens was identified as a blood source of Ny. antunesi, which is congruent with previous records documenting its anthropophilic behavior [16], while there are also reports of its affinity for other mammals such as B. taurus, Choloepus didactylus (sloth), D. leporina, S. scrofa, and T. tetradactyla and birds such as Pteroglossus aracari (toucan) [16]. H. sapiens was also the only blood meal source associated with Ps. panamensis in this study, however other investigations have found a variety of blood traces in this sand fly, including Marmosa robinsoni (robinson’s mouse) [107] and G. gallus [19,107]. In addition, this species has been reported with mixed feedings of H. sapiens/Equus asinus, H. sapiens/B. taurus, B. taurus/C. familiaris, and S. scrofa/H. sapiens [108,109].
The anthropophilic behavior of Ev. (Ald.) walkeri collected in the two departments studied was evidenced by the detection of blood traces of H. sapiens. Traces of C. lugens and S. scrofa were also identified in this species; previously in Brazil it was found with blood traces of G. gallus [13] and S. scrofa [110], while the presence of Leishmania sp. in the specimens collected from Caquetá is not strange, as there are records of natural Leishmania sp. infection in Peru [93] and L. (V.) braziliensis infection in Brazil [13]. Although information on the ecology of Th. cellulana is scarce, this study identified the presence of DNA from H. sapiens, S. scrofa and F. catus, suggesting anthropophilic and eclectic behavior. In particular, the presence of F. catus DNA is relevant, as these felids have been reported naturally infected and categorized as active carriers of L. (L.) infantum in several regions of Brazil, where the transmissibility of the parasite to the vector Lu. (Lut.) longipalpis has categorized them as key players in the VL cycle [111,112]. In addition, Leishmania sp. DNA was detected, reinforcing the connection between sources of ingestion and possible infection. Although not previously recorded in this species, this finding underscores the importance of focusing future research on ecoepidemiological aspects that will provide accurate information on the possible role of Th. cellulana in Leishmania transmission.
The presence of Leishmania in Pi. (Pif.) nevesi is consistent with previous studies that identified L. (V.) braziliensis as the pathogen involved in the infection [13]. Although blood traces of H. sapiens and G. gallus have been detected previously in this species [13,80], this study highlights the ability of this species to feed even on different blood sources. Blood traces found in Ny. umbratilis, a recognized vector of L. (V.) guyanensis in several countries including Brazil [113], corresponded with H. sapiens, demonstrating its anthropophilic behavior. Although no Leishmania DNA was detected in this study, previously, parasites of L. (V.) guyanensis were isolated from females collected in the department of Amazonas [98], In addition, Ny. umbratilis, has been detected with DNA from Endotrypanum sp. [80] and with blood traces of Plecturocebus bernhardi and S. scrofa mainly in the Brazilian Amazon [110].
Ps. davisi previously detected with DNA from L. (V.) braziliensis [13] and Endotrypanum sp. [80], was not found positive with parasite DNA in this study, although its blood source preferences include Pecari tajacu, S. scrofa and T. tetradactyla as evidenced in other studies [110], in this research only blood traces from H. sapiens were found. The finding of DNA of Leishmania not genotyped in understudied species such as Ny. yuilli pajoti and Ny. fraihai provides valuable information on their ecology, highlighting their anthropophilic habits and their interaction with humans in peri and extra-domiciliary areas. The selective preference of some sand flies species for certain hosts, such as S. scrofa in the Jericó locality, raises questions about their role in the dynamics of the pathogen, especially when DNA of the parasite was detected in sand flies associated with this vertebrate.
Although in this study it was not possible to perform consistent sequencing of the Leishmania amplicons, possibly due to DNA degradation or low concentration. Future studies contemplate the use of more sensitive techniques, such as ITS1–2 markers, nested PCR, qPCR and NGS, for accurate characterization of circulating species.
5. Conclusions
The results presented provide a solid information for future research on how the tripartite interactions between sand flies, hosts, and pathogens including the emergence and increased abundance of opportunistic hematophagous species (S3 Fig). Furthermore, these findings underscore the urgent need to protect remaining habitats and mitigate anthropogenic impacts that threaten to significantly alter the ecological dynamics of these areas.
In conclusion, the tripartite relationship among insects, hosts, and Leishmania is crucial for understanding transmission dynamics, especially in poorly studied areas of the Amazon with a high prevalence of leishmaniasis. These findings provide valuable information on zoonotic cycles in the region. The identification of new blood sources suggests that some animals may be underestimated as potential reservoirs of Leishmania (Fig 14). This study reinforces the relevance of the “One Health” approach, which integrates human, animal, and environmental health, emphasizing the need to monitor sand flies while preserving biodiversity.
Fig 14. The ecological dynamics of the tripartite relationship among sand flies, blood meal sources and Leishmania in forest and peridomicialiary areas in the Amazon region from Colombia.
Supporting information
(TIF)
(TIF)
(TIF)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We gratefully acknowledge the communities of the different localities, to Lina Marcela Manjarrez and her team of assistants trained in community work of the Health of Florencia, Caquetá, to Dr. Luz Mila Murcia Montaño, from the Amazonas Government and the Amazonas Public Health Study Group (GESPA), to Biologist and Master’s student Jennifer Danitza Viafara and Biological Engineer, to Master’s student Alejandro Castañeda M, for their invaluable support during sand flies collections, and the access to their laboratories. We express our special gratitude to the Program for the Programa de Estudio y Control de Enfermedades Tropicales (PECET), to its director Dr. Sara M. Robledo and Dr. Laura Cristina Posada-López for providing the space for the identification of sand flies, as well as for the donation of insects from the Ae. aegypti colony Rockefeller and Leishmania strains used as controls in this study.
Data Availability
Sequence data associated with the COI marker is available in the GenBank database as follows: PQ783849, PQ783850, PQ783851, PQ783852, PQ783853, PQ783854, PQ783855, PQ783856, PQ783857, PQ783858, PQ783859, PQ783860, PQ783861, PQ783862, PQ783863, PQ783864, PQ783865, PQ783866, PQ783867, PQ783868, PQ783869, PQ783870, PQ783871, PQ783872, PQ783873, PQ783874, PQ783875, PQ783876, PQ783877, PQ783878, PQ783879, PQ783880, PQ783881, PQ783882, PQ783883, PQ783884, PQ783885, PQ783886, PQ783887, PQ783888, PQ783889, PQ783890, PQ783891, PQ783892, PQ783893, PQ783894, PQ783895, PQ783896, PQ783897, PQ783898, PQ783899, PQ783900, PQ783901, PQ783902. Sequence data associated with the Cytb marker is available in the GenBank database as follows: PQ963791, PQ963792, PQ963793 Sequence data associated with the 12S marker is available in the GenBank database as follows: PQ895496, PQ895497, PQ895498, PQ895499, PQ895500.
Funding Statement
This study received financial support from project Hermes 57545 of the Universidad Nacional de Colombia, "Alianza estratégica interdisciplinaria Leticia, Medellín y La Paz para el estudio del microbioma de insectos vectores de enfermedades tropicales y su relación con el cambio climático y la sociedad", Convocatoria Nacional para el Fomento de Alianzas Estratégicas interdisciplinarias que articulen los procesos misionales de la Universidad Nacional de Colombia 2022–2024, awarded to CXMH and Scholarship Program of Ministerio de Ciencia, Tecnología e Innovación, Call 15, for Human Capital Development in the context of the Bicentennial and the 2021–2022 Biennial Plan, awarded to KCT. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Instituto Nacional de Salud (INS). Public health surveillance protocol: Leishmaniasis. Version 6. Bogotá; 2023 May. 10.33610/IMYH4569 [DOI]
- 2.World Health Organization (WHO). Leishmaniasis. 2020. Available: https://www.paho.org/en/topics/leishmaniasis
- 3.Instituto Nacional de Salud (INS). Weekly epidemiological report. Bogotá; 2024 Nov. Available: https://www.ins.gov.co/buscador-eventos/Paginas/Vista-Boletin-Epidemilogico.aspx
- 4.Instituto Nacional de Salud (INS). Events of interest in public health. Bogotá; 2023. Available: https://portalsivigila.ins.gov.co/Paginas/Buscador.aspx
- 5.Rodrigues MG de A, Sousa JD de B, Dias ÁLB, Monteiro WM, Sampaio V de S. The role of deforestation on American cutaneous leishmaniasis incidence: spatial-temporal distribution, environmental and socioeconomic factors associated in the Brazilian Amazon. Trop Med Int Health. 2019;24(3):348–55. doi: 10.1111/tmi.13196 [DOI] [PubMed] [Google Scholar]
- 6.Portella TP, Sudbrack V, Coutinho RM, Prado PI, Kraenkel RA. Bayesian spatio-temporal modeling to assess the effect of land-use changes on the incidence of Cutaneous Leishmaniasis in the Brazilian Amazon. Sci Total Environ. 2024;953:176064. doi: 10.1016/j.scitotenv.2024.176064 [DOI] [PubMed] [Google Scholar]
- 7.Moreno M, Guzmán-Rodríguez L, Valderrama-Ardila C, Alexander N, Ocampo CB. Land use in relation to composition and abundance of phlebotomines (Diptera: Psychodidae) in five foci of domiciliary transmission of cutaneous leishmaniasis in the Andean region of Colombia. Acta Trop. 2020;203:105315. doi: 10.1016/j.actatropica.2019.105315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellwanger JH, Kulmann-Leal B, Kaminski VL, Valverde-Villegas JM, Veiga ABGD, Spilki FR, et al. Beyond diversity loss and climate change: Impacts of Amazon deforestation on infectious diseases and public health. An Acad Bras Cienc. 2020;92(1):e20191375. doi: 10.1590/0001-3765202020191375 [DOI] [PubMed] [Google Scholar]
- 9.INDEPAZ I for D and PS. Livestock farming and deforestation in the Amazon. Bogotá; 2024 Feb. Available: https://indepaz.org.co/ganaderia-y-deforestacion-en-la-amazonia/
- 10.Tidman R, Abela-Ridder B, de Castañeda RR. The impact of climate change on neglected tropical diseases: a systematic review. Trans R Soc Trop Med Hyg. 2021;115(2):147–68. doi: 10.1093/trstmh/traa192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García Leal YJ, Carrero Sarmiento DA, Hoyos López RO. Diversidad Del Género Lutzomyia (Diptera: Psychodidae) En Municipios Del Departamento De Córdoba – Colombia. Acta biol Colomb. 2022;27(3). doi: 10.15446/abc.v27n3.90684 [DOI] [Google Scholar]
- 12.do Socorro Carvalho Miranda C, Costa de Souza B, Carvalho Garcia Miranda Filgueiras T, de Sousa AM, Cibelle da Silva Peixoto M, Carvalho Garcia Miranda Filgueiras T, et al. Visceral Leishmaniasis and Land Use and Cover in the Carajás Integration Region, Eastern Amazon, Brazil. Trop Med Infect Dis. 2022;7(10):255. doi: 10.3390/tropicalmed7100255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ávila MM, Brilhante AF, de Souza CF, Bevilacqua PD, Galati EAB, Brazil RP. Ecology, feeding and natural infection by Leishmania spp. of phlebotomine sand flies in an area of high incidence of American tegumentary leishmaniasis in the municipality of Rio Branco, Acre, Brazil. Parasit Vectors. 2018;11(1):64. doi: 10.1186/s13071-018-2641-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulloa GM, Vásquez-Achaya F, Gomes C, Del Valle LJ, Ruiz J, Pons MJ, et al. Molecular Detection of Bartonella bacilliformis in Lutzomyia maranonensis in Cajamarca, Peru: A New Potential Vector of Carrion’s Disease in Peru?. Am J Trop Med Hyg. 2018;99(5):1229–33. doi: 10.4269/ajtmh.18-0520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.da Silva MS, Picelli AM, Pereira de França K, Galati EAB, Andrade Filho JD, Julião GR, et al. Entomological inferences highlight the risk of Leishmania transmission in the urban area of Porto Velho, Rondônia, Brazil. PLoS One. 2024;19(8):e0309168. doi: 10.1371/journal.pone.0309168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pimentel AC, Sánchez Uzcátegui YDV, de Lima ACS, Silveira FT, Vasconcelos Dos Santos T, Ishikawa EAY. Blood Feeding Sources of Nyssomyia antunesi (Diptera: Psychodidae): A Suspected Vector of Leishmania (Kinetoplastida: Trypanosomatidae) in the Brazilian Amazon. J Med Entomol. 2022;59(5):1847–52. doi: 10.1093/jme/tjac108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roque ALR, Jansen AM. Wild and synanthropic reservoirs of Leishmania species in the Americas. Int J Parasitol Parasites Wildl. 2014;3(3):251–62. doi: 10.1016/j.ijppaw.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandoval-Ramírez CM, Hernández C, Teherán AA, Gutierrez-Marin R, Martínez-Vega RA, Morales D, et al. Complex ecological interactions across a focus of cutaneous leishmaniasis in Eastern Colombia: novel description of Leishmania species, hosts and phlebotomine fauna. R Soc Open Sci. 2020;7(7):200266. doi: 10.1098/rsos.200266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Posada-López L, Velez-Mira A, Cantillo O, Castillo-Castañeda A, Ramírez JD, Galati EAB, et al. Ecological interactions of sand flies, hosts, and Leishmania panamensis in an endemic area of cutaneous leishmaniasis in Colombia. PLoS Negl Trop Dis. 2023;17(5):e0011316. doi: 10.1371/journal.pntd.0011316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bejarano EE, Estrada LG. Family Psychodidae. Zootaxa. 2016;4122(1):187–238. doi: 10.11646/zootaxa.4122.1.20 [DOI] [PubMed] [Google Scholar]
- 21.Scarpassa VM, Cunha-Machado AS, Alencar RB. Multiple evolutionary lineages for the main vector of Leishmania guyanensis, Lutzomyia umbratilis (Diptera: Psychodidae), in the Brazilian Amazon. Sci Rep. 2021;11(1):15323. doi: 10.1038/s41598-021-93072-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vásquez-Trujillo A, Santamaría-Herreño E, González-Reina AE, Buitrago-Alvarez LS, Góngora-Orjuela A, Cabrera-Quintero OL. Lutzomyia antunesi as suspected vector of cutaneous leishmaniasis in the Orinoquian region of Colombia. Rev Salud Publica (Bogota). 2008;10(4):625–32. doi: 10.1590/s0124-00642008000400012 [DOI] [PubMed] [Google Scholar]
- 23.Azpurua J, De La Cruz D, Valderama A, Windsor D. Lutzomyia sand fly diversity and rates of infection by Wolbachia and an exotic Leishmania species on Barro Colorado Island, Panama. PLoS Negl Trop Dis. 2010;4(3):e627. doi: 10.1371/journal.pntd.0000627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molina JA, Hildebrand P, Olano VA, Muñoz de Hoyos P, Barreto M, Guhl F. Fauna de insectos hematófagos del sur del Parque Natural Nacional Chiribiquete, Caquetá, Colombia. Biomedica. 2000;20(4):314. doi: 10.7705/biomedica.v20i4.1075 [DOI] [Google Scholar]
- 25.Martínez-Pérez L. Frecuencia de uso de animales domésticos como fuente de alimentación de Lutzomyia spp. (Diptera: Psychodidae) en la vereda Toro, San Cayetano, Bolívar. Universidad de Sucre 2017. 2018. Available: http://repositorio.unisucre.edu.co/handle/001/624 [Google Scholar]
- 26.Ferro C, Marín D, Góngora R, Carrasquilla MC, Trujillo JE, Rueda NK, et al. Phlebotomine vector ecology in the domestic transmission of American cutaneous leishmaniasis in Chaparral, Colombia. Am J Trop Med Hyg. 2011;85(5):847–56. doi: 10.4269/ajtmh.2011.10-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morales A, Minter DM. Estudio sobre Flebotomineos en Araracuara Caquetá, Colombia S.A Incluyendo la descripción de Lutzomyia Araracuarensis (Diptera, Psychodidae). biomedica. 1981;1(3):94. doi: 10.7705/biomedica.v1i3.1790 [DOI] [Google Scholar]
- 28.Posada-López L, Galvis-Ovallos F, Galati EAB. Description of Trichophoromyia velezbernali, a New Sand Fly Species (Diptera: Psychodidae: Phlebotominae) from Colombian Amazonia. J Med Entomol. 2018;55(1):122–7. doi: 10.1093/jme/tjx180 [DOI] [PubMed] [Google Scholar]
- 29.Posada-López L, Rodrigues BL, Velez ID, Uribe S. Improving the COI DNA barcoding library for Neotropical phlebotomine sand flies (Diptera: Psychodidae). Parasit Vectors. 2023;16(1):198. doi: 10.1186/s13071-023-05807-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posada-López L. Inventario de especies y diversidad haplotípica MtCOI de Phlebotominae (Diptera: Psychodidae) en zonas de importancia para la transmisión de leishmaniasis en Colombia. Facultad de Ciencias Escuela de Biociencias, Universidad Nacional de Colombia Sede Medellín. 2016. Available: https://repositorio.unal.edu.co/handle/unal/58710 [Google Scholar]
- 31.Contreras Gutiérrez MA, Vivero RJ, Vélez ID, Porter CH, Uribe S. DNA barcoding for the identification of sand fly species (Diptera, Psychodidae, Phlebotominae) in Colombia. PLoS One. 2014;9(1):e85496. doi: 10.1371/journal.pone.0085496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodrigues BL, Galati EAB. Molecular taxonomy of phlebotomine sand flies (Diptera, Psychodidae) with emphasis on DNA barcoding: A review. Acta Trop. 2023;238:106778. doi: 10.1016/j.actatropica.2022.106778 [DOI] [PubMed] [Google Scholar]
- 33.Hoyos LR, Uribe SS, Vélez I. Tipificación de especímenes colombianos de Lutzomyia longipalpis (Diptera: Psychodidae) mediante “Código de Barras.”. Rev Colomb Entomol. 2002;38(N1):1–7. [Google Scholar]
- 34.Gutierrez MAC, Lopez ROH, Ramos AT, Vélez ID, Gomez RV, Arrivillaga-Henríquez J, et al. DNA barcoding of Lutzomyia longipalpis species complex (Diptera: Psychodidae), suggests the existence of 8 candidate species. Acta Trop. 2021;221:105983. doi: 10.1016/j.actatropica.2021.105983 [DOI] [PubMed] [Google Scholar]
- 35.Holdridge LR. Life zone ecology. Tropical Science Center. 1967: 10–149.
- 36.Corpoamazonia. Agenda ambiental departamento del Amazonas. 2008. Available: https://www.corpoamazonia.gov.co/files/Ordenamiento/agendas/01_DMarco_Agenda_Amazonas.pdf
- 37.Gobernación del Caquetá. Plan de Desarrollo Departamental 2020 - 2023. Caquetá; 2020 Feb. Available: https://www.caqueta.gov.co/noticias/p-lan-de-desarrollo-departamental-2020--2023
- 38.World Health Organization (WHO). Leishmaniasis: Number of cases of cuteneous leishmaniasis reported: 2022. 2024. Available: https://apps.who.int/neglected_diseases/ntddata/leishmaniasis/leishmaniasis.html
- 39.Departamento Administrativo Nacional de Estadística (DANE). Página para la descarga de datos geoestadísticos: Versión MGN2024-Nivel Departamento. In: Geoportal DANE [Internet]. 2024 [cited 18 Dec 2024]. Available: https://geoportal.dane.gov.co/servicios/descarga-y-metadatos/datos-geoestadisticos/
- 40.SIAT-AC - Instituto SINCHI. Límite de la Amazonia colombiana. Escala: 1:100.000. In: Datos Abiertos - Instituto SINCHI [Internet]. 2022 [cited 18 Dec 2024]. Available: https://datos.siatac.co/datasets/7fbe5e6357354c7c9baf8a44369ceef7/about
- 41.Departamento Administrativo Nacional de Estadística (DANE). Página para la descarga de datos geoestadísticos: Versión MGN2024-Nivel Municipio. In: Geoportal DANE [Internet]. 2024 [cited 18 Dec 2024]. Available: https://geoportal.dane.gov.co/servicios/descarga-y-metadatos/datos-geoestadisticos/
- 42.Departamento Administrativo Nacional de Estadística (DANE). Página para la descarga de datos geoestadísticos: Versión MGN2024-Nivel Sector urbano. In: Geoportal DANE [Internet]. 2024 [cited 18 Dec 2024]. Available: https://geoportal.dane.gov.co/servicios/descarga-y-metadatos/datos-geoestadisticos/
- 43.QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation Project. 2023. [Google Scholar]
- 44.Vazquez-Prokopec GM, Galvin WA, Kelly R, Kitron U. A new, cost-effective, battery-powered aspirator for adult mosquito collections. J Med Entomol. 2009;46(6):1256–9. doi: 10.1603/033.046.0602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galati E. Morfologia e terminologia de Phlebotominae (Diptera: Psychodidae). Classificação e identificação de táxons das Américas. Vol I. Apostila da Disciplina Bioecologia e Identificação de Phlebotominae do Programa de Pós-Graduação em Saúde Pública. São Paulo; 2024. Available: http://www.fsp.usp.br/egalati
- 46.Young D, Duncan M. Guide to the identification and geographic distribution of Lutzomyia sand flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae). Gainesville. Flodda: Associated Publishers; 1994. [Google Scholar]
- 47.Marcondes CB. A proposal of generic and subgeneric abbreviations for Phlebotomine sandflies (Diptera: Psychodidae: Phlebotominae) of the world. Entomological News. 2007;118(4):351–6. doi: 10.3157/0013-872x(2007)118[351:apogas]2.0.co;2 [DOI] [Google Scholar]
- 48.Porter CH, Collins FH. Species-diagnostic differences in a ribosomal DNA internal transcribed spacer from the sibling species Anopheles freeborni and Anopheles hermsi (Diptera:Culicidae). Am J Trop Med Hyg. 1991;45(2):271–9. doi: 10.4269/ajtmh.1991.45.271 [DOI] [PubMed] [Google Scholar]
- 49.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–9. [PubMed] [Google Scholar]
- 50.Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, et al. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989;86(16):6196–200. doi: 10.1073/pnas.86.16.6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitano T, Umetsu K, Tian W, Osawa M. Two universal primer sets for species identification among vertebrates. Int J Legal Med. 2007;121(5):423–7. doi: 10.1007/s00414-006-0113-y [DOI] [PubMed] [Google Scholar]
- 52.Stevens L, Dorn PL, Hobson J, de la Rua NM, Lucero DE, Klotz JH, et al. Vector blood meals and Chagas disease transmission potential, United States. Emerg Infect Dis. 2012;18(4):646–9. doi: 10.3201/eid1804.111396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montalvo AM, Fraga J, Maes I, Dujardin J-C, Van der Auwera G. Three new sensitive and specific heat-shock protein 70 PCRs for global Leishmania species identification. Eur J Clin Microbiol Infect Dis. 2012;31(7):1453–61. doi: 10.1007/s10096-011-1463-z [DOI] [PubMed] [Google Scholar]
- 54.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–9. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.NCBI (National Center for Biotechnology Information). Basic Local Alignment Search Tool (BLAST). [cited 19 Dec 2024]. Available: https://blast.ncbi.nlm.nih.gov/Blast.cgi
- 56.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol Biol Evol. 2017;34(12):3299–302. doi: 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- 57.Roberts DR, Hsi BP. An Index of Species Abundance for Use with Mosquito Surveillance Data 12. Environmental Entomology. 1979;8(6):1007–13. doi: 10.1093/ee/8.6.1007 [DOI] [Google Scholar]
- 58.Silveira Neto S, Monteiro RC, Zucchi RA, Moraes RCB de. Uso da análise faunística de insetos na avaliação do impacto ambiental. Sci agric (Piracicaba, Braz). 1995;52(1):9–15. doi: 10.1590/s0103-90161995000100003 [DOI] [Google Scholar]
- 59.Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003;270 Suppl 1(Suppl 1):S96-9. doi: 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flores BM, Montoya E, Sakschewski B, Nascimento N, Staal A, Betts RA, et al. Critical transitions in the Amazon forest system. Nature. 2024;626(7999):555–64. doi: 10.1038/s41586-023-06970-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osorno-Mesa E, Morales-Alarcón A, de Osorno F, Ferro-Vela C. Phlebotominae de Colombia (Diptera, Psychodidae) IX. Distribución geográfica de especies de Brumptomyia y Lutzomyia Franca 1924, encontradas en Colombia. 1972.
- 62.Young D. A review of the bloodsucking psychodid flies of Colombia (Diptera: Phlebotominae and Sycoracinae). Institute of Food and Agricultural Sciences, University of Florida, Gainesville. Gainesville; 1979 Apr.
- 63.Cabrera OL, Mosquera L, Santamaría E. Flebótomos (Diptera: Psychodidae) del departamento de Guaviare, Colombia, con nuevos registros para el país. biomedica. 2009;29(1):73. doi: 10.7705/biomedica.v29i1.43 [DOI] [PubMed] [Google Scholar]
- 64.Barreto M, Burbano ME, Barreto P. Lutzomyia sand flies (Diptera: Psychodidae) from middle and lower Putumayo Department, Colombia, with new records to the country. Mem Inst Oswaldo Cruz. 2000;95(5):633–9. doi: 10.1590/s0074-02762000000500009 [DOI] [PubMed] [Google Scholar]
- 65.Fernandez R, Lopez V, Cardenas R, Requena E. Description of Lutzomyia (Trichophoromyia) nautaensis n. sp. (Diptera: Psychodidae) from the Peruvian Amazon Basin. J Med Entomol. 2015;52(4):622–5. doi: 10.1093/jme/tjv057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexander JB, Takaoka H, Eshita Y, Gomez EA, Hashiguchi Y. New records of phlebotomine sand flies (Diptera: Psychodidae) from Ecuador. Mem Inst Oswaldo Cruz. 1992;87(1):123–30. doi: 10.1590/s0074-027619920001000191343789 [DOI] [Google Scholar]
- 67.Santos TVD, Silveira FT. Increasing putative vector importance of Trichophoromyia phlebotomines (Diptera: Psychodidae). Mem Inst Oswaldo Cruz. 2020;115:e190284. doi: 10.1590/0074-02760190284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silveira FT, Souza AA, Lainson R, Shaw JJ, Braga RR, Ishikawa EE. Cutaneous leishmaniasis in the Amazon region: natural infection of the sandfly Lutzomyia ubiquitalis (Psychodidae: Phlebotominae) by Leishmania (Viannia) Lainsoni in Pará State, Brazil. Mem Inst Oswaldo Cruz. 1991;86(1):127–30. doi: 10.1590/s0074-02761991000100021 [DOI] [PubMed] [Google Scholar]
- 69.Pereira Júnior AM, Teles CBG, de Azevedo dos Santos AP, de Souza Rodrigues M, Marialva EF, Pessoa FAC, et al. Ecological aspects and molecular detection of Leishmania DNA Ross (Kinetoplastida: Trypanosomatidae) in phlebotomine sandflies (Diptera: Psychodidae) in terra firme and várzea environments in the Middle Solimões Region, Amazonas State, Brazil. Parasit Vectors. 2015;8:180. doi: 10.1186/s13071-015-0789-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silva TRR, Assis MDG, Freire MP, Rego FD, Gontijo CMF, Shimabukuro PHF. Molecular Detection of Leishmania in Sand Flies (Diptera: Psychodidae: Phlebotominae) Collected in the Caititu Indigenous Reserve of the Municipality of Lábrea, State of Amazonas, Brazil. J Med Entomol. 2014;51(6):1276–82. doi: 10.1603/ME14025 [DOI] [PubMed] [Google Scholar]
- 71.Quiroga C, Cevallos V, Morales D, Baldeón ME, Cárdenas P, Rojas-Silva P, et al. Molecular Identification of Leishmania spp. in Sand Flies (Diptera: Psychodidae, Phlebotominae) From Ecuador. J Med Entomol. 2017;54(6):1704–11. doi: 10.1093/jme/tjx122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valdivia HO, De Los Santos MB, Fernandez R, Baldeviano GC, Zorrilla VO, Vera H, et al. Natural Leishmania infection of Lutzomyia auraensis in Madre de Dios, Peru, detected by a fluorescence resonance energy transfer-based real-time polymerase chain reaction. Am J Trop Med Hyg. 2012;87(3):511–7. doi: 10.4269/ajtmh.2012.11-0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teles CBG, Santos AP de AD, Freitas RA, Oliveira AFJ de, Ogawa GM, Rodrigues MS, et al. Phlebotomine sandfly (Diptera: Psychodidae) diversity and their Leishmania DNA in a hot spot of American Cutaneous Leishmaniasis human cases along the Brazilian border with Peru and Bolivia. Mem Inst Oswaldo Cruz. 2016;0(7):0. doi: 10.1590/0074-02760160054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silveira FT, Ishikawa EAY, De Souza AAA, Lainson R. An outbreak of cutaneous leishmaniasis among soldiers in Belém, Pará State, Brazil, caused by Leishmania (Viannia) lindenbergi n. sp. A new leishmanial parasite of man in the Amazon region. Parasite. 2002;9(1):43–50. doi: 10.1051/parasite/200209143 [DOI] [PubMed] [Google Scholar]
- 75.Carneiro ACG, de Souza EA, Barroso EP, de Ávila MM, Melchior LAK, Rocha R da C, et al. Phlebotomine Fauna (Diptera: Psychodidae) and Infection by Leishmania spp. in Forest Fragments of a University Campus, Western Amazon. J Med Entomol. 2023;60(1):218–23. doi: 10.1093/jme/tjac162 [DOI] [PubMed] [Google Scholar]
- 76.Vásquez Trujillo A, González Reina AE, Góngora Orjuela A, Prieto Suárez E, Palomares JE, Buitrago Alvarez LS. Seasonal variation and natural infection of Lutzomyia antunesi (Diptera: Psychodidae: Phlebotominae), an endemic species in the Orinoquia region of Colombia. Mem Inst Oswaldo Cruz. 2013;108(4):463–9. doi: 10.1590/S0074-0276108042013011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silva ANR, Júnior AMP, de Paulo PFM, da Silva MS, Castro TS, Costa G da S, et al. Detection of Leishmania species (Kinetoplastida, Trypanosomatidae) in phlebotomine sand flies (Diptera, Psychodidae) from Porto Velho, Northern Brazil. Acta Trop. 2021;213:105757. doi: 10.1016/j.actatropica.2020.105757 [DOI] [PubMed] [Google Scholar]
- 78.Braga RR, Lainson R, Ishikawa EAY, Shaw JJ. Leishmania (Viannia) utingensis n. sp., a parasite from the sandfly Lutzomyia (Viannamyia) tuberculata in Amazonian Brazil. Parasite. 2003;10(2):111–8. doi: 10.1051/parasite/2003102111 [DOI] [PubMed] [Google Scholar]
- 79.Vasconcelos Dos Santos T, Prévot G, Ginouvès M, Duarte R, Silveira FT, Póvoa MM, et al. Ecological aspects of Phlebotomines (Diptera: Psychodidae) and the transmission of American cutaneous leishmaniasis agents in an Amazonian/ Guianan bordering area. Parasit Vectors. 2018;11(1):612. doi: 10.1186/s13071-018-3190-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Araujo-Pereira T de, Pita-Pereira D de, Baia-Gomes SM, Boité M, Silva F, Pinto I de S, et al. An overview of the sandfly fauna (Diptera: Psychodidae) followed by the detection of Leishmania DNA and blood meal identification in the state of Acre, Amazonian Brazil. Mem Inst Oswaldo Cruz. 2020;115:e200157. doi: 10.1590/0074-02760200157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Costa JCR, Marchi GH, Santos CS, Andrade MCM, Chaves Junior SP, Silva MAN, et al. First molecular evidence of frogs as a food source for sand flies (Diptera: Phlebotominae) in Brazilian caves. Parasitol Res. 2021;120(5):1571–82. doi: 10.1007/s00436-021-07154-3 [DOI] [PubMed] [Google Scholar]
- 82.Dutra-Rêgo F, Binder C, Capucci DC, Vaz TP, Andrade Filho JD, Fontes G, et al. Diversity, Leishmania detection, and blood meal sources of sand flies from Iguatama, Minas Gerais, Brazil. PLoS One. 2024;19(5):e0302567. doi: 10.1371/journal.pone.0302567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guimarães-E-Silva AS, Silva S de O, Ribeiro da Silva RC, Pinheiro VCS, Rebêlo JMM, Melo MN. Leishmania infection and blood food sources of phlebotomines in an area of Brazil endemic for visceral and tegumentary leishmaniasis. PLoS One. 2017;12(8):e0179052. doi: 10.1371/journal.pone.0179052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fonteles RS, Pereira Filho AA, Moraes JLP, Pereira SRF, Rodrigues BL, Rebêlo JMM. Detection of Leishmania DNA and Blood Meal Identification in Sand Flies (Diptera: Psychodidae) From Lençois Maranhenses National Park Region, Brazil. J Med Entomol. 2018;55(2):445–51. doi: 10.1093/jme/tjx230 [DOI] [PubMed] [Google Scholar]
- 85.Da Silva YY, Sales KGDS, Miranda DEDO, Figueredo LA, Brandão-Filho SP, Dantas-Torres F. Detection of Leishmania DNA in Sand Flies (Diptera: Psychodidae) From a Cutaneous Leishmaniasis Outbreak Area in Northeastern Brazil. J Med Entomol. 2020;57(2):529–33. doi: 10.1093/jme/tjz189 [DOI] [PubMed] [Google Scholar]
- 86.Pereira-Filho AA, Fonteles RS, Bandeira M da CA, Moraes JLP, Rebêlo JMM, Melo MN. Molecular Identification of Leishmania spp. in Sand Flies (Diptera: Psychodidae: Phlebotominae) in the Lençóis Maranhenses National Park, Brazil. J Med Entomol. 2018;55(4):989–94. doi: 10.1093/jme/tjy014 [DOI] [PubMed] [Google Scholar]
- 87.Alexander B, Ferro C, Young DG, Morales A, Tesh RB. Ecology of phlebotomine sand flies (Diptera: Psychodidae) in a focus of Leishmania (Viannia) braziliensis in northeastern Colombia. Mem Inst Oswaldo Cruz. 1992;87(3):387–95. doi: 10.1590/s0074-02761992000300009 [DOI] [PubMed] [Google Scholar]
- 88.Contreras-Gutiérrez MA, Vélez ID, Porter C, Uribe SI. Lista actualizada de flebotomíneos (Diptera: Psychodidae: Phlebotominae) de la región cafetera colombiana. biomedica. 2014;34(3). doi: 10.7705/biomedica.v34i3.2121 [DOI] [PubMed] [Google Scholar]
- 89.Rangel EF, Lainson R. Proven and putative vectors of American cutaneous leishmaniasis in Brazil: aspects of their biology and vectorial competence. Mem Inst Oswaldo Cruz. 2009;104(7):937–54. doi: 10.1590/s0074-02762009000700001 [DOI] [PubMed] [Google Scholar]
- 90.Young DG, Morales A, Kreutzer RD, Alexander JB, Corredor A, Tesh RB, et al. Isolations of Leishmania braziliensis (Kinetoplastida: Trypanosomatidae) from cryopreserved Colombian sand flies (Diptera: Psychodidae). J Med Entomol. 1987;24(5):587–9. doi: 10.1093/jmedent/24.5.587 [DOI] [PubMed] [Google Scholar]
- 91.da Silva MS, Júnior AMP, Costa NVC, Costa G da S, Rodrigues MM de S, Medeiros JF. Use of light emitting diodes (LEDs) are effective and useful for sand fly ecoepidemiology studies in an Amazonian environment. Acta Trop. 2022;233:106550. doi: 10.1016/j.actatropica.2022.106550 [DOI] [PubMed] [Google Scholar]
- 92.Santos WS, Ortega FD, Alves VR, Garcez LM. Flebotomíneos (Psychodidae: Phlebotominae) de área endêmica para leishmaniose cutânea e visceral no nordeste do estado do Pará, Brasil. Revista Pan-Amazônica de Saúde. 2019;10(0). doi: 10.5123/s2176-6223201900059 [DOI] [Google Scholar]
- 93.Zorrilla V, De Los Santos MB, Espada L, Santos RDP, Fernandez R, Urquia A, et al. Distribution and identification of sand flies naturally infected with Leishmania from the Southeastern Peruvian Amazon. PLoS Negl Trop Dis. 2017;11(11):e0006029. doi: 10.1371/journal.pntd.0006029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Young DG, Morales A. New species and records of phlebotomine sand flies from Colombia (Diptera: Psychodidae). J Med Entomol. 1987;24(6):651–65. doi: 10.1093/jmedent/24.6.651 [DOI] [PubMed] [Google Scholar]
- 95.Kato H, Calvopiña M, Criollo H, Hashiguchi Y. First human cases of Leishmania (Viannia) naiffi infection in Ecuador and identification of its suspected vector species. Acta Trop. 2013;128(3):710–3. doi: 10.1016/j.actatropica.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 96.Cantanhêde LM, Cupolillo E. Leishmania (Viannia) naiffi Lainson & Shaw 1989. Parasit Vectors. 2023;16(1):194. doi: 10.1186/s13071-023-05814-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Resadore F, Júnior AMP, de Paulo PFM, Gil LHS, Rodrigues MM de S, Araújo M da S, et al. Composition and Vertical Stratification of Phlebotomine Sand Fly Fauna and the Molecular Detection of Leishmania in Forested Areas in Rondônia State Municipalities, Western Amazon, Brazil. Vector Borne Zoonotic Dis. 2019;19(5):347–57. doi: 10.1089/vbz.2018.2372 [DOI] [PubMed] [Google Scholar]
- 98.Posada-Lopez L, Galati EA, Shaw J, Galvis-Ovallos F. Incriminating leishmaniases vectors in Colombia: An overview and roadmap for future research. Acta Trop. 2024;260:107409. doi: 10.1016/j.actatropica.2024.107409 [DOI] [PubMed] [Google Scholar]
- 99.Santamaría E, Ponce N, Zipa Y, Ferro C. Presencia en el peridomicilio de vectores infectados con Leishmania (Viannia) panamensis en dos focos endémicos en el occidente de Boyacá, piedemonte del valle del Magdalena medio, Colombia. biomedica. 2012;26(1):82. doi: 10.7705/biomedica.v26i1.1503 [DOI] [PubMed] [Google Scholar]
- 100.López M, Erazo D, Hoyos J, León C, Fuya P, Lugo L, et al. Measuring spatial co-occurrences of species potentially involved in Leishmania transmission cycles through a predictive and fieldwork approach. Sci Rep. 2021;11(1):6789. doi: 10.1038/s41598-021-85763-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Soares O de SR, Rodrigues BL, Shimabukuro PHF. Accessing the sand fly diversity of Tocantins, Northern Brazil: species delimitation using COI DNA barcoding. Biota Neotrop. 2024;24(4). doi: 10.1590/1676-0611-bn-2024-1680 [DOI] [Google Scholar]
- 102.Rodrigues BL, Baton LA, Shimabukuro PHF. Single-locus DNA barcoding and species delimitation of the sandfly subgenus Evandromyia (Aldamyia). Med Vet Entomol. 2020;34(4):420–31. doi: 10.1111/mve.12458 [DOI] [PubMed] [Google Scholar]
- 103.Di Muccio T, Marinucci M, Frusteri L, Maroli M, Pesson B, Gramiccia M. Phylogenetic analysis of Phlebotomus species belonging to the subgenus Larroussius (Diptera, psychodidae) by ITS2 rDNA sequences. Insect Biochem Mol Biol. 2000;30(5):387–93. doi: 10.1016/s0965-1748(00)00012-6 [DOI] [PubMed] [Google Scholar]
- 104.Vivero RJ, Contreras-Gutiérrez MA, Bejarano EE. Cambios en el extremo carboxilo terminal de citocromo b como carácter taxonómico en Lutzomyia (Diptera: Psychodidae). Rev Colomb Entomol. 2009;35(1):83–8. doi: 10.25100/socolen.v35i1.9194 [DOI] [Google Scholar]
- 105.Dupuis JR, Roe AD, Sperling FAH. Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Mol Ecol. 2012;21(18):4422–36. doi: 10.1111/j.1365-294X.2012.05642.x [DOI] [PubMed] [Google Scholar]
- 106.Liu J, Jiang J, Song S, Tornabene L, Chabarria R, Naylor GJP, et al. Multilocus DNA barcoding - Species Identification with Multilocus Data. Sci Rep. 2017;7(1):16601. doi: 10.1038/s41598-017-16920-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.González C, León C, Paz A, López M, Molina G, Toro D, et al. Diversity patterns, Leishmania DNA detection, and bloodmeal identification of Phlebotominae sand flies in villages in northern Colombia. PLoS ONE. 2018;13(1):e0190686. doi: 10.1371/journal.pone.0190686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Paternina LE, Verbel-Vergara D, Romero-Ricardo L, Pérez-Doria A, Paternina-Gómez M, Martínez L, et al. Evidence for anthropophily in five species of phlebotomine sand flies (Diptera: Psychodidae) from northern Colombia, revealed by molecular identification of bloodmeals. Acta Trop. 2016;153:86–92. doi: 10.1016/j.actatropica.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 109.Cera-Vallejo Y, Ardila MM, Herrera L, Martínez L, Pérez-Doria A. Phlebotomine (Diptera: Psychodidae) species and their blood meal sources in a new leishmaniasis focus in Los Montes de María, Bolívar, in northern Colombia. Biomedica. 2024;44(2):248–57. doi: 10.7705/biomedica.6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Costa G da S, Júnior AMP, Castro TS, de Paulo PFM, Ferreira GEM, Medeiros JF. Sand fly fauna and molecular detection of Leishmania species and blood meal sources in different rural environments in western Amazon. Acta Trop. 2021;224:106150. doi: 10.1016/j.actatropica.2021.106150 [DOI] [PubMed] [Google Scholar]
- 111.Dos Santos NS, de Pinho FA, Hlavac NRC, Nunes TL, Almeida NR, Solcà MS, et al. Feline Leishmaniasis Caused by Leishmania infantum: Parasite Sequencing, Seropositivity, and Clinical Characterization in an Endemic Area From Brazil. Front Vet Sci. 2021;8:734916. doi: 10.3389/fvets.2021.734916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dantas da Silva M, Nakaghi ACH, Galvis-Ovallos F, Leonel JAF, Vioti G, Galati EAB, et al. Infectiousness to sand flies of a cat naturally infected with Leishmania infantum at the moment of diagnosis and after three different courses of treatment. Rev Bras Parasitol Vet. 2025;34(1):e016524. doi: 10.1590/S1984-29612025006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ready PD, Arias JR, Freitas RA. A pilot study to control Lutzomyia umbratilis (Diptera:Psychodidae), the major vector of Leishmania braziliensis guyanensis, in a peri-urban rainforest of Manaus, Amazonas State, Brazil. Mem Inst Oswaldo Cruz. 1985;80(1):27–36. doi: 10.1590/s0074-02761985000100005 [DOI] [PubMed] [Google Scholar]