Abstract
Compared to yeast esterase, fungal cutinase degraded butyl benzyl phthalate (BBP) far more efficiently; i.e., almost 60% of the BBP disappeared within 7.5 h. Also, the final chemical composition significantly depended on the enzyme used. Toxicity monitoring using bioluminescent bacteria showed that butyl methyl phthalate, a major product of degradation by esterase, was an oxidative toxic hazard.
Phthalates are plasticizers used in the manufacture of polyvinyl chloride and often in paints, lacquers, and cosmetics (18, 26, 31). Phthalates found in sediment, water, and air (13) have also been detected in foods, as they can migrate out of food packaging materials (29, 36). n-Butyl benzyl phthalate (BBP) is a phthalic ester in papers and paperboards used as packaging materials for aqueous, fatty, and dry foods (19, 37). BBP exerted estrogenic activities in several in vitro tests (2, 18, 20, 43). During the past 10 years, there have been a series of reports on the developmental toxicity of BBP in rats (1, 11, 12, 39), and the teratogenic effects have also been observed in mice and chicks (4, 32).
Cutinase, a hydrolytic enzyme that degrades cutin (a polyester consisting of hydroxy and epoxy fatty acids, usually with n-C16 and n-C18 of higher plants), can, unlike other lipases, show enzymatic activity without interfacial activation. Some microorganisms such as Fusarium oxysporum f. sp. pisi live on cutin as their sole carbon source producing extracellular cutinases, and several bacterial cutinases (Pseudomonas putida, Pseudomonas mendocina, and Corynebacterium spp.) have been isolated and characterized (8, 21, 23, 25, 30, 34, 35). These enzymes have been largely exploited for esterification and transesterification in chemical synthesis (16) and have also been applied in the composition of laundry or dishwashing detergent (14, 28, 38). Other potential uses for cutinase include its application in the oleochemistry industry (6) and in the degradation of plastic (22).
In the present study, we investigated the efficacies of fungal cutinase and yeast esterase in the degradation of BBP. During the enzymatic degradation of BBP, time-course compositional changes of several BBP-derived compounds were monitored. The cellular toxicity of degradation products was also measured by using various bioluminescent bacteria.
Enzymatic degradation of BBP by cutinase and esterase.
Purified cutinase from F. oxysporum f. sp. pisi (kindly provided by C. M. J. Sagt, Utrecht University, Utrecht, The Netherlands) and commercial Candida cylindracea esterase (Boehringer Mannheim GmbH, Mannheim, Germany) were dissolved in Tris-HCl buffer (10 mM, pH 8.0), and the enzyme concentration was adjusted to 10 or 100 mg liter−1. The enzymatic degradation of BBP (98% purity; Aldrich, Milwaukee, Wisc.) was begun by adding 50 μl of the concentrated BBP solution (500 g liter−1 in pure methanol [99.9%; Merck, Darmstadt, Germany]) to 50 ml of the enzyme solution described above in a 250-ml flask and was continued for 3 days in the dark in a shaking incubator (30°C, 200 rpm).
Analyses of BBP-derived degradation products.
Each time-course sample (500 μl) was mixed with 500 μl of n-hexane (>99% purity; Sigma) for 3 h. After phase separation had been completed, we confirmed that the nonpolar compounds, including BBP and its derivatives, were successfully extracted into the hexane phase with almost 100% efficiency. To recover residual compounds in the aqueous phase (0.1% methanol), the separated aqueous phase was lyophilized after filtering with a YM-3 membrane (Amicon, Cambridge, Mass.) to remove enzymes, followed by the addition of 500 μl of methanol. All chemicals in n-hexane or methanol were analyzed using gas chromatography-mass spectrometry (GC/MS) by injecting a 4-μl sample into an HP6890 series gas chromatograph-mass selective detector with helium as the carrier gas at 1 ml min−1. The temperature at the injection port was 280°C, and the oven temperature was programmed to increase from 80°C (for 4 min) to 310°C (for 5 min) at 7°C min−1. Data collection and processing were performed with HP MSD ChemStation software containing the Wiley chemical library.
Toxicity monitoring of degradation products by using bioluminescent bacteria.
Escherichia coli strain GC2 (lac::luxCDABE) has the luxCDABE gene from Xenorhabdus luminescens under the control of the lac promoter. Several other bioluminescent E. coli strains were used for evaluating possible modes of toxicity: DPD2794 (recA::luxCDABE), which can be used to detect genotoxicity (42); TV1016 (grpE::luxCDABE), which is sensitive to protein damage (41); DPD2511 (katG::luxCDABE), which is sensitive to oxidative damage (3); and DPD2540 (fabA::luxCDABE), which is sensitive to membrane damage (7). Each strain was first cultured in 100 ml of Luria-Bertani medium (37°C and 250 rpm). When the optical density at 600 nm reached 0.8, 0.2-ml portions of the culture broth were mixed with various test samples (25 μl), i.e., Tris-HCl buffer (10 mM, pH 8.0) containing neither enzyme nor BBP, Tris-HCl buffer containing enzyme only (cutinase [10 mg liter−1] or esterase [100 mg liter−1]), Tris-HCl buffer containing only BBP (500 mg liter−1), or the final samples after enzymatic degradation of BBP for 3 days. The recombinant culture and test sample mixtures were prepared in triplicate in a highly sensitive 96-well microplate luminometer (Microlite; Dynex Technologies, Chantilly, Va.) at 30°C, and the emitted bioluminescence (BL) was measured at a regular time interval, 6 h. The relative BL, i.e., the maximum BL measured with the final degradation products divided by the maximum BL measured with only buffer solution containing neither BBP nor enzyme, was used as a parameter indicating the toxicity of each test sample.
Enzymatic degradation of BBP: fungal cutinase versus yeast esterase.
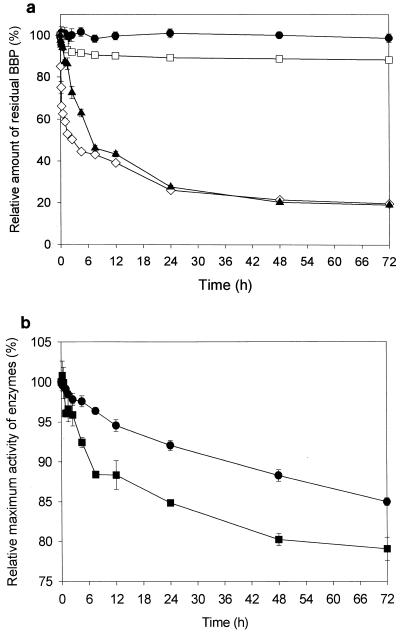
With the fungal cutinase (10 mg liter−1) from F. oxysporum f. sp. pisi, almost 60% of the initial BBP was decomposed within 7.5 h, while with the yeast esterase (from C. cylindracea), more than 90% of the initial BBP remained nondegraded even after 3 days of treatment (Fig. 1a). When esterase that was tenfold more concentrated (100 mg liter−1) was applied, the BBP concentration decreased significantly after 3 days (Fig. 1a). The cutinase or esterase activity in various time-course samples was estimated by measuring the initial maximum activity with PNB (p-nitrophenyl butyrate) as the substrate. During the 72-h process in 0.1% methanol solution, cutinase (10 mg liter−1) and esterase (100 mg liter−1) maintained more than 85% and 80% of the initial hydrolytic activities, respectively (Fig. 1b). As shown in Fig. 1b, the stability of the cutinase activity was apparently greater than that of esterase throughout the 3 days of degradation.
FIG. 1.
(a) Time-course variation in the residual amount of BBP, expressed in terms of relative amount (i.e., initial BBP amount = 100%), during the enzymatic degradation of BBP. •, BBP (500 mg liter−1) in Tris-HCl buffer (10 mM, pH 8.0) without enzyme; □, BBP (500 mg liter−1) degraded by esterase (10 mg liter−1); ▴, BBP (500 mg liter−1) degraded by esterase (100 mg liter−1); ⋄, BBP (500 mg liter−1) degraded by cutinase (10 mg liter−1). (b) PNB hydrolysis activities of cutinase (•) and esterase (▪), expressed in terms of relative activity (i.e., maximum PNB hydrolysis activity by the initial sample = 100%), contained in various time-course samples taken during the enzymatic degradation of BBP.
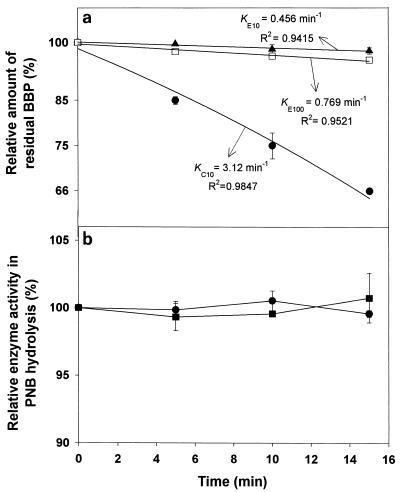
The initial BBP breakdown (for 15 min), shown in the semilogarithmic plot in Fig. 2a, is presumed to follow a first-order kinetics, i.e., dN/dt = −KN, where N and K represent the residual BBP amount and the degradation constant, respectively. The estimation of the degradation constants, KC10 (for cutinase, 10 mg liter−1) and KE100 (for esterase, 100 mg liter−1), demonstrated that the initial BBP degradation by cutinase seemed to proceed more than 4 times faster than that by esterase (Fig. 2a). This significant difference in the initial degradation rates seems to have no correlation with the enzyme stability, because both enzymes showed almost 100% stability for the initial 15 min (Fig. 2b). It is noteworthy that more than 30% of the initial BBP was degraded by cutinase only after 15 min (Fig. 2a).
FIG. 2.
(a) Semilogarithmic plot of time-course variation in the amount of relative residual BBP during the initial 15 min after the start of enzymatic degradation by cutinase (10 mg liter−1) (•) and esterase (10 mg liter−1 [▴] or 100 mg liter−1 [□]). KC10, KE10, and KE100 represent the degradation constants in the first-order degradation kinetics of BBP resulting from the action of 10-mg/liter cutinase and 10- and 100-mg/liter esterase, respectively. (b) Time-course variation, for the initial 15 min, in the PNB hydrolysis activities of cutinase (10 mg liter−1) (•) and esterase (100 mg liter−1) (▪).
Formation of BBP-derived chemicals in enzymatic decomposition process.
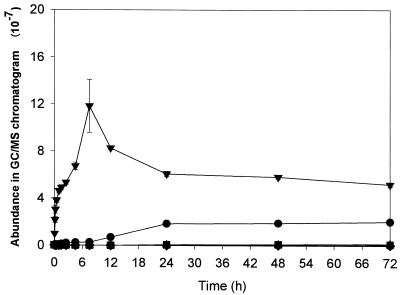
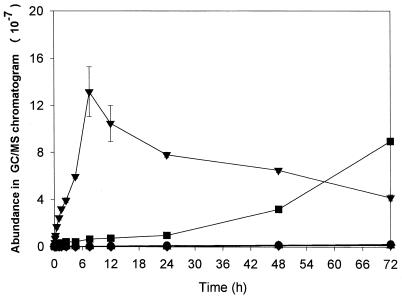
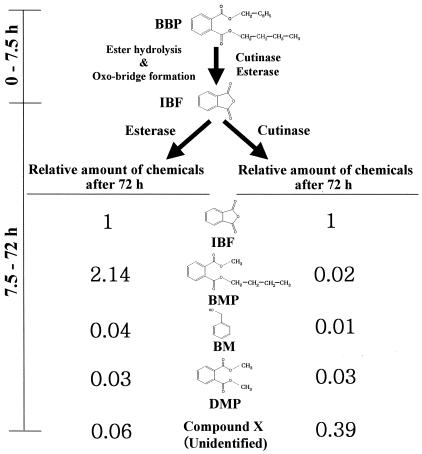
During biodegradation, GC/MS analysis detected five different chemicals: butyl methyl phthalate (BMP), dimethyl phthalate (DMP), benzene methanol (BM), 1,3-isobenzofurandione (IBF), and an unidentified chemical (X) (Fig. 3 and 4). As shown in Fig. 3 and 4, the formation of the decomposition products significantly depended on the enzyme used. In both enzymatic processes, IBF was rapidly formed as a major product for the initial 7.5 h but then progressively decreased. While the amount of IBF continued to decrease after 7.5 h, the amount of BMP continued to increase in the esterase process and became the most abundant chemical after 3 days (Fig. 4). In the cutinase process (Fig. 3), the degradation compounds other than IBF were produced at very low concentrations.
FIG. 3.
Time-course variation in the amounts of chemical compounds produced during BBP degradation by cutinase, as analyzed by GC/MS (BBP and the other chemical compounds formed in the BBP degradation by cutinase were first extracted by n-hexane, and residual compounds in aqueous phase were dissolved in methanol after water was completely removed through lyophilization). ▾, IBF; ▴, DMP; ▪, BMP; ♦, BM; •, compound X.
FIG. 4.
Time-course variation in the amount of BBP and the other chemical compounds produced during BBP degradation by esterase, as analyzed by GC/MS (BBP and the other chemical compounds formed in the BBP degradation by esterase were first extracted by n-hexane, and residual compounds in aqueous phase were dissolved in methanol after water was completely removed through lyophilization). ▾, IBF; ▴, DMP; ▪, BMP; ♦, BM; •, compound X.
The important characteristics of the enzymatic degradation of BBP are schematically summarized in Fig. 5. Approximately 75% of the total degraded amount of BBP was degraded during the initial 7.5 h and was simultaneously converted to IBF via ester hydrolysis of BBP, followed by spontaneous oxo bridge formation. After 7.5 h, the conversion of IBF to other compounds was clearly differentiated depending on the enzyme used. Apparently, BM was an ester hydrolysis product of BBP. The production of BMP and DMP likely occurred via the transesterification reaction of esterase or cutinase in 0.1% methanol. This is not surprising because cutinase and esterase have been widely used as biocatalysts catalyzing esterification and transesterification in organic solvents (5, 6, 34). The methyl and butyl esters in BMP and DMP seem to originate from methanol and one of the hydrolysis products of BBP, respectively. Except for the unidentified chemical (X) produced by cutinase, the BBP-derived chemicals (BMP, DMP, BM, and IBF) are not currently classified as endocrine-disrupting chemicals associated with effects on the endocrine system in animals and humans or in vitro (http://www.kfda.go.kr/webzine/endocrine/endocrine/).
FIG. 5.
Schematic summary showing significant differences between the pathways of BBP degradation by cutinase and esterase, including the relative chemical compositions after 72 h of BBP degradation.
Toxicity estimation of BBP-derived degradation products by using recombinant bioluminescent bacteria.
Toxicity detection using recombinant bioluminescent E. coli is not directly related to a toxic effect in animals and/or humans, and it is now generally accepted as a potential method for environmental monitoring of various industrial wastes and pollutants, including genotoxicants and stress inducers (9, 24, 27, 33, 40).
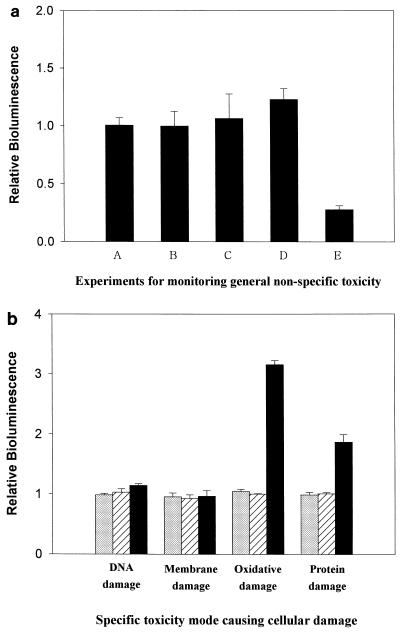
First, general toxicity due to nonspecific cellular stresses was analyzed by using strain GC2. Under increased cellular stress that represses cell growth, the BL emitted by the GC2 strain is subject to being decreased. As shown in Fig. 6a, the final products of degradation by C. cylindracea esterase significantly decreased the relative BL, and therefore, the presence of a toxic component(s) seems evident. Supplementing the bioluminescent culture with BBP (500 ppm) did not decrease the BL (Fig. 6a); hence, BBP did not seem to be toxic to bacterial cells, although endocrine-disrupting chemicals like BBP have been known to potentially interfere with human endogenous hormones (10, 15, 17). As is evident in Fig. 6a, no harmful effect was detected with the enzyme itself, cutinase (100 mg liter−1), or esterase (100 mg liter−1). In the final products of both enzymatic decomposition processes, the amounts of BM and DMP were typically negligible, although they were comparable to the amounts of IBF. Consequently, BMP, a major product of degradation by esterase, seems to be the toxic compound causing the significant cellular stresses.
FIG. 6.
(a) Results of estimation of relative BL by using strain GC2, showing the general toxicity (i.e., nonspecific cellular stress) of the test samples: A, Tris-HCl buffer containing BBP (500 ppm); B, Tris-HCl buffer containing cutinase (100 mg liter−1); C, Tris-HCl buffer containing esterase (100 mg liter−1); D, final products of BBP degradation by cutinase; and E, final products of BBP degradation by esterase. (b) Results of the estimation of relative BL, using strains DPD2794, TV1016, DPD2511, and DPD2540, showing the specific mode of toxicity of the test samples, i.e., Tris-HCl buffer containing BBP (500 ppm) (gray columns) and the final products of BBP degradation by cutinase and esterase (striped and black columns, respectively).
We investigated which type of cellular damage was related to BMP toxicity. As shown in Fig. 6b, BBP (500 ppm) and the products of its degradation by cutinase never caused any cellular damage, which is consistent with the results shown in Fig. 6a. Figure 6b shows that BMP exerted oxidative damage as well as protein damage on the bacteria. Cellular stress by oxidative hazards like active oxygen species may be a great threat to the structures and functions of proteins, nucleic acids, lipids, and membranes, whether they are added externally or produced intracellularly. Also, the increase of BL due to the induction of the grpE promoter (a heat shock promoter) indicated that defective protein synthesis took place in the presence of BMP.
For in situ BBP degradation with cutinase, the addition of the purified cutinase or fungus itself to BBP-contaminated sites does not seem to be practical. A plausible approach to the practical application of cutinase for BBP degradation may be to develop a microbial gene expression system for producing recombinant cutinase. The recombinant yeast system may have a great advantage in that extracellular production of recombinant enzyme is possible, and therefore, the cell-free medium containing a large amount of recombinant cutinase can be applied directly to the in situ degradation of BBP without costly purification.
Acknowledgments
This work was supported by a Korea University Grant.
We thank Jin-Soo Park and Byoung-Chan Kim at Kwangju Institute of Science and Technology for their technical assistance in the utilization of GC/MS and the bioluminescent bacteria.
REFERENCES
- 1.Aldert, H., P. A. Verhoef, J. T. Biesebeek, M. N. Pieters, and W. Slob. 2000. Developmental toxicity of butyl benzyl phthalate in the rat using a multiple dose study design. Toxicology 14:417-425. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, H. R., A. M. Andersson, S. F. Arnold, H. Autrup, M. Barfoed, N. A. Beresford, P. Bjerregaard, L. B. Christiansen, B. Gissel, R. Hummel, E. B. Jorgensen, B. Korsgaard, R. Le Guevel, H. Leffers, J. McLachlan, A. Moller, J. B. Nielsen, N. Olea, A. Oles-Karasko, F. Pakdel, K. L. Pedersen, P. Perez, N. E. Skakkeboek, C. Sonnenschein, A. M. Soto, J. P. Sumpter, S. M. Thorpe, and P. Grandjean. 1999. Comparison of short-term estrogenicity tests for identification of hormone-disrupting chemicals. Environ. Health Perspect. 107:89-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belkin, S., D. R. Smulski, A. C. Vollmer, T. K. Van Dyk, and R. A. LaRossa. 1996. Oxidative stress detection with Escherichia coli harboring a katG′::lux fusion. Appl. Environ. Microbiol. 62:2252-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bower, R. K., S. Haberman, and P. D. Minton. 1970. Terotogenic effects in the chick embryo caused by esters of phthalic acid. J. Pharmacol. Exp. Ther. 171:314-324. [PubMed] [Google Scholar]
- 5.Cartwright, C. D., S. A. Owen, I. P. Thompson, and R. G. Burns. 2000. Biodegradation of diethyl phthalate in soil by a novel pathway. FEMS Microbiol. Lett. 186:27-34. [DOI] [PubMed] [Google Scholar]
- 6.Carvalho, C. M., M. R. Aires-Barros, and J. M. Cabral. 1999. Cutinase: from molecular level to bioprocess development. Biotechnol. Bioeng. 66:17-34. [DOI] [PubMed] [Google Scholar]
- 7.Choi, S. H., and M. B. Gu. 2001. Phenolic toxicity—detection and classification through the use of a recombinant bioluminescent Escherichia coli. Environ. Toxicol. Chem. 20:248-255. [PubMed] [Google Scholar]
- 8.Dantzig, A. H., S. H. Zuckerman, and M. M. Andonov-Roland. 1986. Isolation of a Fusarium mutant reduced in cutinase activity and virulence. J. Bacteriol. 168:911-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidov, Y., R. Rozen, D. R. Smulski, T. K. Van Dyk, A. C. Vollmer, D. A. Elsemore, R. A. LaRossa, and S. Belkin. 2000. Improved bacterial SOS promoter::lux fusions for genotoxicity detection. Mutat. Res. 466:97-107. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Ferrero, J., M. C. Rodriguez-Larena, L. Comellas, and B. Jimenez. 1997. Bioanalytical methods applied to endocrine disrupting polychlorinated bisphenyls, polychlorinated dibenzo-ρ-dioxins and polychlorinated dibensofurans: a review. Trends Anal. Chem. 16:563-573. [Google Scholar]
- 11.Ema, M., E. Miyawaki, and K. Kawashima. 1999. Developmental effects of plasticizer butyl benzyl phthalate after a single administration in rats. J. Appl. Toxicol. 19:357-365. [DOI] [PubMed] [Google Scholar]
- 12.Ema, M., R. Kurosaka, H. Amano, and Y. Ogawa. 1995. Comparative developmental toxicity of n-butyl benzyl phthalate and di-n-butyl phthalate in rats. Arch. Environ. Contam. Toxicol. 28:223-228. [DOI] [PubMed] [Google Scholar]
- 13.Fatoki, O. S., and F. Vernon. 1990. Phthalate esters in rivers of the Greater Manchester area, U.K. Sci. Total Environ. 95:227-232. [Google Scholar]
- 14.Flipsen, J. A. C., A. C. M. Appel, H. T. W. M. Van der Hijden, and C. T. Verrips. 1998. Mechanism of removal of immobilized triacylglycerol by lipolytic enzymes in a sequential laundry wash process. Enzyme Microb. Technol. 23:274-280. [Google Scholar]
- 15.Gascon, J., A. Qubina, and D. Barcelo. 1997. Detection of endocrine-disrupting pesticides by enzyme-linked immunosorbent assay (ELISA): application to atrazine. Trends Anal. Chem. 16:554-562. [Google Scholar]
- 16.Gerard, H. C., W. F. Fett, S. F. Osman, and R. A. Moreau. 1993. Evaluation of cutinase activity of various industrial lipases. Biotechnol. Appl. Biochem. 17:181-189. [Google Scholar]
- 17.Gu, M. B., J. Min, and E. J. Kim. 2002. Toxicity monitoring and classification of endocrine-disrupting chemicals (EDCs) using recombinant bioluminescent bacteria. Chemosphere 46:289-294. [DOI] [PubMed] [Google Scholar]
- 18.Harris, C. A., P. Henttu, M. G. Parker, and J. P. Sumpter. 1997. The estrogenic activity of phthalate esters in vitro. Environ. Health Perspect. 105:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Agency for Research on Cancer. 1982. Butyl benzyl phthalate. IARC Monogr. Eval. Carcinog. Risks Hum. 29:193-201. [PubMed] [Google Scholar]
- 20.Jobling, S., T. Reynolds, R. White, M. G. Parker, and J. P. Sumpter. 1995. A variety of environmentally persistent chemicals, including some phthalates plasticizers, are weakly estrogenic. Environ. Health Perspect. 103:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolattukudy, P. E. 1984. Cutinase from fungi and pollen, p. 471-504. In B. Borgstrom and T. Brockman (ed.), Lipases. Elsevier, Amsterdam, The Netherlands.
- 22.Kolattukudy, P. E., R. E. Purdy, and I. B. Maiti. 1981. Cutinase from fungi and pollen. Methods Enzymol. 71:652-664. [Google Scholar]
- 23.Lin, T.-S., and P. E. Kolattukudy. 1980. Structural studies on cutinase, a glycoprotein containing novel amino acids and glucuronic acid amide at the N terminus. Eur. J. Biochem. 106:341-351. [DOI] [PubMed] [Google Scholar]
- 24.Min, J., E. J. Kim, R. A. LaRossa, and M. B. Gu. 1999. Distinct responses of a recA::luxCDABE Escherichia coli strain to direct and indirect DNA damaging agents. Mutat. Res. 442:61-68. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, C. A., J. A. Cameron, S. J. Huang, and R. T. Vinopal. 1996. Fusarium polycaprolactone depolymerase is cutinase. Appl. Environ. Microbiol. 62:456-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niazi, J. H., D. T. Prasad, and T. B. Karegoudar. 2001. Initial degradation of dimethylphthalate by esterases from Bacillus species. FEMS Microbiol. Lett. 196:201-205. [DOI] [PubMed] [Google Scholar]
- 27.Nunoshiba, T., and H. Nishioka. 1991. ‘Rec-lac test' for detecting SOS-inducing activity of environmental genotoxic substances. Mutat. Res. 254:71-77. [DOI] [PubMed] [Google Scholar]
- 28.Okkels, J. S., M. Thellersen, D. A. Petersen, A. Svendsen, S. A. Patkar, and K. Borch. February1997. Novel lipolytic enzymes. Patent WO97/07202.
- 29.Petersen, J. H. 1991. Survey of di-(2-ethylhexyl) phthalate plasticizer contamination of retail Danish milks. Food Addit. Contam. 8:701-706. [DOI] [PubMed] [Google Scholar]
- 30.Petersen, M. T. N., P. Martel, E. I. Petersen, F. Drablos, and S. B. Petersen. 1997. Surface and electrostatics of cutinases. Methods Enzymol. 284:130-154. [DOI] [PubMed] [Google Scholar]
- 31.Picard, K., J.-C. Lhuguenot, M.-C. Lavier-Canivenc, and M.-C. Chagnon. 2001. Estrogenic activity and metabolism of n-butyl benzyl phthalate in vitro: identification of the active molecule(s). Toxicol. Appl. Pharmacol. 172:108-118. [DOI] [PubMed] [Google Scholar]
- 32.Price, C., E. Field, M. Marr, C. Myers, and R. Morrissey. 1990. Final report on the developmental toxicity of butyl benzyl phthalate (CAS no. 85-68-7) in CD-1 Swiss mice. NTP report 90-14. National Toxicology Program, Research Triangle Park, N.C.
- 33.Ptitsyn, L. R., G. Horneck, O. Komova, S. Kozubek, E. A. Krasavin, M. Bonev, and P. Rettberg. 1997. A biosensor for environmental genotoxin screening based on an SOS lux assay in recombinant Escherichia coli cells. Appl. Environ. Microbiol. 63:4377-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purdy, R. E., and P. E. Kolattukudy. 1975. Hydrolysis of plant cuticle by plant pathogens. Purification, amino acid composition, and molecular weight of two isozymes of cutinase and a nonspecific esterase from Fusarium solani f. pisi. Biochemistry 14:2824-2831. [DOI] [PubMed] [Google Scholar]
- 35.Sebastian, J., A. K. Chandra, and P. E. Kolattukudy. 1987. Discovery of a cutinase-producing Pseudomonas sp. cohabiting with an apparently nitrogen-fixing Corynebacterium sp. in the phyllosphere. J. Bacteriol. 169:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharman, M., W. A. Read, L. Castle, and J. Gilbert. 1994. Levels of di-(2-ethylhexyl)phthalate and total phthalate esters in milk, cream, butter and cheese. Food Addit. Contam. 11:375-385. [DOI] [PubMed] [Google Scholar]
- 37.Tetsuji, N., R. Ohta, H. Marumo, T. Shindo, S. Yoshimura, and H. Ono. 2000. Effect of butyl benzyl phthalate in Sprague-Dawley rats after gavage administration: a two-generation reproductive study. Reprod. Toxicol. 14:513-532. [DOI] [PubMed] [Google Scholar]
- 38.Unilever. March 1994. Enzyme-containing surfactant compositions. U.S. patent 94-04771.
- 39.Uriu-Adams, J. Y., C. K. Reece, L. K. Nguyen, B. J. Horvath, R. Nair, R. A. Barter, and C. L. Keen. 2001. Effect of butyl benzyl phthalate on reproduction and zinc metabolism. Toxicology 159:55-68. [DOI] [PubMed] [Google Scholar]
- 40.Van Dyk, T. K., W. R. Majarian, K. B. Konstantinov, R. M. Young, P. S. Dhurjati, and R. A. LaRossa. 1994. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl. Environ. Microbiol. 60:1414-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Dyk, T. K., D. R. Smulski, T. R. Reed, S. Belkin, A. C. Vollmer, and R. A. LaRossa. 1995. Responses to toxicants of an Escherichia coli strain carrying a uspA′::lux genetic fusion and an E. coli strain carrying a grpE′::lux fusion are similar. Appl. Environ. Microbiol. 61:4124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vollmer, A. C., S. Belkin, D. R. Smulski, T. K. Van Dyk, and R. A. LaRossa. 1997. Detection of DNA damage by use of Escherichia coli carrying recA′::lux, uvrA′::lux, or alkA′::lux reporter plasmids. Appl. Environ. Microbiol. 63:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zacharewski, T., K. L. Bondy, P. McDonnell, and Z. F. Wu. 1998. Identification and assessment of endocrine disruptors: limitations of in vivo and in vitro assays. Environ. Health Perspect. 106:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]