Abstract
In haploid strains of Saccharomyces cerevisiae, glucose depletion causes invasive growth, a foraging response that requires a change in budding pattern from axial to unipolar-distal. To begin to address how glucose influences budding pattern in the haploid cell, we examined the roles of bud-site-selection proteins in invasive growth. We found that proteins required for bipolar budding in diploid cells were required for haploid invasive growth. In particular, the Bud8p protein, which marks and directs bud emergence to the distal pole of diploid cells, was localized to the distal pole of haploid cells. In response to glucose limitation, Bud8p was required for the localization of the incipient bud site marker Bud2p to the distal pole. Three of the four known proteins required for axial budding, Bud3p, Bud4p, and Axl2p, were expressed and localized appropriately in glucose-limiting conditions. However, a fourth axial budding determinant, Axl1p, was absent in filamentous cells, and its abundance was controlled by glucose availability and the protein kinase Snf1p. In the bud8 mutant in glucose-limiting conditions, apical growth and bud site selection were uncoupled processes. Finally, we report that diploid cells starved for glucose also initiate the filamentous growth response.
INTRODUCTION
Cells of the yeast S. cerevisiae can undergo a developmental switch from a yeast form of growth to a filamentous form of growth (Gimeno et al., 1992; for reviews, see Kron, 1997; Madhani and Fink, 1998). In haploid cells, one of the triggers for the switch to the filamentous form is glucose starvation (Cullen and Sprague, 2000). The developmental switch to filamentation has at least three components. First, cells change their budding pattern. For example, haploid cells switch from an axial budding pattern, in which new buds emerge at sites adjacent to the birth scar or the site of the preceding bud, to a unipolar-distal budding pattern, in which new buds emerge at the pole distal to the birth scar (Roberts and Fink, 1994; Cullen and Sprague, 2000). Second, the cells become elongated. Third, the cell surface changes, enabling the cells to adhere to each other and to invade the agar substratum. In this article, we investigate the requirements for the change in budding pattern associated with filamentation in haploid cells. Are proteins required for bud site selection in yeast-form cells necessary for the budding pattern observed during filamentation? If so, which ones, and how are their activities modulated to affect the unipolar pattern during filamentation?
In vegetative cells, the budding pattern is controlled by cell type (Freifelder, 1960; Hicks et al., 1977; Chant and Pringle, 1995; for reviews, see Pringle et al., 1995; Herskowitz, 1997; Mata and Nurse, 1998; Madden and Snyder, 1998; Chant, 1999; Hales et al., 1999; Pruyne and Bretscher, 2000a,b). As described above, haploid cells bud in an axial pattern. Diploid cells, on the other hand, bud in a bipolar pattern. A new bud can emerge from either the birth scar pole or the distal pole, although there is a bias for distal pole budding in the first bud formed (Chant and Pringle, 1995). A GTPase module is required for cells to display either of these budding patterns; in its absence, cells bud in a random pattern (Bender and Pringle, 1989; Chant and Herskowitz, 1991; Chant et al., 1991). The module is composed of a RAS-like GTPase, Rsr1p/Bud1p; its GTPase-activating protein, Bud2p; and its guanine nucleotide exchange factor, Bud5p (Bender, 1993; Park et al., 1993). Bud2p and Bud5p are localized to axial positions in haploid cells and bipolar positions in diploid cells (Park et al., 1999; Kang et al., 2001; Marston et al., 2001), where they direct bud emergence, in part through interaction with polarity establishment proteins (Chant et al., 1991; Ruggieri et al., 1992; Zheng et al., 1995; Park et al., 1997).
The recruitment of the GTPase module to the appropriate site is controlled by other bud-site-selection proteins. In haploid cells, axial budding requires Bud3p, Bud4p, and Axl2p/Bud10p/Sro4p (Chant and Herskowitz, 1991; Chant and Pringle, 1995; Roemer et al., 1996; Halme et al., 1996). These proteins are localized to the mother-bud neck and together recruit Bud5p to the axial position (Chant et al., 1995; Sanders and Herskowitz, 1996; Kang et al., 2001). In addition, Axl1p is a haploid-specific protein required for axial budding (Fujita et al., 1994; Adames et al., 1995). Loss of Bud3p, Bud4p, Axl2p, or Axl1p causes bipolar budding in haploid cells, but does not affect budding pattern in diploid cells.
A different set of factors is required to orchestrate bipolar, rather than axial, budding in diploid cells. Genetic studies suggest that Bud8p and Bud9p mark the poles distal and proximal to the birth scar, respectively (Zahner et al., 1996; Harkins et al., 2001; Schenkman et al., 2002). For example, mutants deleted for BUD8 bud exclusively from the proximal pole. Moreover, green fluorescent protein (GFP)-tagging and immunofluorescence studies reveal that Bud8p is located at the distal pole and Bud9p at the proximal pole, implying that they may comprise part of the marks that identify these poles to the GTPase module (Harkins et al., 2001). In addition, Bud6p and Bni1p, which form a protein complex (Evangelista et al., 1997), are also required for bipolar budding (Amberg et al., 1997; Zahner et al., 1996; Sheu et al., 2000). Loss of any of these four proteins disrupts bipolar budding in diploid cells, but does not affect axial budding in haploid cells. Pea2p and Spa2p are also components of the Bud6p/Bni1p protein complex and are important determinants of bipolar budding and of polarized growth (Chenevert et al., 1994; Fujiwara et al., 1998; Sheu et al., 1998).
The switch in budding pattern during filamentous growth is particularly striking in the case of the axial-to-unipolar transition of haploid cells deprived of glucose (Cullen and Sprague, 2000). We show herein that Bud8p is localized to the distal tip of haploid cells, and under glucose-limiting conditions it directs bud emergence to the distal pole. Glucose depletion results in the Snf1p-dependent disappearance of Axl1p, providing one mechanism by which glucose modulates budding pattern in haploid cells.
MATERIALS AND METHODS
Strains, Plasmids, and Microbiological Techniques
The yeast strains used in this study are listed in Table 1. All of the strains were derived from HYL333 and HYL334 of the filamentous Σ1978b background (provided by G. Fink, Whitehead Institute for Biomedical Research, Cambridge, MA); these strains exhibit particularly robust filamentous growth compared with other strains from the Σ1978b background (H. Madhani, UCSF, San Francisco, CA; personal communication). To construct SY3687 and SY3688, HYL334 was made his3::URA3 or leu2::URA3, by using a polymerase chain reaction (PCR)-based method (Wach et al., 1994, and references therein) and plasmid pRS306 as a template (Sikorski and Hieter, 1989). The resulting strains were then made Ura3− by selection on 5-fluoroorotic acid (Biovectra, Oxford, CT). Disruption of BNI1 was performed using plasmid p321, provided by C. Boone (Evangelista et al., 1997). Disruptions of PEA2 and SPA2 were performed using plasmids pNV44 and p210, provided by I. Herskowitz (Valtz and Herskowitz, 1996). The plasmid used to disrupt GRR1, pBM2101, was provided by M. Johnston (Flick and Johnston, 1991). Other gene disruptions were performed by PCR-based methods (Wach et al., 1994, and references therein) to remove the entire open reading frame with plasmids described by Longtine et al. (1998), or other plasmids containing auxotrophic markers from Candida glabrata (for LEU2 and HIS3) and Kluyveromyces lactis (for URA3) and that were provided by I. Herskowitz. Integrated GFP fusions and GAL1 promoter fusions were made by PCR-based methods with plasmids provided by J. Pringle (Longtine et al., 1998). Gene disruptions and integrated promoter and protein fusions were confirmed by PCR analysis and by phenotype. All of the GFP- and hemagglutinin (HA)-tagged fusion proteins used in this study were functional with respect to bud site selection and invasive growth phenotypes.
Table 1.
Yeast strains
| Strain | Relevant genotype | Reference |
|---|---|---|
| HYL333 | MATaura3 | G. Fink, Cullen and Sprague, 2000 |
| HYL334 | MATα ura3 | G. Fink, Cullen and Sprague, 2000 |
| SY3687 | HYL334 his3∷ura3a | This study |
| SY3688 | HYL334 leu2∷ura3 | This study |
| SY3689 | SY3687 bud8∷KlURA3 | This study |
| SY3690 | SY3687 bud7∷KlURA3 | This study |
| SY3691 | SY3687 bud7∷KlURA3 bud8∷CgHIS3 | This study |
| SY3692 | SY3687 bud9∷KlURA3 | This study |
| SY3693 | HYL333 bud8∷Klura3 bud9∷KlURA3 | This study |
| SY3694 | SY3687 bud7∷KlURA3 bud9∷CgHIS3 | This study |
| SY3695 | SY3688 ura3 leu2 YEpGFP*-BUD8 | This study |
| SY3696 | SY3688 ura3 leu2 bud6∷URA3 YEpGFP*-BUD8 | This study |
| SY3697 | SY3688 ura3 leu2 bni1∷URA3 YEpGFP*-BUD8 | This study |
| SY3698 | SY3688 ura3 leu2 pea2∷URA3 YEpGFP*-BUD8 | This study |
| SY3699 | SY3688 ura3 leu2 spa2∷URA3 YEpGFP*-BUD8 | This study |
| SY3707 | SY3687 bud2∷CgHIS3 pHP726 | This study |
| SY3708 | SY3687 bud2∷CgHIS3 bud8∷Klura3 pHP726 | This study |
| SY3709 | HYL333 BUD3-GFP∷KanMX6 | This study |
| SY3710 | HYL333 BUD4-GFP∷KanMX6 | This study |
| SY3711 | HYL333 AXL2-GFP∷KanMX6 | This study |
| SY3712 | SY3687 bud3∷KlURA3 | This study |
| SY3713 | SY3687 bud4∷KlURA3 | This study |
| SY3714 | SY3687 axl2∷KlURA3 | This study |
| SY3715 | SY3687 bud3∷KlURA3 bud8∷CgHIS3 | This study |
| SY3716 | SY3687 bud4∷KlURA3 bud8∷CgHIS3 | This study |
| SY3717 | SY3687 axl2∷KlURA3 bud8∷CgHIS3 | This study |
| SY3718 | HYL333 p151 | This study |
| SY3720 | SY3688 snf1∷CgLEU2 p151 | This study |
| SY3721 | SY3687 axl1∷KlURA3 | This study |
| SY3722 | SY3687 axl1∷KlURA3 bud8∷CgHIS3 | This study |
| SY3723 | HYL333 GAL-AXL1∷KanMX6 | This study |
| SY3724 | SY3687 rsr1∷CgHIS3 | This study |
| SY3725 | SY3687 rsr1∷CgHIS3 bud8∷KlURA3 | This study |
| SY3726 | SY3687 grr1∷URA3 | This study |
| SY3727 | SY3687 rsr1∷CgHIS3 grr1∷URA3 | This study |
| SY3728 | SY3687 ste20∷URA3 | This study |
| SY3729 | SY3687 flo11∷KanMX6 | This study |
| SY3730 | SY3687 flo11∷KanMX6 bud8∷CgHIS3 | This study |
| SY3731 | SY3687 flo11∷KanMX6 pea2∷URA3 | This study |
| SY3732 | SY3687 pea2∷URA3 bud8∷CgHIS3 | This study |
| SY3733 | SY3687 pea2∷URA3 bud8∷CgHIS3 flo11∷KanMX6 | This study |
| SY3734 | HYL333/HYL334 | This study |
Strains containing the designation ura3 were made URA3 at the designated chromosomal locus and then ura3 by selection on 5-FOA as described in MATERIALS AND METHODS.
Yeast and bacterial strains were propagated using standard methods (Rose et al., 1990). Yeast peptone dextrose (YPD) and synthetic complete dextrose (SCD) media have been described previously (Rose et al., 1990). Yeast transformations were performed as described previously (Gietz et al., 1995). Bacterial transformations, bacterial DNA preparations, and plasmid constructions were performed by standard methods (Sambrook et al., 1989). Genes controlled by a galactose-inducible promoter were induced in SC or YP medium containing 2% galactose (Gal) as indicated. Geneticin (Biovectra) selection was performed as described previously (Longtine et al., 1998).
Protein Localization
The localization of Bud8p was determined using plasmid YEpGFP*-BUD8 (provided by J. Pringle; Harkins et al., 2001), in which the GFP-Bud8p fusion was expressed from its own promoter. Wild-type cells containing YEpGFP*-BUD8 were grown in synthetic medium lacking leucine (SCD-LEU) to stationary phase and spread onto SCD-LEU or SC-LEU medium for 16 h at 25°C. A coverslip was placed directly onto the plates, and GFP-Bud8p was visualized by fluorescence microscopy with a fluorescein isothiocyanate (FITC) filter at 100×. The localization of GFP-Bud2p was determined using plasmid pHP726 (provided by H.-O. Park; Park et al., 1999) carried in bud2 and bud2 bud8 strains. The localizations of Bud3p, Bud4p, and Axl2p were determined using C-terminal GFP fusions that were integrated into the genome. Actin staining was performed as described previously (Rose et al., 1990). Cells were incubated in SCD or SC medium and fixed in 3.7% formaldehyde for 1 h. Fixed cells were incubated with rhodamine (Rh)-phalloidin (Molecular Probes, Eugene, OR). Cells were washed twice and visualized by florescence microscopy at 100× by using an Rh filter.
Invasive Growth Assays
The single cell invasive growth assay was performed as described previously (Cullen and Sprague, 2000). For some experiments, cells were scraped from plates by using 4 ml of distilled water, concentrated by centrifugation, resuspended in 20 μl of water, and visualized by microscopy. For other experiments, a coverslip was placed directly on the agar medium and cells were visualized directly by microscopy. The plate-washing assay was performed essentially as described previously (Roberts and Fink, 1994). Equal concentrations of cells were spotted onto YPD or YPGal medium as specified, invasion was allowed to proceed for 2 d at 30°C, and then plates were washed vigorously with water and rubbed with a wet finger to remove cells that did not invade the agar. In some cases, invasion was allowed to proceed for 5 d, during which time the cells became more than twice as elongated as observed in the single cell assay. Cell-cell adhesion was assessed by a standard flocculation assay, as described previously (Guo et al., 2000).
Microscopy
Differential interference contrast (DIC) and fluorescence microscopy with Rh and FITC filter sets were performed using an Axioplan 2 microscope (Zeiss, Jena, Germany), a black-and-white Orca II digital camera (Hamamatsu, San Jose, CA), and the Openlab software program (Improvision, Coventry, UK). Only brightness and contrast digital adjustments were performed on photographs.
Budding Pattern Analysis
Budding pattern determination was performed as described previously (Chant and Pringle, 1995), with the following modifications. Equal concentrations of cells were spotted onto YPD medium and incubated for 2 d at 30°C. Plates were washed, and invaded cells were excised from the agar by using a toothpick. Cells were resuspended in water containing 1 μg/ml calcofluor (Sigma-Aldrich, St. Louis, MO), and after a 10-min incubation, bud scars were visualized directly by fluorescence microscopy. The enhanced cohesion of cells in the filamentous background facilitated the distinction between proximal and distal bud scars by their position relative to the cell-cell orientation. A bud scar was scored as distal if it was at the pole opposite to the birth scar, or if it was present at the distal pole of a cell that comprised a filament whose growth direction was obvious. A bud scar was scored as proximal if it was at the same pole as the birth scar or at the same pole as the attached parent cell. Bud scars in the middle third of the cell were scored as equatorial. At least 200 bud scars were scored for each experiment. Previously, wild-type cells in glucose-limiting conditions were shown to bud at the distal pole for 95% of all first buds (Cullen and Sprague, 2000). Subsequent buds were more frequently observed at the proximal pole. In the bud scar counts in the present work, all budding events were considered, resulting in the lower percentage of bud scars observed at the distal pole (∼70%).
Budding patterns were corroborated by using the single cell invasive growth assay (Cullen and Sprague, 2000). Equal concentrations of cells were spread onto SCD or SC medium, and budding pattern was assessed directly by microscopic examination. Microcolonies at the 10-cell stage or less were chosen for analysis. For some experiments cells were placed onto SCD or SC medium by micromanipulation and allowed to grow to the 10-cell stage, which showed the exact lineage of cells within the microcolony. The precise position of bud placement was determined for a subset of experiments by photographing cells and aligning the photographs to an arbitrary model cell.
Western Blot Analysis and Determination of Axl1p Abundance
Western blots were performed as described previously (Cullen et al., 2000). Proteins were separated by 10% SDS-PAGE, transferred to nitrocellulose, and visualized by probing with antibodies specific to GFP (Roche Applied Science, Indianapolis, IN), HA, or Dpm1p (provided by Tom Stevens, Institute of Molecular Biology, University of Oregon, Eugene, OR), which served as a loading control. Band intensity was determined using ImageQuant software (Amersham, Piscataway, NJ), and, where indicated, the values reported were normalized to Dpm1p levels. The abundance of Axl1p was measured using plasmid p151 (provided by C. Boone; Adames et al., 1995), which expresses a functional Axl1p-HA fusion protein expressed from the AXL1 promoter. In some experiments, Axl1p-HA abundance in yeast-form and filamentous cells was determined by incubating wild-type cells containing p151 (SY3718) on SCD-URA or SC-URA solid agar medium. Cells were harvested from plates, resuspended in water, and adjusted to equal density by measuring optical density. Proteins were then extracted and subjected to Western analysis. In other experiments, Axl1p-HA abundance was measured through the course of a growth cycle in cells incubated in SD-URA liquid medium for various times. In addition, the glucose-limited disappearance of Axl1p-HA was measured in p151-containing wild-type (SY3718) and snf1 mutant (SY3720) cells. Cells were grown to early log phase in liquid SCD-URA medium at 30°C, and each culture was split, washed twice with water, and incubated in liquid SC-URA or SCD-URA medium prewarmed to 30°C for various times.
RESULTS
Bud8p Is Required for Haploid Invasive Growth, whereas Bud9p Impedes Invasion
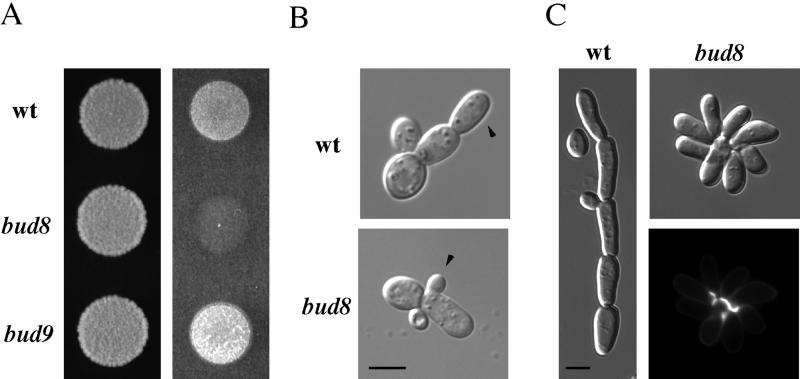
Disruption of BUD8 in a haploid strain of the filamentous background caused an invasive growth defect in the plate-washing assay (Figure 1A). In addition, both the single cell invasive growth assay (Cullen and Sprague, 2000) and bud scar staining demonstrated that the bud8 mutant was defective in budding at the distal pole (Figure 1B; Table 2). The mutant cells budded at the distal pole at a frequency of only 11%, in contrast to 70% for wild-type cells. The bud8 mutant cells were as elongated as wild-type cells (Figure 1C), suggesting that in the bud8 mutant, apical growth and the selection of budding sites were independent. The morphology of the bud8 mutant microcolony was a rosette, a morphology that contrasted strikingly with the linear form of wild-type filamentous cells (Figure 1C).
Figure 1.
Bud8p is required for agar invasion and distal pole budding in haploid cells; Bud9p impedes invasion. (A) Plate washing assay. Equal concentrations of wild-type (SY3687), bud8 (SY3689), and bud9 (SY3692) cells were spotted onto YPD medium and incubated for 2 d at 30°C. The plate was photographed (left), washed, and photographed again (right). (B) Single cell invasive growth assay. Equal concentrations of wild-type (top) or bud8 mutant (bottom) cells were spread onto SC medium, incubated for 16 h at 25°C, scraped from the plates, and photographed. Arrows indicate the first bud produced by the first daughter cell. (C) Prolonged incubation illustrates the difference between wild-type (left) and the bud8 mutant (right). Equal concentrations of cells were spotted onto YPD medium and grown for 5 d at 30°C. The lower right panel shows bud scar (calcofluor) staining for the bud8 mutant (Table 2). Bars, 5 μm.
Table 2.
Budding patterns of mutants known to have defects in bipolar bud site selection during haploid invasive growth
| Straina | Distal | Equatorial | Proximal |
|---|---|---|---|
| Wild type | 70 | 4 | 26 |
| bud8 | 11 | 2 | 87 |
| bud7 | 17 | 5 | 78 |
| bud8 bud7 | 4 | 3 | 93 |
| bud9 | 86 | 0 | 14 |
| bud8 bud9 | 10 | 2 | 88 |
| bud7 bud9 | 18 | 6 | 76 |
| bud6 | 45 | 36 | 19 |
| bni1 | 40 | 45 | 15 |
| pea2 | 71 | 12 | 17 |
| spa2 | 73 | 15 | 12 |
Strains are as indicated: wild type (SY3687), bud8 (SY3689), bud7 (SY3690), bud7 bud8 (SY3691), bud9 (SY3692), bud8 bud9 (SY3693), bud7 bud9 (SY3694), bud6 (SY3696), bni1 (SY3697), pea2 (SY3698), and spa2 (SY3699). Invaded cells were recovered, stained, and counted as described in MATERIALS AND METHODS
Disruption of BUD7 caused an invasive growth defect similar to that of the bud8 mutant in the plate-washing assay, although the bud7 mutant was slightly more invasive (our unpublished data). The bud7 mutant also exhibited a distal pole bud site selection defect (Table 2). Disruption of bud7 and bud8 together had an invasive growth defect equivalent to either single mutant, and the double mutant had a similar (although slightly more severe) budding-pattern defect than either single mutant (Table 2; our unpublished data). Taken together, these data suggest that Bud7p and Bud8p may be components of the same genetic pathway.
In contrast to the noninvasive phenotype of bud7 and bud8 mutants, the bud9 mutant exhibited hyperinvasive growth (Figure 1A). The bud9 mutant also had a higher percentage of distal pole buds compared with wild-type cells (Table 2), which may account for its hyperinvasive growth phenotype. Disruption of BUD8 in the bud9 mutant caused invasive-growth and distal-pole budding defects equivalent to those of the bud8 single mutant (Table 2), consistent with the budding pattern observed in bud8 bud9 homozygous diploid cells during vegetative growth (Zahner et al., 1996). Disruption of BUD7 also suppressed the hyperinvasive growth of the bud9 mutant but to a lesser extent than did disruption of BUD8, consistent with the phenotypes of the bud7 and bud8 single mutants (Table 2). In glucose-rich conditions, haploid bud7, bud8, and bud9 mutants did not show a budding pattern defect (Zahner et al., 1996; Harkins et al., 2001; our unpublished data). In summary, genes identified by virtue of their role in diploid cell budding pattern determination also have a role in haploid cells, specifically during invasive growth that occurs under glucose limitation.
Bud8p Is Localized to Distal Pole in Haploid Cells
Bud8p is localized to the distal pole of diploid cells (Taheri et al., 2000; Harkins et al., 2001). We examined the localization of Bud8p in haploid cells by using a plasmid containing a functional GFP-Bud8p fusion under the control of the BUD8 promoter (Harkins et al., 2001). GFP-Bud8p was observed at the distal pole of haploid cells grown in glucose-limiting conditions (Figure 2). GFP-Bud8p was also observed at the distal pole in glucose-rich conditions (Figure 2), a situation in which distal pole budding does not occur. Western blot analysis confirmed that the level of Bud8p was equivalent in glucose-rich and limiting conditions (our unpublished data). Thus, the Bud8p at the distal pole of haploid cells is apparently recognized only under glucose-limiting conditions.
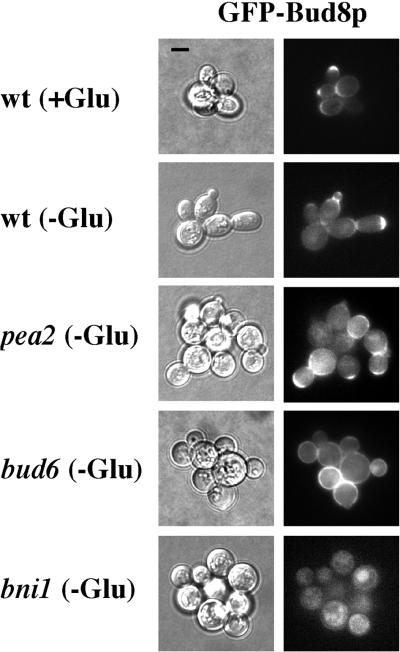
Figure 2.
Bud8p is localized to the distal tip of haploid cells, and its localization is dependent upon Bud6p and Bni1p. Strain backgrounds (wild-type, SY3695; pea2, SY3698; bud6, SY3696; and bni1, SY3697) are as shown. Growth on SCD (+Glu) or SC medium (−Glu) is as indicated. Left, DIC. Right, FITC filter. All pictures were taken at the same scale; bar, 5 μm.
In diploid cells, Bud8p's localization to the distal pole is dependent upon Bni1p (Ni and Snyder, 2001; Harkins et al., 2001). We examined the localization of Bud8p in bni1 and related mutants in haploid cells. Distal-pole localization of GFP-Bud8p was not observed in the bni1 mutant (<0.2% of cells had GFP-Bud8p at the distal pole compared with >50% for wild-type cells), but GFP-Bud8p was observed throughout the cell periphery (Figure 2). In the bud6 mutant, distal-pole localization of GFP-Bud8p was observed in a lower percentage of cells than wild type (5% of cells had GFP-Bud8p at the distal pole), and in bud6 cells in which GFP-Bud8p was at the distal pole, the fluorescence intensity was reduced (Figure 2). The abundance of GFP-Bud8p was equivalent in bud6, bni1, and wild-type cells (by Western blot). Consistent with the peripheral localization pattern of Bud8p, bud scar staining of invaded cells showed that bud6 and bni1 mutants had random budding patterns (Table 2). No budding pattern defects were detected in the mutants in glucose-rich conditions, under which Bud8p was also mislocalized; the cells showed normal axial budding. The Pea2p and Spa2p proteins were not required for distal-pole localization of Bud8p (Figure 2; our unpublished data), and pea2 and spa2 mutants maintained the unipolar budding pattern (Table 2). However, the pea2, spa2, bud6, and bni1 mutants were all defective in the extended apical growth that results in elongated cells during haploid invasion (Figure 2). Consequently, the four mutants all exhibited an invasive growth defect.
Bud8p Is Required for Distal Pole Localization of Bud2p and Actin in Glucose-limiting Conditions
Bud2p, the GTPase-activating protein for Rsr1p, has been shown to localize to the incipient bud site, where it presumably recruits Rsr1p to the bud site (Park et al., 1999). In diploid cells, this localization is dependent upon Bud8p (Kang et al., 2001). We examined the localization of a functional GFP-Bud2p fusion (provided by Hay-Oak Park, Ohio State University, Columbus, OH) expressed from a high-copy plasmid in bud2 and bud2 bud8 strains. In glucose-limiting conditions, Bud2p was observed at the distal pole of the cell, directly underneath the emerging bud, and at the mother-bud neck of small distal buds (Figure 3). In contrast, GFP-Bud2p localization in a bud2 bud8 mutant was mostly at the proximal pole (Figure 3), a result consistent with the proximal budding pattern observed in the bud8 mutant. In glucose-rich conditions, Bud2p was observed adjacent to the previous bud site (our unpublished observations; Park et al., 1999).
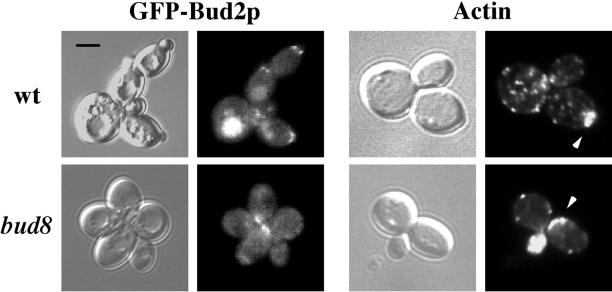
Figure 3.
Bud8p is required for localization of Bud2p and actin to the distal pole in glucose-limiting conditions. Left four panels, GFP-Bud2p localization. bud2 (top, SY3707) and bud2 bud8 (bottom, SY3708) strains containing a GFP-BUD2 fusion plasmid were grown in glucose-limiting conditions. Cells were photographed using DIC (left) or FITC filters (right). Right four panels, actin localization. Wild-type (SY3687) and bud8 mutant (SY3689) cells grown in glucose-limiting conditions were stained with rhodamine-phalloidin and photographed using DIC (left) or Rh filters (right). Arrows indicate regions of highly localized actin patches. All pictures were taken at the same scale; bar, 5 μm.
Bud site selection components (e.g., Bud8p and Bud2p) are known to recruit actin to the incipient bud site, an event required for bud emergence (Pruyne and Bretscher, 2000b). We examined actin localization in filamentous cells and found that in glucose-limiting conditions, actin localized to the distal tip of daughter cells (Figure 3). In contrast, actin accumulation was observed at the proximal pole in the bud8 mutant (Figure 3) and in wild-type cells grown in glucose-rich conditions (our unpublished data). Thus, glucose limitation caused the localization of Bud2p and bud emergence machinery (e.g., actin) to the distal pole of the cell, an event that was dependent upon Bud8p.
Proteins Required for Axial Budding Are Localized Appropriately in Filamentous Cells
We hypothesized that the disappearance of an axial cue in glucose-limiting conditions might result in Bud8p-dependent distal pole budding. In particular, the axial cues Bud3p and Bud4p are transient and are reported to disappear in nutrient-limiting conditions, because they are cell cycle regulated and absent in the Go phase of the cell cycle (Chant et al., 1995; Sanders and Herskowitz, 1996). The localization of the known axial cues (Bud3p, Bud4p, and Axl2p) was examined in cells grown in both glucose-rich and glucose-limiting conditions. Functional Bud3p-GFP, Bud4p-GFP, and Axl2p-GFP fusions were expressed from chromosomal loci and were found to be localized to the mother-bud neck in cells grown under both conditions (Figure 4A). Thus, an explanation other than axial cue disappearance must be invoked to explain the change in budding pattern to Bud8p-dependent bud site selection.
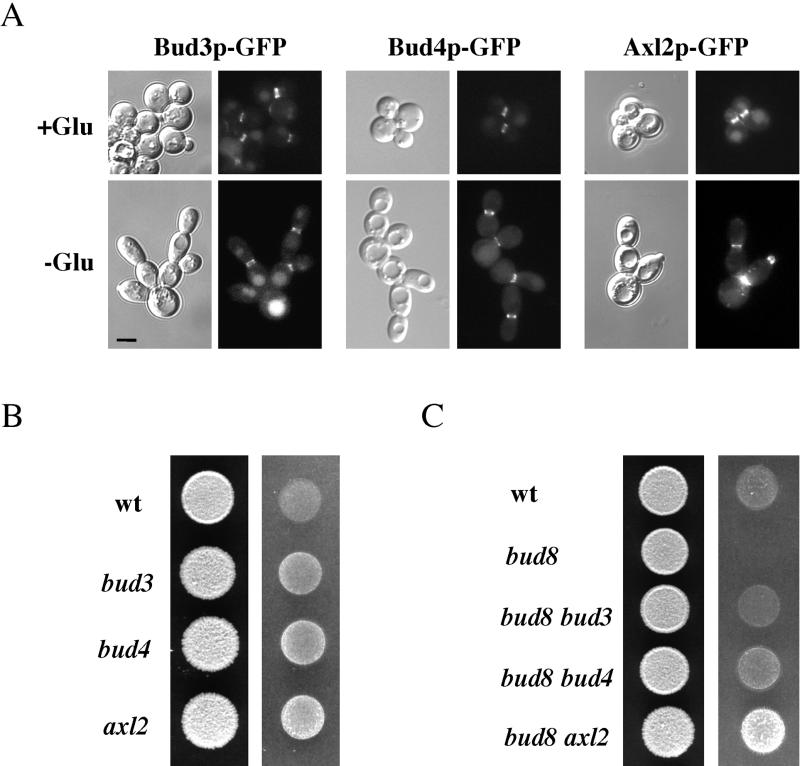
Figure 4.
Localization and role of axial cues in haploid filamentous cells. (A) Localization of axial cues in cells grown in glucose-rich and glucose-limiting conditions. Equal concentrations of cells (Bud3p-GFP, SY3709; Bud4p-GFP, SY3710; and Axl2p-GFP, SY3711) were spread onto either SCD (+Glu) or SC (−Glu) medium, incubated for 16 h at 25°C, scraped from plates, and photographed using DIC (left) or FITC filters (right). Bar (all panels), 5 μm. (B) Loss of axial cues causes hyperinvasive growth. Strains were spotted onto YPD medium and incubated for 2 d at 30°C. The plate was photographed (left), washed, and photographed again (right). Strains: wt (SY3687), bud3 (SY3712), bud4 (SY3713), and axl2 (SY3714). (C) Suppression of the bud8 invasive growth defect by disruption of axial cues. Strains were spotted onto YPD medium and incubated for 2 d at 30°C. The plate was photographed (left), washed, and photographed again (right). Strains: wt (SY3687), bud8 (SY3689), bud8 bud3 (SY3715), bud8 bud4 (SY3716), and bud8 axl2 (SY3717).
Although Bud8p is the primary bud site cue in haploid cells undergoing filamentous growth, genetic evidence suggests that axial cues are used to some degree. First, disruption of BUD3, BUD4, or AXL2 caused hyperinvasive growth (Figure 4B), due to a significant decrease in bud site selection at the proximal pole (Table 3). Second, disruption of axial cues in the bud8 mutant ablated the proximal budding (Table 3), correlating with partial suppression of the invasive growth defect (Figure 4C). The increased percentage of distal pole budding in the axl2 bud8 mutant, compared with the bud3 bud8 and bud4 bud8 mutants, may be due to the enhanced apical growth that was observed in axl2 mutant cells (our unpublished data).
Table 3.
Budding patterns of mutants lacking axial-specific cues or both axial cues and Bud8p
| Straina | Distal | Equatorial | Proximal |
|---|---|---|---|
| Wild type | 69 | 4 | 27 |
| bud3 | 87 | 9 | 4 |
| bud4 | 95 | 4 | 1 |
| axl2 | 93 | 4 | 3 |
| bud8 | 10 | 2 | 88 |
| bud8 bud3 | 37 | 29 | 34 |
| bud8 bud4 | 37 | 31 | 32 |
| bud8 axl2 | 59 | 36 | 5 |
Strains are as indicated: wild type (SY3687), bud3 (SY3712), bud4 (SY3713), axl2 (SY3714), bud8 (SY3689), bud8 bud3 (SY3715), bud8 bud4 (SY3716), and bud8 axl2 (SY3717). Growth and bud scar staining were performed as described in Table 2.
Control of Axl1p Abundance by Glucose and Snf1p
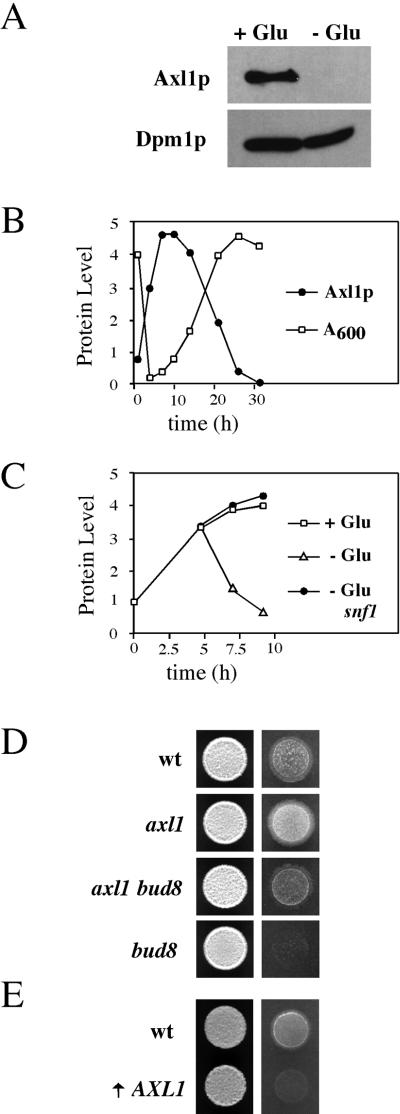
Because AXL1 is transcriptionally repressed in diploid cells, an event sufficient to prevent axial budding in wild-type cells (Fujita et al., 1994), we considered the possibility that Axl1p protein abundance is regulated by glucose in haploid cells. Axl1p protein levels were measured in cells expressing an Axl1p-HA fusion from the AXL1 promoter (provided by C. Boone). We initially observed that Axl1p-HA was not present in filamentous cells (Figure 5A). Axl1p-HA abundance was examined in cells grown throughout a culture growth cycle. In reference to a control protein, the amount of Axl1p-HA increased steadily during early log phase and was highest in mid-log phase (Figure 5B). Axl1p-HA abundance declined as growth rate slowed and was significantly reduced in stationary phase (Figure 5B). To confirm that glucose influenced Axl1p abundance, cells containing the Axl1p-HA fusion were grown to early log phase and shifted to medium lacking glucose. As expected, the level of Axl1p-HA declined markedly upon the shift to glucose-limited medium (Figure 5C).
Figure 5.
Axl1p abundance is controlled by glucose and Snf1p, and Axl1p is an inhibitor of invasive growth. (A) Axl1p is absent in filamentous cells. Western blot of protein extracts from wild-type cells containing p151 (SY3718) incubated on SCD-URA (+Glu) or SC-URA (−Glu) solid agar medium for 16 h at 25°C and probed with antibodies against HA (to detect Axl1p-HA) or Dpm1p (see MATERIALS AND METHODS). (B) Abundance of Axl1p correlates with the culture growth cycle. Wild-type cells containing p151 (SY3718) were grown through a growth cycle in SCD-URA liquid medium at 30°C, and protein extracts were harvested from cells at the times indicated. Quantitation of Western blots of Axl1p levels (adjusted to Dpm1p levels) is shown (filled circles). Optical density at 600 nm is also shown (open squares). (C) Decrease in Axl1p abundance upon a shift to glucose-limited medium is dependent upon Snf1p. Wild-type cells containing p151 (SY3718) were grown to early log phase and shifted to SCD-URA (open squares, +Glu) or SC-URA (open triangles, −Glu) medium. A snf1 mutant containing p151 (SY3720) was grown in the same way (closed circles, −Glu). Quantitation of Western blots of Axl1p levels (adjusted to Dpm1p levels) is shown. (D) Hyperinvasive growth in axl1 mutants. Equal concentrations of wild-type (SY3687), axl1 (SY3721), axl1 bud8 (SY3722), and bud8 (SY3689) cells were spotted onto YPD medium and incubated for 2 d at 30°C. The plate was photographed (left), washed, and photographed again (right). The colonies shown are all from the same plate. (E) Overproduction of Axl1p prevents invasive growth. Wild-type (SY3687) and GAL1-AXL1 (↑ AXL1, SY3723) cells were grown to saturation in YPGal medium, and equal concentrations of cells were spotted onto YPGal medium for 2 d at 30°C. The plates were photographed (left), washed, and photographed again (right).
The Snf1p protein kinase is a global regulator of glucose response (Carlson 1999), and we showed previously that Snf1p is required for unipolar budding during haploid invasive growth (Cullen and Sprague, 2000). We investigated the role of Snf1p in the glucose-dependent regulation of Axl1p. In contrast to the observations reported above for wild-type cells, Axl1p-HA protein abundance remained high in a snf1 mutant after a shift to glucose-limited medium (Figure 5C). Thus, Snf1p is required for the disappearance of Axl1p in glucose-limiting conditions.
Genetic analysis also supports a role for Axl1p in the transition to filamentous growth. Disruption of AXL1 caused hyperinvasive growth (Figure 5D) due to distal pole budding in both glucose-rich and glucose-limiting conditions (Table 4). These phenotypes were largely suppressed by disruption of BUD8 (Figure 5D and Table 4). The axl1 bud8 double mutant invades better than the bud8 single mutant because it has constitutive nonaxial budding, whereas the bud8 single mutant buds almost exclusively from the proximal pole under both glucose-rich and glucose-limiting conditions. In contrast, overexpression of AXL1 suppressed agar invasion by wild-type cells (Figure 5E) due to an increase in proximal budding (Table 4). Thus, the disappearance of the Axl1p protein in glucose-limiting conditions is sufficient to explain the Bud8p-dependent budding during haploid invasive growth.
Table 4.
Budding patterns of strains deleted for AXL1 and/or BUD8 or overexpressing AXL1
| Genotype | Distal | Equatorial | Proximal |
|---|---|---|---|
| Wild-type (+Glu)a | 4 | 1 | 95 |
| axl1 (+Glu) | 55 | 4 | 41 |
| axl1 bud8 (+Glu) | 16 | 37 | 47 |
| Wild-type (−Glu)b | 69 | 1 | 30 |
| axl1 (−Glu) | 82 | 10 | 8 |
| axl1 bud8 (−Glu) | 25 | 9 | 66 |
| bud8 (−Glu) | 10 | 1 | 89 |
| Wild type (YPGal)c | 30 | 0 | 70 |
| GAL1-AXL1 (YPGal) | 12 | 1 | 87 |
For +Glu, cells were grown in YPD to mid-log phase, and bud scars were stained and counted as described in MATERIALS AND METHODS. Strains are as indicated: wild type (SY3687), axl1 (SY3721), axl1 bud8 (SY3722), bud8 (SY3689), and GAL-AXL1 (SY3723).
For −Glu, invaded cells were collected, stained, and counted as described in MATERIALS AND METHODS.
Cells were spotted onto solid agar YPGal medium for 24 h and prepared as for the −Glu experiments (see footnote b).
Unipolar Budding in rsr1 Mutant Due to Increased Apical Growth during Haploid Invasion
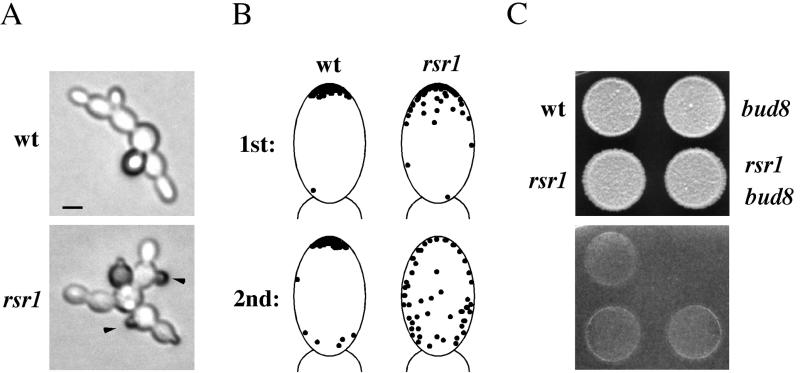
It has been reported that ablation of the general bud-site-selection machinery does not disrupt the ability of haploid cells to undergo agar invasion (Roberts and Fink, 1994; Lo et al., 1997). This result is seemingly at odds with our finding that mutants that are defective for distal-pole budding cannot invade agar. Therefore, we characterized the rsr1 mutant, which is lacking the core bud-site-selection GTPase, in detail. In the single cell invasive growth assay, the rsr1 mutant formed filaments composed of elongated cells emanating away from the mother cell, suggesting a unipolar-distal pattern (Figure 6A); however, buds emerging from the equatorial regions of cells were also observed (Figure 6A, black arrows). To determine more precisely the budding pattern of first and second buds for the rsr1 mutant, cells were placed onto glucose-limited medium by micromanipulation and budding pattern was assessed by microscopic examination. Strikingly, the first bud produced was at the distal pole >90% of the time (Table 5). The second buds emerged uniformly around the entire surface of the cell (Table 5). In glucose-rich conditions, a less dramatic distal-pole bias was observed in the rsr1 mutant (Table 5), consistent with previous reports (Chant and Herskowitz, 1991; Bender, 1993; Michelitch and Chant, 1996).
Figure 6.
Distal pole budding in the rsr1 mutant during haploid invasive growth. (A) Wild-type (SY3687) and rsr1 mutant (SY3724) cells were spread onto SC medium, incubated for 16 h at 25°C, and photographed. Bar, 5 μm. (B) Diagram of precise position of buds as determined by microscopic examination. Microcolonies at the 10-cell stage (or less) were visualized by light microscopy and photographed. Photographs were analyzed for bud position; only buds whose precise locations were unambiguous were chosen for analysis. The position of 50 buds is shown for each depiction. (C) Bud8p is not required for agar-invasion in the rsr1 mutant. Wild-type (SY3687), bud8 (SY3689), rsr1 (SY3724), and rsr1 bud8 mutant (SY3725) cells were spotted onto YPD medium and grown for 2 d at 30°C. Plates were photographed (top), washed, and photographed again (bottom).
Table 5.
Bud position of the rsr1 mutant grown on glucose-rich or glucose-limited medium
| Genotype | Bud | Distal | Equatorial | Proximal |
|---|---|---|---|---|
| Wild type (SC)a | 1st | 97 | 0 | 3 |
| rsr1 (SC) | 1st | 95 | 2.5 | 2.5 |
| Wild type (SC) | 2nd | 88 | 2 | 10 |
| rsr1 (SC) | 2nd | 29 | 35.5 | 35.5 |
| rsr1 bud8 (SC) | 1st | 95 | 2 | 3 |
| Wild type (SCD) | 1st | 5 | 0 | 95 |
| rsr1 (SCD) | 1st | 57 | 29 | 14 |
| grr1 (SCD) | 1st | 14 | 0 | 86 |
| rsr1 grr1 (SCD) | 1st | 85 | 11 | 4 |
Strains are as indicated: wild type (SY3687), rsr1 (SY3724), rsr1 bud8 (SY3725), grr1 (SY3726), and rsr1 grr1 (SY3727). Microcolonies of <10 cells were examined for bud placement by microscopy, and all observed microcolonies were included in the analysis. Bud position was considered distal if the bud was observed in the distal third of the cell; equatorial, for the middle third of the cell; and proximal, for the proximal third of the cell. For each sample, >200 buds were counted, of which <20 were ambiguous; these were not included in the final percentage. Analysis was performed on glucose-limited (SC) or glucose-rich (SCD) medium.
We hypothesized that the distal-pole budding observed in the rsr1 mutant was due to the lengthened period of apical growth that leads to cell elongation during haploid invasion (Ahn et al., 1999). Precise mapping of bud placement showed that in the rsr1 mutant, buds emerged within an arc that included the distal pole, whereas in wild-type cells, bud emergence was confined to the extreme tip of the cell (Figure 6B), suggesting that the mechanism of distal budding in the rsr1 mutant was different than in wild-type cells. Indeed, disruption of BUD8 did not affect distal-pole budding (Table 5) or agar invasion of the rsr1 mutant (Figure 6C), demonstrating that the distal-pole budding in the rsr1 mutant was not due to Bud8p-dependent bud site selection. To gain further support for the idea that enhanced apical growth can influence budding pattern in the rsr1 mutant, we used the grr1 mutation (Flick and Johnston, 1991), which causes hyperpolarized growth (Barral et al., 1995; Sheu et al., 2000). grr1 mutant cells were elongated in glucose-rich conditions, but their budding pattern was axial (Table 5). The rsr1 grr1 double mutant, however, had a clear distal pole bias compared with the rsr1 single mutant in glucose-rich medium (Table 4), supporting the idea that in the rsr1 mutant, hyperpolarized growth leads to distal-pole budding.
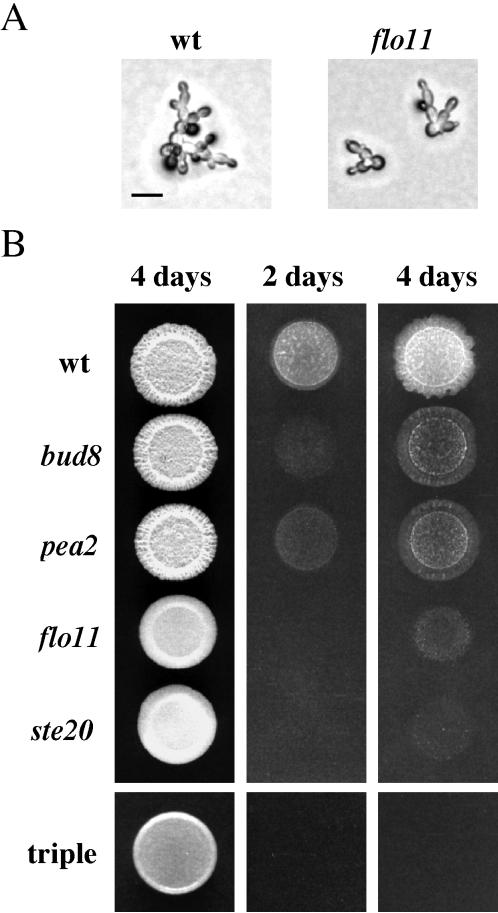
Contribution of Different Aspects of Filamentous Growth to Agar Invasion
Filamentous growth is characterized by several physiological events: a change in budding pattern, an increase in cell length, and enhanced cell-cell adhesion. We found that disruption of FLO11, which is required for cell-cell adhesion and haploid invasive growth (Lo and Dranginis, 1998; Palecek et al., 2000), did not affect cell elongation or unipolar-distal budding (Figure 7A), implicating cell-cell adhesion as the primary defect in the flo11 mutant. We directly compared mutants defective primarily in a single aspect of filamentous growth to a ste20 mutant, which is defective in all three aspects. In particular, we compared a bud8 mutant (defective for distal-pole budding but not elongation or adhesion), a pea2 mutant (defective for elongation but not distal-pole budding or adhesion), and a flo11 mutant (defective for adhesion but not distal-pole budding or elongation). A defect in any single aspect of filamentous growth caused a partial invasive growth defect, although the flo11 mutant was less invasive than the pea2 and bud8 mutants (Figure 7B). These results were extended by examining the phenotypes of double and triple mutants. The bud8 pea2, bud8 flo11, and pea2 flo11 double mutants were less invasive than any single mutant, and in the pea2 bud8 flo11 triple mutant, no agar invasion was observed (Figure 7B), implying that the three physiological events affected by these mutations are the major contributors to agar invasion.
Figure 7.
Genes involved primarily in a single aspect of filamentous growth contribute independently to agar invasion. (A) Flo11p is not required for unipolar-distal budding or cell elongation. Wild-type (SY3687) and flo11 (SY3729) cells were spread onto SC medium, incubated for 16 h at 25°C, and photographed. Bar, 20 μm. (B) Contribution of individual filamentation functions to agar-invasion. Equal concentrations of cells were spotted onto YPD medium and grown for 2 d or 4 d at 30°C, as indicated. Plates were photographed, washed, and photographed again. Strains are as indicated: wild-type (SY3687), bud8 (SY3689), pea2 (SY3698), flo11 (SY3729), ste20 (SY3728), and triple, the pea2 bud8 flo11 triple mutant (SY3733).
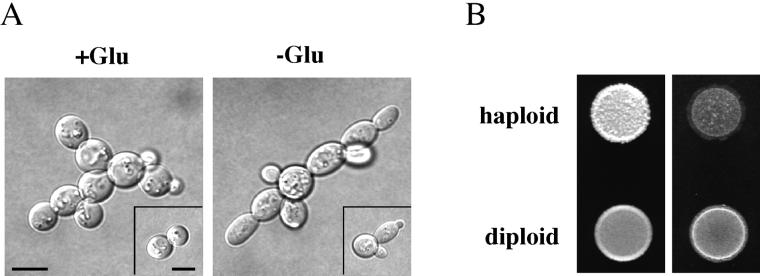
Diploid Cells Exhibit Filamentous Growth in Response to Glucose Depletion
Filamentous growth has been thought to be induced by different cues in haploid cells (glucose limitation; Cullen and Sprague, 2000) and diploid cells (nitrogen limitation; Gimeno et al., 1992). We investigated the effect of glucose depletion on diploid cells and found that on glucose-limited medium, diploid cells were more elongated than on glucose-rich medium (Figure 8A). The elongated phenotype was apparent within the first cell division and was observed for >80% of cells. In addition to the increase in cell length, diploid cells budded in a unipolar-distal pattern upon glucose limitation. Bud scar staining of invaded diploid cells showed unipolar-distal bud scars 85% of the time (15% had scars at both poles); in contrast, diploid cells in glucose-rich medium had unipolar-distal bud scars 51% of the time (45% had scars at both poles and 4% had at least one scar in an equatorial site). Wild-type diploid cells also exhibited robust agar invasion by the plate-washing assay (Figure 8B). Thus, diploid cells can undergo filamentous growth in response to limiting glucose.
Figure 8.
Diploid cells exhibit filamentous growth in response to limiting glucose. (A) Single cell assay. Diploid cells (SY3734) were spread onto SC (−Glu) and SCD (+Glu) medium, incubated for 16 h at 25°C, and photographed. Bars, 10 μm. (B) Equal concentrations of diploid cells (SY3734) and haploid cells (SY3687) were spotted onto YPD medium and grown for 2 d at 30°C. Plates were photographed (left), washed, and photographed again (right).
DISCUSSION
Bipolar Bud-Site-Selection Components Are Required for Haploid Invasive Growth
We investigated the role of bud-site-selection components in haploid invasive growth and found that proteins required for bipolar budding in diploid cells were required in haploid cells for distal pole budding during invasive growth. In particular, our data indicate that Bud8p marks the distal pole of haploid cells and is recognized upon glucose depletion to direct budding to the distal pole. Bud8p had not previously been implicated in haploid invasion (Lorenz et al., 2000; Palecek et al., 2000), perhaps because bud8 mutants exhibit only a partial invasive growth defect. We have shown, however, that Bud8p has a profound role in bud site selection under conditions of glucose limitation, even though it has no known role when glucose is abundant. This contrasts with Bud8p's role in diploid cells, where it controls budding pattern during both yeast form and filamentous growth (Mösch and Fink, 1997; Taheri et al., 2000). Thus, an additional level of regulation is required in haploid cells to prevent Bud8p from directing budding pattern in glucose-rich conditions (see below).
Our results also indicate that Bud7p, Bud9p, Bni1p, and Bud6p are required in glucose-limiting conditions for normal haploid invasive growth. As in diploid cells, Bni1p is required in haploid cells to localize Bud8p to the distal pole. We have also found that Bud6p is important, although not essential, for Bud8p localization in haploid cells, an apparent difference from previously reported results with diploid cells (Harkins et al., 2001); this may either represent a genuine difference between haploid and diploid cells or just be a function of different strain backgrounds. (Note that previous studies have also shown less precise use of the distal pole in bud6 mutant diploids; Zahner et al., 1996; Amberg et al., 1997; Sheu et al., 2000). Two pieces of evidence in our study support the conclusion that the bipolar budding machinery is in place, albeit dormant, in haploid cells. First, Bud8p is present and localized appropriately in haploid cells in glucose-rich conditions. Second, Bni1p is required for localization of Bud8p to the distal pole in glucose-rich conditions. Because these proteins are not required for bud site selection during yeast-form growth in haploid cells, we suggest that the bipolar budding machinery constitutes a default program in haploid cells that becomes active in glucose-limiting conditions.
Indirect evidence from a number of studies has indicated that haploid cells have properly located proximal and distal cues (Chant and Herskowitz, 1991; Chant et al., 1995; Chant and Pringle, 1995; Roemer et al., 1996; Sanders and Herskowitz, 1996; Madden and Snyder, 1992; Erdman and Snyder, 2001), and we have provided direct evidence for this conclusion. In glucose-rich environments, the axial Bud3p/Bud4p/Axl2p cues are chosen, whereas glucose limitation causes the distal cue Bud8p to be chosen in preference to Bud3p/Bud4p/Axl2p. The presence of both sets of cues enables an individual cell to reorient bud growth rapidly in response to changing nutrient availability.
Glucose Controls Bud Site Selection by the Snf1p-dependent Regulation of Axl1p
How is unipolar-distal budding prevented in haploid cells growing in glucose-rich conditions? We show here that the abundance of the axial-promoting factor Axl1p is controlled by glucose. Axl1p levels decline sharply upon glucose limitation (Figure 5), an event that is concomitant with the appearance of unipolar buds (our unpublished data). Thus, it is reasonable to speculate that the disappearance of Axl1p is a trigger for distal-pole budding. We found that the Snf1p protein kinase, which plays a role in the derepression of glucose-repressed genes (Carlson, 1999), is required for the disappearance of Axl1p in glucose-limiting conditions. Snf1p may exert its effect on Axl1p indirectly, by allowing derepression of a gene whose product regulates Axl1p. Alternatively, Snf1p may directly phosphorylate Axl1p and target it for degradation. Irrespective of the mechanism by which Snf1p regulates Axl1p abundance, the genetic evidence that we have presented identifies Axl1p as an important regulator of haploid invasive growth. Loss of Axl1p permits Bud8p-dependent unipolar-distal budding, whereas overexpression of Axl1p suppresses unipolar-distal budding and agar invasion in glucose-limiting conditions. How Axl1p functions to direct budding to sites marked by Bud3p/Bud4p/Axl2p is not known but is a question that is crucial to the ultimate understanding of bud site determination in yeast.
Coordination of Unipolar Budding and Polarized Growth
A lengthened period of polarized, apical growth promotes bipolar budding in diploid cells (Sheu et al., 2000) and also promotes unipolar-distal budding in haploid cells during invasive growth (Ahn et al., 1999). In fact, in this latter case, we showed that an extended period of apical growth could confer distal-pole budding to a mutant lacking a functional bud-site-selection system (rsr1 mutant). In wild-type cells, polarized growth and localization of the bud site machinery are coordinated. For example, Bni1p, which is involved in polarized growth, is also required for localization of Bud8p to the distal tip of the daughter cell in both haploids (Figure 2) and diploids (Harkins et al., 2001). These two processes are not inextricably linked, however. Pea2p (and Spa2p) were shown to be required for cell elongation during invasive growth but not for Bud8p localization or distal-pole budding. Conversely, loss of Bud8p disrupted the budding pattern but did not affect cell elongation. Thus, two distinct cues mark the distal pole of daughter cells: a bud site cue (Bud8p) to direct bud site selection and a second cue to direct polarized growth.
The notion that the components of invasive growth could be genetically isolated was extended to cell-cell adhesion. Disruption of FLO11 prevented adhesion but had no effect on cell elongation or the budding pattern. Thus, haploid invasive growth can be divided into three separate processes: cell elongation, unipolar-distal budding, and cell adhesion. Loss of any one of these processes partially compromises invasive growth; loss of all three prevents it entirely.
Diploid Cells Starved for Glucose Initiate the Filamentous Growth Response
Filamentous growth was first characterized in diploid cells and was described as a response to nitrogen limitation (Gimeno et al., 1992). Mutations in nitrogen-sensing and nitrogen utilization pathways confirmed that pseudohyphal growth was a response to low levels of environmental fixed nitrogen (Mösch and Fink, 1997). Subsequently, haploid cells were also shown to undergo a filamentation-like process in which they invaded the agar substratum (Roberts and Fink, 1994). Diploid pseudohyphal growth and haploid invasive growth were presumed to be similar processes because they required some of the same signal transduction pathways, but differences are apparent, in particular in the degree of agar invasiveness. Recently, we showed that glucose depletion was a trigger for haploid invasive growth (Cullen and Sprague, 2000). Hence, it was proposed that diploid cells initiate filamentous growth in response to limiting nitrogen, whereas glucose depletion triggers haploid invasive growth (Madhani, 2000). However, we have shown here that diploid cells manifest all of the characteristics of filamentous growth in response to glucose limitation. Perhaps variation within the Σ1278b background has caused confusion as to the cues that underlie filamentous growth. For example, strains of yeast capable of starch degradation have been reported to initiate filamentous growth upon either carbon or nitrogen source depletion (Lambrechts et al., 1996). Diploid cells sporulate upon limitation of both carbon and nitrogen cues (Mitchell, 1994); depletion of either single cue, however, triggers filamentous growth.
ACKNOWLEDGMENTS
We thank John Pringle, Gerald Fink, Michael Snyder, Hiten Madhani, Hans-Ulrich Mösch, Charlie Boone, Ira Herskowitz, Hay-Oak Park, Tom Stevens, Mark Johnston, Henry Baker, and April Goehring for providing advice, strains, plasmids, and/or antibodies. We also thank Dave Mitchell, Greg Smith, Hilary Kemp, Megan Keniry, David Rivers, Elizabeth Monika, and Hailey Rose for comments and suggestions. This work was supported by research (GM-30027 to G.F.S.) and training (GM-19188 to P.J.C.) grants from the U.S. Public health service and by a fellowship (AHA-120635Z) from the American Heart Association.
Abbreviations used:
- DIC
differential interference contrast
- FITC
fluorescein
- GFP
green fluorescent protein
- HA
hemagglutinin
- Rh
rhodamine
- SC
synthetic complete
- URA
uracil
- YPD
yeast peptone dextrose
- wt
wild-type
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–03–0151. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–03–0151.
REFERENCES
- Adames N, Blundell K, Ashby MN, Boone C. Role of yeast insulin-degrading enzyme homologs in propheromone processing and bud site selection. Science. 1995;270:464–467. doi: 10.1126/science.270.5235.464. [DOI] [PubMed] [Google Scholar]
- Ahn SH, Acurio A, Kron SJ. Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol Biol Cell. 1999;10:3301–3316. doi: 10.1091/mbc.10.10.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- Bender A, Pringle JR. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene. RSR1. Proc Natl Acad Sci USA. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A. Genetic evidence for the roles of the bud-site-selection genes BUD5 and BUD2 in control of the Rsr1p (Bud1p) GTPase in yeast. Proc Natl Acad Sci USA. 1993;90:9926–9929. doi: 10.1073/pnas.90.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- Chant J. Cell polarity in yeast. Annu Rev Cell Dev Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- Chant J, Corrado K, Pringle JR, Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene. BEM1. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Chant J, Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert J, Valtz N, Herskowitz I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics. 1994;136:1287–1296. doi: 10.1093/genetics/136.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Schultz J, Horecka J, Stevenson BJ, Jigami Y, Sprague GF., Jr Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics. 2000;155:1005–1018. doi: 10.1093/genetics/155.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF., Jr Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci USA. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S, Snyder M. A filamentous growth response mediated by the yeast mating pathway. Genetics. 2001;159:919–928. doi: 10.1093/genetics/159.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Flick JS, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder D. Bud position in Saccharomyces cerevisiae. J Bacteriol. 1960;80:567–568. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A, Oka C, Arikawa Y, Katagai T, Tonouchi A, Kuhara S, Misumi Y. A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature. 1994;372:567–570. doi: 10.1038/372567a0. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci USA. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A, Michelitch M, Mitchell EL, Chant J. Bud10p directs axial cell polarization in budding yeast and resembles a transmembrane receptor. Curr Biol. 1996;6:570–579. doi: 10.1016/s0960-9822(02)00543-2. [DOI] [PubMed] [Google Scholar]
- Hales KG, Bi E, Wu J-Q, Adam JC, Yu I-C, Pringle JR. Cytokinesis: an emerging unified theory for eukaryotes? Curr Opin Cell Biol. 1999;11:717–725. doi: 10.1016/s0955-0674(99)00042-3. [DOI] [PubMed] [Google Scholar]
- Harkins HA, Page′ N, Schenkman LR, De Virgilio C, Shaw S, Bussey H, Pringle JR. Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol Biol Cell. 2001;12:2497–2518. doi: 10.1091/mbc.12.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Building organs and organisms: elements of morphogenesis exhibited by budding yeast. Cold Spring Harb Symp Quant Biol. 1997;62:57–63. [PubMed] [Google Scholar]
- Hicks JB, Strathern JN, Herskowitz I. Interconversion of yeast mating types. III. Action of the homothallism (HO) gene in cells homozygous for the mating type locus. Genetics. 1977;85:395–405. doi: 10.1093/genetics/85.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang PJ, Sanson A, Lee B, Park H-O. A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science. 2001;292:1376–1378. doi: 10.1126/science.1060360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ. Filamentous growth in budding yeast. Trends Microbiol. 1997;5:450–454. doi: 10.1016/S0966-842X(97)01131-1. [DOI] [PubMed] [Google Scholar]
- Lambrechts MG, Bauer FF, Marmur J, Pretorius IS. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Dranginis AM. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Raitses EI, Dranginis AM. Development of pseudohyphae by embedded haploid and diploid yeast. Curr Genet. 1997;32:197–202. doi: 10.1007/s002940050266. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, DeMarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Cutler NS, Heitman J. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Snyder M. Specification of sites for polarized growth in Saccharomyces cerevisiae and the influence of external factors on site selection. Mol Biol Cell. 1992;3:1025–1035. doi: 10.1091/mbc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- Madhani HD. Interplay of intrinsic and extrinsic signals in yeast differentiation. Proc Natl Acad Sci USA. 2000;97:13461–13463. doi: 10.1073/pnas.011511198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- Marston AL, Chen T, Yang MC, Belhumeur P, Chant J. A localized GTPase exchange factor, Bud5, determines the orientation of division axes in yeast. Curr Biol. 2001;11:803–807. doi: 10.1016/s0960-9822(01)00230-5. [DOI] [PubMed] [Google Scholar]
- Mata J, Nurse P. Discovering the poles in yeast. Trends Cell Biol. 1998;8:163–167. doi: 10.1016/s0962-8924(98)01224-0. [DOI] [PubMed] [Google Scholar]
- Michelitch M, Chant J. A mechanism of Bud1p GTPase action suggested by mutational analysis and immunolocalization. Curr Biol. 1996;6:446–454. doi: 10.1016/s0960-9822(02)00512-2. [DOI] [PubMed] [Google Scholar]
- Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch H-U, Fink GR. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek SP, Parikh AS, Kron SJ. Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics. 2000;156:1005–1023. doi: 10.1093/genetics/156.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-O, Bi E, Pringle JR, Herskowitz I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci USA. 1997;94:4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-O, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- Park H-O, Sanson A, Herskowitz I. Localization of Bud2p, a GTPase-activating protein necessary for programming cell polarity in yeast to the presumptive bud site. Genes Dev. 1999;13:1912–1917. doi: 10.1101/gad.13.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Bi E, Harkins HA, Zahner JE, De Virgilio C, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harb Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci. 2000a;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J Cell Sci. 2000b;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Roemer T, Madden K, Chang J, Snyder M. Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev. 1996;10:777–793. doi: 10.1101/gad.10.7.777. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY.: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Ruggieri R, Bender A, Matsui Y, Powers S, Takai Y, Pringle JR, Matsumoto K. RSR1, a ras-like gene homologous to Krev-1 (smg21A/rap1A): role in the development of cell polarity and interactions with the Ras pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:758–766. doi: 10.1128/mcb.12.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sanders SL, Herskowitz I. The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J Cell Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkman LR, Caruso C, Page′ N, Pringle JR. The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast. J Cell Biol. 2002;156:829–841. doi: 10.1083/jcb.200107041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Barral Y, Snyder M. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5235–5247. doi: 10.1128/mcb.20.14.5235-5247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevissiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri N, Köhler T, Braus GH, Mösch H-U. Asymmetrically localized Bud8p and Bud9p proteins control yeast cell polarity and development. EMBO J. 2000;19:6686–6696. doi: 10.1093/emboj/19.24.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtz N, Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J Cell Biol. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Bender A, Cerione RA. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J Biol Chem. 1995;270:626–630. doi: 10.1074/jbc.270.2.626. [DOI] [PubMed] [Google Scholar]