Abstract
Saccharomyces cerevisiae Pkh1 and Pkh2 are functionally redundant homologs of mammalian protein kinase, phosphoinositide-dependent protein kinase-1. They activate two closely related, functionally redundant enzymes, Ypk1 and Ykr2 (homologs of mammalian protein kinase, serum- and glucocorticoid-inducible protein kinase). We found that Ypk1 has a more prominent role than Ykr2 in mediating their shared essential function. Considerable evidence demonstrated that Pkh1 preferentially activates Ypk1, whereas Pkh2 preferentially activates Ykr2. Loss of Pkh1 (but not Pkh2) reduced Ypk1 activity; conversely, Pkh1 overexpression increased Ypk1 activity more than Pkh2 overexpression. Loss of Pkh2 reduced Ykr2 activity; correspondingly, Pkh2 overexpression increased Ykr2 activity more than Pkh1 overexpression. When overexpressed, a catalytically active C-terminal fragment (kinase domain) of Ypk1 was growth inhibitory; loss of Pkh1 (but not Pkh2) alleviated toxicity. Loss of Pkh2 (but not Pkh1) exacerbated the slow growth phenotype of a ypk1Δ strain. This Pkh1-Ypk1 and Pkh2-Ykr2 dichotomy is not absolute because all double mutants (pkh1Δ ypk1Δ, pkh2Δ ypk1Δ, pkh1Δ ykr2Δ, and pkh2Δ ykr2Δ) were viable. Compartmentation contributes to selectivity because Pkh1 and Ypk1 were located exclusively in the cytosol, whereas Pkh2 and Ykr2 entered the nucleus. At restrictive temperature, ypk1-1ts ykr2Δ cells lysed rapidly, but not in medium containing osmotic support. Dosage and extragenic suppressors were selected. Overexpression of Exg1 (major exoglucanase), or loss of Kex2 (endoprotease involved in Exg1 processing), rescued growth at high temperature. Viability was also maintained by PKC1 overexpression or an activated allele of the downstream protein kinase (BCK1-20). Conversely, absence of Mpk1 (distal mitogen-activated protein kinase of the PKC1 pathway) was lethal in ypk1-1ts ykr2Δ cells. Thus, Pkh1-Ypk1 and Pkh2-Ykr2 function in a novel pathway for cell wall integrity that acts in parallel with the Pkc1-dependent pathway.
INTRODUCTION
A cascade of protein kinases is a commonly used mechanism for amplifying and disseminating signals that control metabolism, growth, survival, and differentiation in eukaryotic cells. In animal cells, recruitment of phosphatidylinositol 3-kinase by growth factor receptors generates 3-phosphoinositides, which stimulate 3-phosphoinositide-dependent protein kinase-1 (PDK1) (for review, see Toker and Newton, 2000; Vanhaesebroeck and Alessi, 2000). Activated PDK1 phosphorylates and activates multiple downstream targets, including protein kinase B/c-Akt (Brazil and Hemmings, 2001; Lawlor and Alessi, 2001), p70 S6 kinase (Alessi et al., 1998; Kozma and Thomas, 2002), protein kinase C (PKC) isoforms (Chou et al., 1998; Le Good et al., 1998), and serum- and glucocorticoid-inducible protein kinase (SGK) isoforms (Kobayashi and Cohen, 1999; Kobayashi et al., 1999), thereby eliciting physiological responses.
We have demonstrated previously that, in budding yeast (Saccharomyces cerevisiae), Pkh1 and Pkh2 are the homologs and functional equivalents of mammalian PDK1. Pkh1 and Pkh2 share an essential function because pkh1Δ and pkh2Δ single mutants are viable, whereas a pkh1Δ pkh2Δ double mutant is inviable. Expression of human PDK1 rescues the lethality of a pkh1Δ pkh2Δ strain (Casamayor et al., 1999). The PDK1 enzymes from Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens all possess a C-terminal pleckstrin homology (PH) domain that binds phosphatidylinositol (PtdIns)(3,4,5)P3 and PtdIns(3,4)P2 (Stephens et al., 1998; Currie et al., 1999; Fruman et al., 1999). However, S. cerevisiae does not produce PtdIns(3,4,5)P3 or PtdIns(3,4)P2 (Hawkins et al., 1993; De Camilli et al., 1996). Moreover, Pkh1 and Pkh2 lack discernible PH domains, and PDK1 lacking its PH domain was sufficient to rescue the growth of pkh1Δ pkh2Δ cells (Casamayor et al., 1999), suggesting that the activity of Pkh1 and Pkh2 in yeast does not depend on phosphoinositides. It was shown subsequently that sphingosine (4-dehydro-sphinganine) can also stimulate mammalian PDK1 autophosphorylation and increase its ability to phosphorylate in vitro known PDK1 substrates, such as c-Akt and PKCβ (King et al., 2000). Correspondingly, it has been reported recently that Pkh1 and Pkh2 can be activated in vitro by nanomolar concentrations of the major sphingoid base in yeast, phytosphingosine (4-hydroxy-sphinganine) (Friant et al., 2001). Moreover, endocytosis in yeast seems to require sphingoid base synthesis and overexpression of Pkh1 or Pkh2 can suppress this requirement (Friant et al., 2001), suggesting that sphingoid bases activate a signaling pathway involving Pkh1 and Pkh2.
Mammalian PDK1 activates its downstream targets by phosphorylating a Thr residue (starred) in a sequence motif, Thr*-Phe-Cys-Gly-Thr-X-Glu-Tyr (where X represents any amino acid), that lies within the “activation loop” of their catalytic domains (Hanks and Hunter, 1995) and is unique to and conserved in all known PDK1 substrates. Full activation of c-Akt/PKB and other PDK1 targets also seems to require phosphorylation at a second site (starred) situated in a hydrophobic motif, Phe-X-X-Ar-Ser*/Thr*-Ar (where Ar represents an aromatic residue), that is located near the C terminus of each of these enzymes (Toker and Newton, 2000; Vanhaesebroeck and Alessi, 2000). In S. cerevisiae, four previously characterized protein kinases possess both of these motifs, suggesting that they are physiological substrates of Pkh1 and/or Pkh2. These four protein kinases are the products of the following genes: YPK1 (Maurer, 1988), YKR2/YPK2 (Maurer, 1988; Chen et al., 1993), PKC1 (Levin et al., 1990), and SCH9 (Toda et al., 1988). Studies from this laboratory have demonstrated that Ypk1 is a direct substrate of Pkh1 (Casamayor et al., 1999) and that Ykr2 is phosphorylated by Pkh2 (Torrance, 2000). Similarly, it has been shown that Pkc1 can also be phosphorylated by Pkh1 and Pkh2 (Inagaki et al., 1999; Friant et al., 2001). Reduced Pkc1 activity was observed in a pkh1-1ts pkh2Δ strain, and the temperature sensitivity of this strain was partially suppressed by a dominant PKC1(R398P) allele, suggesting that Pkh1 and Pkh2 are required for Pkc1 function in vivo (Inagaki et al., 1999).
The catalytic domains of Ypk1 and Ykr2 are 88% identical and these proteins also share extensive homology across their N- and C-terminal extensions. Moreover, the catalytic domains of Ypk1 and Ykr2 closely resemble (55% identity) that of mammalian SGK. Indeed, cells lacking Ypk1 or Ykr2 are viable, whereas cells lacking both Ypk1 and Ykr2 are inviable (Chen et al., 1993; Schnieders, 1996), and expression of mammalian SGK rescues this inviability (Casamayor et al., 1999). Furthermore, both purified PDK1 and purified Pkh1 phosphorylate the same residue (Thr504) in the consensus motif in purified Ypk1, and Ypk1 phosphorylation is significantly diminished in vivo in cells lacking Pkh1 (Casamayor et al., 1999). Thus, just as SGK is a downstream target of PDK1 in animal cells, Ypk1 and Ykr2 seem to act downstream of Pkh1 and Pkh2 in yeast. Moreover, lipid-derived signals are required as upstream activators in both pathways, 3-phosphoinositides and sphingosine in the case of PDK1 and closely related sphingoid bases in the case of Pkh1 and Pkh2. Consistent with this view, overexpression of Ypk1 confers resistance to myriocin (ISP-1), an antibiotic that specifically inhibits serine C-palmitoyltransferase (product of the LCB1 gene), which is the enzyme responsible for sphinganine biosynthesis (Sun et al., 2000).
Herein, we describe experiments that address the genetic and biochemical interrelationships between Pkh1 and Pkh2 and Ypk1 and Ykr2, which we undertook to try to understand the reason for the redundancies within these protein kinase cascades. To provide further insight, we also investigated the subcellular localization of all four proteins. Finally, as two independent approaches for discerning the physiological function of the Ypk1 and Ykr2 enzymes, we selected for dosage suppressors and also for chromosomal mutations that suppress the lysis phenotype of ypk1-1ts ykr2Δ cells.
MATERIALS AND METHODS
Strains and Growth Conditions
Yeast strains used in this study are listed in Table 1. Standard rich (YP) and defined minimal (SC) media (Sherman et al., 1986), containing either 2% glucose (Glc), 2% raffinose (Raf), or 2% galactose (Gal) as the carbon source and supplemented with appropriate nutrients to maintain selection for plasmids, were used for yeast cultivation. For gene expression from the galactose-inducible GAL1 promoter in liquid media, cells were pregrown to mid-exponential phase in SC containing 2% raffinose-0.2% sucrose (Raf/Suc) and then Gal was added to a final concentration of 2% and incubation continued for 2 h. In experiments involving growth on solid medium containing 5-fluoroorotic acid, 5-fluoroorotic acid was used at a concentration of 0.5 mg/ml (Boeke et al., 1984). Cells were grown routinely at 30°C, except for strains carrying temperature-sensitive mutations, which were propagated at their permissive temperature (26°C).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| YPH499 | MATaade2-101oc his3-Δ200 leu2-Δ1 lys2-801am trp1-Δ1 ura3-52 | Sikorski and Hieter (1989) |
| YPH500 | MATα ade2-101oc his3-Δ200 leu2-Δ1 lys2-801am trp1-Δ1 ura3-52 | Sikorski and Hieter (1989) |
| W303-1A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Sherman et al. (1986) |
| W303-1B | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Sherman et al. (1986) |
| YES3a | YPH499 ypk1-Δ1∷HIS3 | This study |
| YES5b | YPH500 ypk1-Δ1∷HIS3 | This study |
| YFR107c | YPH500 ypk1-Δ1∷his3∷LEU2 | This study |
| YES1d | YPH499 ykr2-Δ1∷TRP1 | This study |
| YFR64e | YPH500 ykr2-Δ1∷TRP1 | This study |
| YFR119f | YPH500 ykr2-Δ1∷trp1∷LEU2 | This study |
| AC301 | W303-1B pkh1-Δ1∷TRP1 | Casamayor et al. (1999) |
| AC303 | W303-1A pkh2-Δ1∷HIS3 | Casamayor et al. (1999) |
| YPT67g | W303-1B pkh2-Δ1∷HIS3 | This study |
| AC394 | W303 pkh1-Δ1∷TRP1/PKH1 pkh2-Δ1∷HIS3/PKH2 [pYES2-PKH2] | Casamayor et al. (1999) |
| YFR105h | YPH499 pkh1-Δ1∷TRP1 | This study |
| YFR106i | YPH499 pkh2-Δ1∷HIS3 | This study |
| INA106 | pkh1D398G pkh2∷LEU2 ura3 trp1 his2 ade1 | Inagaki et al. (1999) |
| YPT40 | YPH499 ypk1-1ts∷HIS3 ykr2-Δ1∷TRP1 | Casamayor et al. (1999) |
| YAN2j | YPH500 ypk1-1ts∷his3∷URA3 ykr2-Δ1∷TRP1 | This study |
| KRY24 | W303-1A kex2-Δ2∷LEU2 | Fuller et al. (1989) |
| YFR84k | YPH500 exg1Δ∷HIS3 | This study |
| YFR127l | YPH499 mpk1Δ∷HIS3 | This study |
| YFR128m | YPH500 mpk1Δ∷HIS3 | This study |
| YFRM4An | YPH500 ypk1-1ts∷his3∷URA3 ykr2-Δ1∷TRP1 rot2∷Tn3∷LEU2 | This study |
| YFR129o | YPH500 rot2∷Tn3∷LEU2 | This study |
| YFR66p | MATa/MATα ypk1-1ts∷his3∷URA3/YPK1 ykr2Δ∷TRP1/YKR2 kex2Δ∷LEU2/KEX2 | This study |
| YFR101q | MATa/MATα ypk1-1ts∷his3∷URA3/YPK1 ykr2Δ∷TRP1/YKR2 kex2Δ∷LEU2/KEX2 mpk1Δ∷HIS3/MPK1 | This study |
Derived from YPH499 by transforming with the 3.6-kb XbaI-SalI fragment of pESB5 (pypk1-Δ1∷HIS3) containing a deletion-insertion allele of YPK1 in which the majority of the open reading frame (NsiI-EcoRI fragment) was replaced with the selectable marker HIS3.
Derived from YPH500 by transforming with the 3.6-kb XbaI-SalI fragment of pESB5 (pypk1-Δ1∷HIS3) containing a deletion-insertion allele of YPK1 in which the majority of the ORF (NsiI-EcoRI fragment) was replaced with the selectable marker HIS3.
Derived from YES5 by transforming with plasmid pHL3 (Cross, 1997) digested with ApaI and PstI and selecting for Leu+ His− colonies.
Derived from YPH499 by transforming with the 2.5-kb Kpn1-Sac1 fragment of pESB2 (pykr2-Δ1∷TRP1) containing a deletion-insertion allele of YKR2 in which the majority of the open reading frame (Pst1-Pst1 fragment) was replaced with the selectable marker TRP1.
Segregant of a diploid (YES5 × YES1).
Derived from YFR64 by transforming with plasmid pTL7 (Cross, 1997) digested with XhoI and SmaI and selecting for Leu+ Trp− colonies.
Segregant of a diploid AC394.
Derived from YPH499 by transforming with a deletion-insertion allele of PKH1 obtained by PCR amplification by using genomic DNA of AC301.
Derived from YPH499 by transforming with a deletion-insertion allele of PKH2 obtained by PCR amplification by using genomic DNA of AC303.
Derived from YFR93 by transforming with plasmid pHU10 (Cross, 1997) digested with XhoI and SmaI and selecting for Ura+ His− colonies.
Derived from YPH500 by transforming with a PCR fragment obtained by amplifying the HIS3 gene from pRS313 (Sikorski and Hieter, 1989) with primers containing 45-bp of homology with the sequence upstream or downstream of EXG1.
Derived from YPH499 by transforming with a PCR fragment obtained by amplifying the HIS3 gene from pRS313 (Sikorski and Hieter, 1989) with primers containing 45 bp of homology with the sequence upstream or downstream of MPK1.
Derived from YPH500 by transforming with the same PCR fragment described in footnote l.
Segregant of a diploid obtained by crossing an isolate of the transposon insertion screen corresponding to an insertion in the ROT2 gene with YAN2.
Segregant of a diploid (YPH499 × YFRM4A).
Diploid derived from a cross of YAN2 against KRY24.
Derived by transformation of YFR66 with the same PCR fragment described in footnote l.
Recombinant DNA Methods
Escherichia coli strain DH5α (Hanahan, 1983) was used for the construction and propagation of plasmids. Conventional recombinant DNA methods were used for the construction of plasmids (Sambrook et al., 1989). The sequences of constructs that contained DNA fragments amplified by polymerase chain reaction (PCR) were verified by the dideoxy chain termination-sequencing method (Sanger et al., 1977). Native and Turbo Pfu polymerases (Stratagene, La Jolla, CA) were used for PCR, unless noted otherwise.
Plasmids
Plasmids pYPK1, pYKR2, pGAL-YPK1, pGAL-YKR2 (pAM1), pGAL-Ypk1-Myc (pAM54), pADH-YPK1, pRS316-YKR2 (pAM12), pGAL-PKH1 (pAM73), and pGAL-PKH2 (pAM79) have been described previously (Maurer, 1988; Kubo et al., 1989; Casamayor et al., 1999). To create plasmid pADH-YKR2 (pAM4), which constitutively overexpresses YKR2 from the ADH1 promoter, a 2.4-kb XhoI (blunt)-SalI fragment containing the entire YKR2 gene was excised from pYKR2, gel purified, and inserted into vector pAD4 M (Martin et al., 1990) that had been linearized with SmaI/SalI. To generate a version of Ykr2 tagged at its C-terminal end with the c-Myc epitope (Evans et al., 1985), a PCR-based method for precise gene fusion (Yon and Fried, 1989) was performed using the YKR2 sequence cloned in pUC18 as one template (pYKR2), and as the other template, pOGFP (E. Swartzman, this laboratory), which contains a sequence encoding the 16-residue version of the c-Myc epitope followed by a (His)6 tag cloned in pBluescript (Stratagene); with three appropriate synthetic oligonucleotide primers: T3 (Stratagene); 5′-GGA CAT ATT GCA CTG TGT G-3′ (RMN5), corresponding to sequences in YKR2 overlapping a DraIII site near the C terminus; and a “joiner” primer, 5′-TTC AGA AAT CAA CTT TTG TTC ACT AAT GCT TCT CCC CTG-3′ (RMC), corresponding to the 3′ end of the YKR2 coding sequence and the first several residues of the c-Myc epitope. An ∼1.6-kb DraIII/KpnI fragment of the resulting PCR product was used to replace the corresponding segment in pYKR2, yielding pYkr2-Myc (pAM24). An ∼3-kb NcoI/HindIII fragment from pYkr2-Myc was gel purified and used to replace the corresponding ∼2.2-kb NcoI/HindIII segment in pGAL-YKR2 to create a 2-μm DNA-containing, LEU2-marked plasmid, pGAL-Ykr2-Myc (pAM59), that overexpresses Ykr2-Myc upon galactose induction. To generate a catalytically inactive (“kinase-dead”) version of Ypk1, a PCR-based method for site-directed mutagenesis was performed using pYPK1 as the template and three appropriate synthetic oligonucleotide primers: 5′-CTT GAA CAC AGT AAG TAA CGG-3′ (PKC2), corresponding to the flanking genomic sequence commencing 68-base pairs downstream of the stop codon; 5′-CAC AAA AAG TAT ACG CCT TGG CGG CAA TCA G-3′ (PKD), where the underlined nucleotide is a silent mutation to introduce a BglI site, and the bold nucleotides correspond to an introduced alanine codon (GCG) in place of the native lysine codon (AAG); and 5′-GTC CAT CGA TGA TTT CGA TC-3′ (Pseq2), corresponding to the coding strand of YPK1 starting at nucleotide position 1024. The resulting ∼1.1-kb PCR product was digested with ClaI and NcoI, and the resulting ∼850-base pair fragment was used to replace the corresponding segment in pYPK1, yielding pYPK1(K376A-KD) (pAM46). Conversion of the Lys residue at the equivalent position in all other protein kinases examined to date eliminates their catalytic activity (Hanks and Hunter, 1995). To generate a catalytically inactive (kinase-dead) version of Ykr2, a similar PCR-based approach for site-directed mutagenesis was performed using pYKR2 as the template, and three appropriate synthetic oligonucleotide primers: 5′-AGT ATA GCC CTG CCC CAA C-3′ (Rseq2), corresponding to the noncoding strand of YKR2 commencing at nucleotide position 1544; 5′-CCC AAA AGA TTT ACG CCT TGG CGG CTC TGA G-3′ (RKD), where the underlined nucleotide is a silent mutation to introduce a BglI site, and the bold nucleotides correspond to an introduced alanine codon (GCG) in place of the native lysine codon (AAG); and 5′-CGT GGG GTA ATG GCC TG-3′ (Rseq3), corresponding to the coding strand of YKR2 starting at nucleotide position 66. The resulting ∼1.4-kb PCR product was digested with NcoI and DraIII and used to replace the corresponding segment in pYKR2, yielding pYKR2(K373A-KD) (pAM47). Plasmid pYPK1(K376A-KD) was digested with AlwnI, converted to flush ends by treatment with T4 polymerase (NEB) and all four dNTPs then digested with SalI. The resulting 3.3-kb YPK1(K376A-KD)–containing fragment was gel purified and ligated into YEp351GAL that had been linearized by digestion with XbaI, converted to flush ends by incubation with T4 polymerase and all four dNTPs, and then digested with SalI. The resulting plasmid, pGAL-YPK1(K376A-KD) (pAM48), expresses a catalytically inactive allele [Ypk1-(K376A-KD)] from a 2-μm DNA-containing, LEU2-marked plasmid under control of the GAL1 promoter. An ∼1.2-kb NcoI/SalI fragment from pYpk1-Myc was gel purified and used to replace the corresponding NcoI/SalI segment in pGAL-YPK1(K376A-KD) to create pGAL-YPK1(K376A-KD)-Myc (pAM49). An ∼2.4-kb XhoI-HindIII fragment containing the entire YKR2(K373A-KD) allele was excised from pYKR2(K373A-KD), gel purified, and inserted into YEp351GAL that had been linearized with SalI and HindIII, to create a 2-μm DNA-containing, LEU2-marked plasmid, pGAL-YKR2(K373A-KD) (pAM50), that overexpresses catalytically inactive Ykr2 upon galactose induction. Galactose-inducible expression vectors that are URA3 based were constructed as follows. An ∼3.3-kb BamHI/HindIII fragment carrying YPK1 was excised from pGAL-YPK1, gel purified, and ligated into YEp352GAL (Benton et al., 1994), which had been linearized with BamHI/HindIII, yielding YEp352GAL-YPK1 (pAM75). An ∼3.8-kb BamHI/HindIII fragment from pGAL-Ypk1-Myc was gel purified and ligated into YEp352GAL that had been linearized with BamHI/HindIII, yielding YEp352GAL-Ypk1-Myc (pAM76). An ∼2.2-kb BamHI/HindIII fragment from p2GAL-YKR2 was gel purified and ligated into YEp352GAL, which had been linearized with BamHI/HindIII, yielding YEp352GAL-YKR2 (pAM77). An ∼3.0-kb BamHI/HindIII fragment from p2GAL-Ykr2-Myc was gel purified and ligated into YEp352GAL, which had been linearized with BamHI/HindIII, yielding YEp352GAL-Ykr2-Myc (pAM78). To generate an amino-terminal truncation of Ypk1, the following two-step approach was taken. First, an ∼1.1-kb fragment corresponding to the last 344 amino acids of Ypk1 was amplified by PCR from pYPK1 with the following oligonucleotides: 5′-GGC GGA TCC ATG TCC AGA AAT AAA CCT TTG TCC-3′ (PCT), corresponding to sequences in the middle of the YPK1 coding sequence, just upstream of the beginning of the catalytic domain, where the underlined nucleotides correspond to an introduced BamHI restriction site, and the bold nucleotides represent an introduced start codon (ATG); and 5′-CTT GAA CAC AGT AAG TAA CGG-3′ (PKC2), corresponding to the flanking genomic sequence commencing 68-base pairs downstream of the stop codon. The resulting PCR product was digested with BamHI and NcoI, gel purified, and used to replace an ∼2.6-kb BamHI/NcoI fragment in pRS315-YPK1(B/H). The resulting CEN-containing, LEU2-marked plasmid encodes an amino-terminal truncation of Ypk1, which contains only the catalytic domain, but essentially no promoter sequence, and is called pRS315-YPK1-ΔN (pAM55). An ∼1.2-kb NcoI/SalI fragment from pRS315-Ypk1-myc was gel purified and used to replace the corresponding NcoI/SalI segment in pRS315-YPK1-ΔN to create a CEN-containing, LEU2-marked plasmid, pRS315-Ypk1-ΔN-myc (pAM56), that encodes a myc-tagged version of the Ypk1 catalytic domain. To insert a promoter, an ∼2.3-kb BamHI/SalI fragment from pRS315-YPK1-ΔN was gel purified and inserted into YEp351GAL that had been linearized with BamHI/SalI to create a 2-μm ΔNA-containing, LEU2-marked plasmid, pGAL-YPK1-ΔN (pAM99), that overexpresses the amino-terminal truncation of Ypk1 upon galactose induction. Likewise, an ∼2.0-kb BamHI/SalI fragment from pRS315-Ypk1-ΔN-Myc was gel purified and inserted into YEp351GAL that had been linearized with BamHI/SalI to create a 2-μm ΔNA-containing, LEU2-marked plasmid, pGAL-Ypk1-ΔN-Myc (pAM100), that overexpresses a myc-tagged version of the amino-terminal truncation of Ypk1 upon galactose induction. To move these truncated Ypk1 derivatives into URA3-marked plasmids, an ∼2.3-kb BamHI/SalI fragment from pGAL-YPK1-ΔN was gel purified and inserted into YEp352GAL that had been linearized with BamHI/SalI to create a 2-μm DNA-containing, URA3-marked plasmid, YEp352GAL-YPK1-ΔN (pAM101), that overexpresses the amino-terminal truncation of Ypk1 upon galactose induction. Similarly, an ∼2.0-kb BamHI/SalI fragment from pGAL-Ypk1-ΔN-Myc was gel purified and inserted into YEp352GAL that had been linearized with BamHI/SalI to create a 2-μm DNA-containing, URA3-marked plasmid, YEp352GAL-Ypk1-ΔN-myc (pAM102), that overexpresses a myc-tagged version of the amino-terminal truncation of Ypk1 upon galactose induction. To generate a catalytically inactive derivative of the Ypk1-ΔN allele, a 2.3-kb ClaI/HindIII fragment from pGAL-YPK1-KD, encoding the carboxy terminus (containing the K376A kinase-dead mutation of Ypk1) was gel purified and used to replace the corresponding segment in YEp352GAL-Ypk1-ΔN-Myc. The resulting plasmid, YEp352GAL-Ypk1-ΔN-KD (pFR30), overexpresses a catalytically inactive derivative of the amino-terminal truncation of Ypk1 upon galactose induction.
Protein Localization by Using Chimeras Containing Green Fluorescent Protein (GFP)
To create vectors for galactose-inducible expression of YPK1, YKR2, and PKH2, each fused to the carboxy terminus of a protein comprising three tandem repeats of an enhanced (S65T V163A) mutant of GFP, the following approach was taken. Two primers, 5′-GCG AGC GGG ATC CAT G, the first 18 bases of the gene-3′ (primer A), where underlined bases correspond to an introduced BamHI site and start codon in bold; and 5′-GGC ACG CGT CGA CTT A, the last 18 bases of the gene-3′ (primer B), where underlined bases correspond to an introduced SalI site and stop codon in bold, were used to amplify the entire open reading frames of the corresponding genes from genomic DNA. The PCR products were digested with BamHI and SalI and ligated into vector pGS836 (YCpGAL-3GFP) (Maurer et al., 2001) that had been digested with BamHI and SalI, yielding plasmids pGAL-3GFP-YPK1 (pFR33), pGAL-3GFP-YKR2 (pER2), and pGAL-3GFP-PKH2 (pER3). To create pGAL-3GFP-PKH1 (pFR37), the same approach was used, but due to the presence of BamHI and SalI restriction sites in the gene, two PCR products were made, one with primer A and a primer corresponding to the sequence 3′ to the ClaI site present in PKH1, and the other with a primer 3′ of the ClaI site and primer B. A three-way ligation was then used to ligate the two PCR products digested with BamHI and ClaI, or ClaI and SalI, into pGS836 that had been digested with BamHI and SalI.
Cells expressing the GFP constructs were grown to mid-exponential phase at 30°C in SC-Leu containing Raf/Suc and then induced with 2% galactose for 3 h. Nuclear DNA was stained with 4,6-diamidino-2-phenylindole (DAPI) by adding the dye directly in the medium at a concentration of 1 μg/ml for the last hour of growth. Rhodamine-labeled phalloidin was purchased from Molecular Probes (Eugene, OR). Samples of each culture were viewed directly with a TE300 fluorescence microscope (Nikon, Melville, NY) equipped with a 100×/1.4 Plan-Apo objective and a 1.4 numerical aperture condenser. Digital images were acquired with a bottom-ported Orca 100 charge-coupled device camera (Hamamatsu, Bridgewater, NJ) and Phase 3 Imaging Systems software (Northern Exposure, Inc., Glen Mills, PA). The fraction of lysed cells in cultures was assessed by direct counting of at least 200 cells after staining with a vital dye (LIVE/DEAD Yeast Viability kit; catalog no. 7009; Molecular Probes).
Antibody Production
To express a glutathione S-transferase (GST)-Ypk1 fusion protein in E. coli, an ∼370-base pair fragment corresponding to the first 114 amino acids of Ypk1 was amplified by PCR from pYPK1 by using the following oligonucleotides: 5′-GGG GGG GGA TCC ATG TAT TCT TGG AAG TCA AAG TTT-3′, where the underlined nucleotides correspond to an introduced BamHI site, and the bold nucleotides correspond to the start codon; and 5′-GGG GGG AAT TCT CAG GTG GCA TCA TTG GGT GTC CC-3′, where the underlined nucleotides correspond to an introduced EcoRI site, and the bold nucleotides correspond to the reverse complement of an introduced stop codon. This PCR fragment was then digested with BamHI and EcoRI, gel purified, and ligated into the pGEX2T vector (Pharmacia, Peapack, NJ), which had been linearized with BamHI and EcoRI to create plasmid pGEX-Ypk1 (pAM5). To express a GST-Ykr2 fusion protein in E. coli, an ∼360-base pair fragment corresponding to the first 111 amino acids of Ykr2 was amplified by PCR from pYKR2 by using the following olignucleotides: 5′-GGG GGG GGA TCC ATG CAT TCC TGG CGA ATA TCC AAG-3′, where the underlined nucleotides correspond to an introduced BamHI site, and the bold nucleotides correspond to the start codon; and 5′-GGG GGG AAT TCT CAA CTC GGT CCC TGC GTC TCA GT-3′, where the underlined nucleotides correspond to an introduced EcoRI site, and the bold nucleotides correspond to the reverse complement of an introduced stop codon. This PCR fragment was then digested with BamHI and EcoRI, gel purified, and ligated into the pGEX2T vector, which had been linearized with BamHI and EcoRI to create plasmid pGEX-Ykr2 (pAM6).
To prepare antigen, expression of GST-Ypk1(1-114) and GST-Ykr2(1-111) fusions from plasmids, pGEX-Ypk1 and pGEX-Ykr2, respectively, were induced in a protease-deficient E. coli strain BL21 (DE3)[pLys] (Studier, 1991) by addition of isopropyl-β-d-thiogalacto-pyranoside to a final concentration of 0.2 mM followed by incubation with aeration for 2 h at 30°C. Cells were harvested, washed once with ice-cold wash buffer (50 mM Tris-HCl pH 8, 0.5 mM dithiothreitol [DTT], 100 mM KCl, 1 mM phenylmethylsulfonyl fluoride, 0.05% NP-40, 1 mM EGTA, and 1 mM EDTA), and resuspended in 1/20 volume of wash buffer containing 1 M NaCl. Cells were disrupted by digestion with lysozyme (final concentration, 2 mg/ml) followed by sonication. Insoluble material was removed by centrifugation at 12,000 × g, and the soluble GST-Ypk1(1-114) or GST-Ykr2(1-111) proteins were purified by adsorption to, and elution from, glutathione-agarose beads (Pharmacia), essentially as directed by the manufacturer, except that elution was performed in the presence of 1 M NaCl and 20 mM glutathione. The purified proteins were used as immunogens to raise polyclonal antisera in adult female New Zealand White rabbits following standard immunization protocols (Harlow and Lane, 1988). The resulting anti-Ypk1 antibodies (serum #1446) and anti-Ykr2 antibodies (serum #1732) are specific to Ypk1 and Ykr2 and do not display any cross-reaction against the incorrect antigen. Anti-GFP antibodies were the generous gift of Roger Tsien and Charles Zuker (Department of Cellular and Molecular Medicine, University of California, San Diego, CA).
Preparation of Cell Extracts and Immunoblot Analysis
Yeast cells were grown at 30°C to mid-exponential phase (A600 nm = 0.5–1), either in SC medium supplemented in a manner appropriate for maintenance of plasmids or in rich medium (YPGlc). If cells required galactose induction for expression from the GAL1 promoter, galactose was added to a final concentration of 2% and the cultures were incubated at 30°C for an additional 2 h. Cells were harvested by brief centrifugation, washed twice by resuspension and resedimentation in ice-cold lysis buffer (50 mM Tris-HCl pH 7.5, 5 mM EDTA, 3 mM DTT and 1 mM phenylmethylsulfonyl fluoride), and resuspended in 200 μl of the same buffer. Prechilled glass beads (0.45–0.6 mm in diameter) were added to the meniscus of the cell suspension, and lysis was achieved by vigorous vortex mixing for six 1-min intervals, with intermittent cooling on ice. To remove the glass beads, the bottom of the Eppendorf tube was punctured with a syringe needle (<0.5 mm in diameter) and inserted into another tube; the lysate was collected into the fresh tube by brief centrifugation in a clinical centrifuge. The crude extract was subjected to centrifugation at 30,000 × g for 15 min to remove unbroken cells and large debris. The protein concentration of the crude extract was measured using a dye-binding method (Bradford, 1976) with a protein assay kit as instructed by the manufacturer (Bio-Rad, Hercules, CA), by using bovine serum albumin (New England Biolabs, Beverly, MA) as the standard.
For immunoblot analysis, samples (50 μg of total protein) were diluted into SDS-PAGE sample buffer (Laemmli, 1970), subjected to electrophoresis in an 8–12% gel, and then transferred to nitrocellulose (Towbin et al., 1979). To detect Ypk1, rabbit polyclonal anti-Ypk1 antiserum #1446 was used at a dilution of 1:3000. To detect Ykr2, rabbit polyclonal anti-Ykr2 antiserum #1732 was used at a dilution of 1:3000. To detect proteins using the anti-c-Myc monoclonal antibody (mAb) 9E10, ascites fluid containing this mAb was used at a dilution of 1:10,000 (Evans et al., 1985). Immobilized immune complexes were detected using a commercial chemiluminescence detection system (Renaissance; PerkinElmer Life Sciences, Boston, MA) and x-ray film (Biomax MR; Eastman Kodak, Rochester, NY).
Immunoprecipitations
Yeast cultures to be used for immunoprecipitation analysis were grown as described above, and then rinsed in ice-cold IP buffer (20 mM Tris-HCl pH 7.5, 125 mM potassium acetate, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 0.1% Triton X-100, and 12.5% glycerol). Glass beads were added to the meniscus of the cell suspension, and lysis was achieved by vigorous vortex mixing for eight 30-s intervals with intermittent cooling on ice. The lysate was clarified by centrifugation at 14,000 × g at 4°C for 30 min. The clarified extract was assayed for protein concentration, and a sample (1 mg of total protein) was diluted to a final volume of 200 μl in IP buffer. An aliquot (20 μl) of protein G/protein A-agarose beads (30% slurry) (Oncogene Science, Cambridge, MA) and a sample of an appropriate control antibody, either 2 μl of preimmune rabbit serum or 1 μg of purified mouse anti-T-cell receptor antibody (gift of James Allison, Department of Molecular and Cell Biology, University of California, Berkeley, CA), were added. The samples were then incubated on a roller drum for 1 h at 4°C to adsorb proteins that bound nonspecifically to the solid support and to rabbit or mouse IgG (preclearing). The beads were removed by centrifugation for 10 min in a microfuge, and the supernatant fraction was transferred to a fresh tube containing another aliquot (15 μl) of protein G/protein A-agarose beads and either 2 μl of anti-Ypk1 (or anti-Ykr2) polyclonal antiserum or 1 μl of anti-c-Myc (mAb 9E10) ascites, and incubated on a roller drum for 1 to 3 h at 4°C. The beads were sedimented by brief centrifugation in a microfuge and washed three times (1 ml each) with ice-cold IP buffer and collected by centrifugation for 1 min in a microfuge on maximum speed. Bead-bound immune complexes were solubilized in SDS-PAGE sample buffer and immediately boiled for 5 min in a water bath and then clarified by brief centrifugation in a Microfuge before resolution by SDS-PAGE. The proteins of interest were visualized as described above.
Immune-Complex Protein Kinase Assays
Cells expressing either wild-type or kinase-dead Ypk1-myc (or Ykr2-myc) under control of the GAL1 promoter were grown in SC containing Raf/Suc to an A600 nm = 0.6, induced by addition of galactose (2% final concentration), incubated with shaking at 30°C for 2 h, collected by centrifugation, washed with ice-cold 1× phosphate-buffered saline, resuspended in 0.2 ml of ice-cold IP buffer, and lysed as described above. The resulting lysates were clarified by centrifugation at 4°C for 30 min at 30,000 × g. Protein concentration in the resulting crude extracts was determined by the Bradford (1976) method. A volume of extract containing 1 mg of total protein was immunoprecipitated with mAb 9E10 as described above. The immunoprecipitates were washed once with ice-cold IP buffer, once with ice-cold IP buffer containing 0.5 M NaCl, and twice with ice-cold buffer A (50 mM Tris-HCl pH 7.5, 0.1 mM EGTA, and 0.1% [by vol] 2-mercaptoethanol). As part of the final wash, the slurry of beads was split into two equal portions. For immunoblot analysis, SDS-PAGE sample buffer (∼15 μl) was added directly to one sample of each bead suspension. For protein kinase assays, the activity of the Ypk1-myc or Ykr2-myc immune complex was assayed by adding 30 μl of a mixture containing 1 μM microcystin-LR, 10 mM Mg-acetate, 100 μM [γ-32P]ATP (200–400 cpm/pmol), and 100 μM Cross-tide (GRPRTSSFAEG) (Cross et al., 1995), which we have documented previously is an excellent peptide phospho-acceptor substrate for Ypk1 and Ykr2 (Casamayor et al., 1999; Torrance, 2000). After incubation for 15 min at 30°C, each reaction was terminated by spotting a portion (45 μl) of the reaction mixture onto small squares of phosphocellulose paper (P81; Whatman, Maidstone, United Kingdom), which were washed and analyzed as described in detail previously (Alessi et al., 1995). In some experiments, samples of the immunoprecipitates were resuspended in an appropriate buffer (20 mM Tris-HCl pH 8.8 and 10 mM MgCl2) and treated with shrimp alkaline phosphatase (0.25 U; US Biochemical, Cleveland, OH) in either the absence or presence of a mixture of inhibitors of this phosphatase (25 μM Na-orthovanadate and 100 μM β-glycerol-phosphate, adjusted to pH 8).
Bioassays for Drug Sensitivity
An agar diffusion (halo) assay (Reneke et al., 1988) was performed to test the relative sensitivity of various strains to rapamycin, valinomycin, hygromycin B, cycloheximide, and polyoxin D. Nascent lawns of the strains to be tested were prepared by mixing ∼2 × 106 cells from a saturated culture with 2 ml of molten (55°C) 1% agar. The cell-containing agar was rapidly mixed and immediately poured evenly onto plates containing an appropriate medium. Various concentrations of rapamycin (50 and 500 ng/μl), hygromycin B (5 and 50 μg/μl), or the other drugs indicated, were spotted in the same volume (10 μl) onto sterile cellulose filter discs (0.6 cm), which were placed on the nascent lawn. The plates were incubated at 30°C, and photographed after 2 d.
Selection and Analysis of Dosage Suppressors
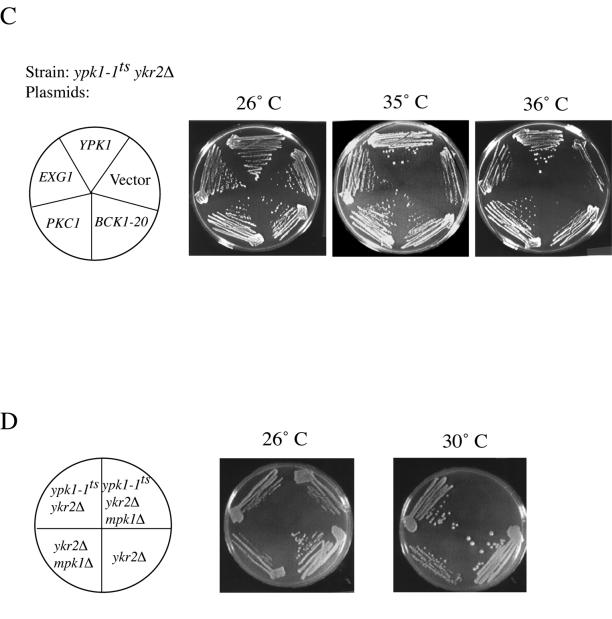
A library of restriction fragments of yeast genomic DNA cloned into a URA3-marked, 2-μm DNA-based vector, YEp352 (Hill et al., 1986), was introduced into strain YPT40 (ypk1-1ts ykr2Δ) (Casamayor et al., 1999) by selecting transformants on SCGlc-Ura medium at 26°C. Temperature-resistant clones were then selected by their ability to grow at 35°C. One suppressor plasmid obtained carried the EXG1 locus as the sole open reading frame. To create a plasmid that expressed EXG1 from a high-level constitutive promoter, two primers, 5′-GCG TCT CGA GAT GCT TTC GCT TAA AA-3′, where the underlined bases correspond to an introduced XhoI site, and the start codon is in bold; and 5′-CGC CGG AGC TC T TAG TTA GAA ATT GTG CC-3′, where the underlined bases correspond to an introduced SacI site, and the stop codon is in bold, were used to amplify the entire open reading frame of EXG1 from the library plasmid that was originally isolated as a dosage suppressor of the ypk1-1ts ykr2Δ strain (see RESULTS). This 1.4-kb PCR product was digested with XhoI and SacI and ligated into vector pAD4 M that had been digested with SalI and SacI, yielding pADH-EXG1 (pAM88). To express PKC1 from its own promoter on a high copy number (2-μm DNA) plasmid, a 4.2-kb NsiI fragment containing the entire open reading frame of PKC1 as well as 560 base pairs upstream of the ATG and 175 base pairs downstream of the stop codon, was cut out of a genomic clone obtained by F. Owen Fields (this laboratory; Fields, 1991; Fields and Thorner, 1991) and ligated into the URA3-based plasmid YEp352 that had been linearized with PstI, yielding plasmid pPKC-PN, or into the LEU2-based plasmid YEp351 that had been linearized with PstI, yielding plasmid pFR32. To express BCK1-20 in a LEU2-based plasmid, the PvuI-PvuI fragment of plasmid pRS314-BCK1–20 (Lee and Levin, 1992) was replaced by the equivalent LEU2-containing fragment of plasmid pRS315, yielding plasmid pRS315-BCK1-20.
Selection and Analysis of Chromosomal Suppressor Mutations
A genomic DNA library containing Tn3::LacZ::LEU2 insertions (generous gift of Michael Snyder, Department of Biology, Yale University) was introduced into strain YPT40 (ypk1-1ts ykr2Δ) (Casamayor et al., 1999) by selecting transformants on SCGlc-Leu medium at 26°C. Temperature-resistant clones were then selected by their ability to grow at 35°C. Plasmids carrying genomic DNA corresponding to the sites of insertion were recovered as described in detail previously (Ross-Macdonald et al., 1999) and characterized by direct nucleotide sequence analysis.
RESULTS
Ypk1 Has a More Prominent Role in Maintaining Viability Than Ykr2
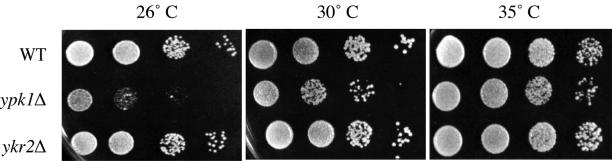
The two protein kinases encoded by the YPK1 and YKR2/YPK2 genes are very similar to each other and functionally redundant at the genetic level (Chen et al., 1993; Casamayor et al., 1999). Yeast cells missing either Ypk1 or Ykr2 are viable, but cells lacking both proteins are inviable, indicating that these enzymes share an essential function. However, several observations suggest that Ypk1 plays the predominant role in executing this essential function. First, ypk1Δ mutants are slow growing at 30°C (Maurer, 1988; Chen et al., 1993), whereas ykr2Δ cells do not display any obvious growth phenotype, compared with otherwise isogenic YPK1+ YKR2+ control cells (Figure 1). Moreover, we found that the slow-growth phenotype of ypk1Δ cells is strongly exacerbated at lower temperatures, even at 26°C (Figure 1). In essence, ypk1Δ mutants are cold sensitive and ykr2Δ mutants are not.
Figure 1.
Phenotypes of ypk1Δ mutants. ypk1Δ cells (but not ykr2Δ cells) grow slowly at 30°C and are cold sensitive. Serial dilutions of exponentially growing wild-type (WT; YPH499) and derived ypk1Δ (YES3) and ykr2Δ (YES1) mutants were spotted on YPGlc plates and grown for 3 d at the indicated temperatures (26, 30, and 35°C).
Among the close mammalian relatives of Ypk1 and Ykr2 is p70 S6 kinase (50% identity in the catalytic domain). Activation of mammalian p70 S6 kinase by mitogens is blocked by rapamycin, an immunosuppressive drug (Chung et al., 1992; Kuo et al., 1992; Thomas, 1993). Therefore, we tested whether loss of either Ypk1 or Ykr2 conferred on yeast cells elevated sensitivity to this agent. Compared with wild-type cells, we found that ypk1Δ cells, but not ykr2Δ cells, were hypersensitive (∼10-fold) to the growth inhibitory effect of rapamycin (Schnieders, 1996; Torrance, 2000), providing a second distinction between ypk1Δ and ykr2Δ mutants. We found that ypk1Δ cells (but not ykr2Δ cells) were also hypersensitive to hygromycin B, valinomycin, polyoxin D, cycloheximide (Torrance, 2000), and caffeine (Bezman, unpublished observations). These results suggested that ypk1Δ cells are generally more permeable to drugs and provided indirect evidence for a defect or perturbation in the cell envelope in ypk1Δ cells (see below). When overexpressed, either YPK1 or YKR2 was able to restore the normal level of drug sensitivity (Roelants, unpublished observations). Given the fact that Pkh1 and Pkh2 are upstream activators of Ypk1 and Ykr2 (Casamayor et al., 1999), it was of interest to test whether loss of either Pkh1 or Pkh2 might also confer a drug-sensitive phenotype. However, neither a pkh1Δ mutant nor a pkh2Δ mutant showed any degree of hypersensitivity to the compounds mentioned above, compared with the parental strain (Torrance, 2000).
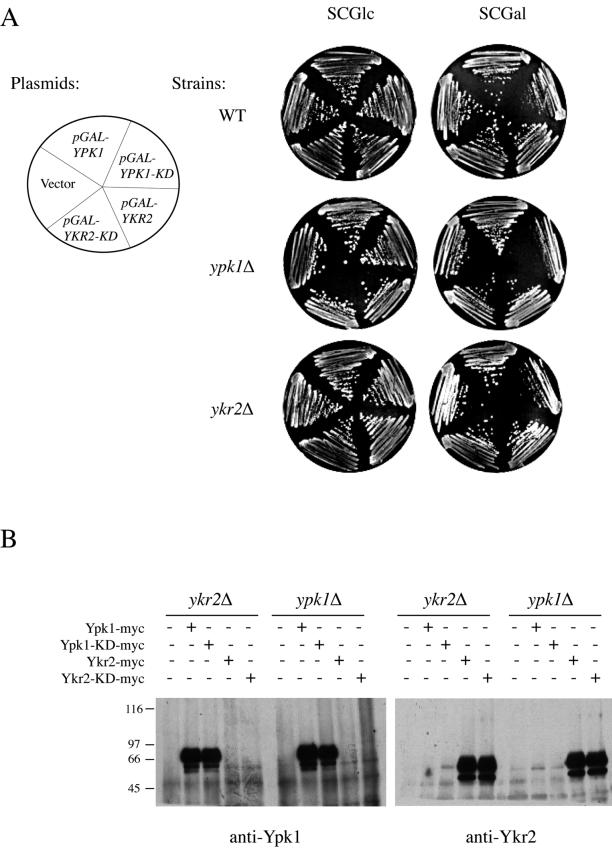
Catalytically inactive (kinase-dead) alleles of protein kinases frequently act as dominant-negatives (Herskowitz, 1987). Hence, we tested whether overexpression of catalytically inactive versions of Ypk1 and Ykr2 would be toxic to cells. We constructed two such derivatives altered in the invariant Lys found in conserved kinase motif II (Hanks and Hunter, 1995). We showed that Ypk1(K376A) and Ykr2(K373A) are indeed catalytically nonfunctional in vitro and unable to complement the lethality of ypk1Δ ykr2Δ cells in vivo (Torrance, 2000). To test whether these alleles behave in a dominant-negative manner when overexpressed from an inducible promoter, plasmids pGAL-YPK1-(K376A-KD) and pGAL-YKR2-(K373A-KD) were introduced into wild-type cells (YPH499) and into ypk1Δ (YES3) and ykr2Δ (YES1) mutants, selecting for transformants on SC-Leu medium containing Glc as the carbon source. The resulting transformants then were streaked onto SC-Leu medium containing Gal/Suc to select for maintenance of the plasmid and to induce expression of either kinase-dead Ypk1 or kinase-dead Ykr2. High-level expression of catalytically inactive Ypk1 was growth inhibitory to all three cell types; however, the strongest growth inhibition was observed in ypk1Δ cells and the mildest was observed in wild-type cells (Figure 2A). In contrast, high-level expression of catalytically inactive Ykr2 had no detectably detrimental effect on growth in any of the strains (Figure 2A). To rule out the possibility that this differential effect was due to a difference in the level of expression of these proteins, Ypk1, Ypk1(K376A), Ykr2, and Ykr2(K373A) were tagged at their C termini with a c-Myc epitope and introduced into ypk1Δ or ykr2Δ strains. Identical amounts of protein from extracts of the resulting transformants were immunoprecipitated with anti-Myc mAb 9E10 antibodies, resolved by SDS-PAGE, and visualized by immunoblotting with polyclonal anti-Ypk1 or anti-Ykr2 antibodies. This analysis verified that the proteins were expressed at equivalent levels (Figure 2B). The fact that kinase-dead Ypk1 was able to effectively impede its own function and that of Ypk2, and the fact the converse was not true, provided a third independent indication that Ypk1 plays the more predominant role.
Figure 2.
Overexpression of catalytically inactive Ypk1 inhibits growth. (A) Dominant-negative effect of inactive Ypk1. Wild-type cells (WT; YPH499), and ypk1Δ (YES3) and ykr2Δ (YES1) mutants were transformed, as indicated, with either an empty vector (YEp352GAL) or the same vector expressing from the GAL1 promoter either full-length Ypk1 (pGAL-YPK1), a catalytically inactive (kinase-dead) ypk1 allele (pGAL-YPK1-KD; pAM48), full-length Ykr2 (pGAL-YKR2; pAM59), or a catalytically inactive (kinase-dead) ykr2 allele (pGAL-YKR2-KD; pAM61). Transformants were selected on glucose-containing medium and then representative isolates were streaked to single colonies on selective medium containing either Glc (left) or Gal (right) as the carbon source and grown for 3 d at 30°C. (B) Equivalent overexpression confirmed by immunoblotting. Cultures of the resulting transformants were grown at 30°C in SCGal/Suc-Leu medium, induced by addition of galactose (2% final concentration) for 3 h. After harvesting the cells by centrifugation, extracts were prepared, and an identical amount of total protein (1 mg) from each extract was subjected to immunoprecipitation with anti-Myc mAb 9E10, as described in MATERIALS AND METHODS. After washing, the immunoprecipitates were solubilized and resolved by electrophoresis on an SDS-slab gel. The species corresponding to Ypk1 and Ypk2 were visualized by immunoblotting with rabbit polyclonal anti-Ypk1 (left) or anti-Ykr2 (right) antibodies, respectively.
Pkh1 Preferentially Activates Ypk1 and Pkh2 Preferentially Activates Ykr2
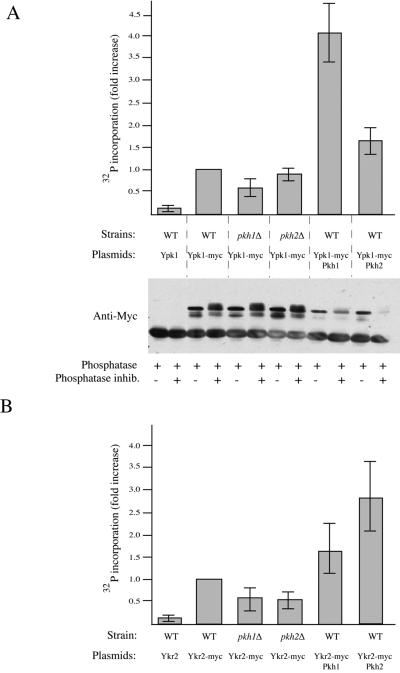
Purified Pkh1 phosphorylates and activates purified Ypk1 in vitro (Casamayor et al., 1999). To examine the state of activation of Ypk1 and Ykr2 in cell extracts and its dependence on the function of Pkh1 and Pkh2, we developed an immune-complex kinase assay. Cell extracts were prepared from strains expressing c-Myc epitope-tagged derivatives of Ypk1 or Ykr2, immunoprecipitated with anti-Myc mAb 9E10, and samples of the resulting immunoprecipitates were examined for protein content by SDS-PAGE and for catalytic activity using a specific peptide substrate (Cross-tide) and [γ-32P]ATP in a filter binding assay (Alessi et al., 1995). As independent negative controls to assess the nonspecific background, extracts were prepared from cells expressing untagged Ypk1 and Ykr2 and from cells expressing kinase-dead derivatives of Ypk1 and Ykr2. Immune complexes from wild-type cells expressing wild-type Ypk1-myc showed ∼10-fold increase in phosphotransferase activity compared with both negative controls: immune complexes from wild-type cells expressing untagged Ypk1 (Figure 3A, top) and immune complexes from wild-type cells expressing catalytically inactive Ypk1-myc (Torrance, 2000). When Ypk1-myc was isolated from pkh1Δ cells, however, the increase in activity was reproducibly 40–60% lower than that observed in wild-type cells, whereas within experimental error, the recovery of Ypk1-myc activity was unaffected in pkh2Δ cells (Figure 3A, top). Most tellingly, activity was greatly increased (≥4-fold) when Ypk1-myc was isolated from cells cooverexpressing Pkh1 but only modestly elevated (∼1.5-fold) when Pkh2 was cooverexpressed (Figure 3A, top). Because Ypk1-myc and Pkh1 (or Pkh2) were both expressed from multicopy plasmids carrying the GAL1 promoter, which compete for a limiting pool of the Gal4 transactivator, the total amount of Ypk1 produced was reduced (Figure 3A, bottom); thus, the increase in specific activity when Ypk1-myc and Pkh1 were co-overexpressed is even more dramatic than the observed increase in total activity. Moreover, immunoprecipitated Ypk1-myc ran as a set of multiple bands that were collapsed into a single band of faster mobility upon treatment with a phosphatase (Figure 3A, bottom). Phosphatase-treated samples were no longer catalytically active (Torrance, unpublished observations). These findings suggest that activation is due to phosphorylation and that activation of Ypk1 is more dependent on Pkh1 than on Pkh2.
Figure 3.
Preferential activation of Ypk1 and Ykr2 by Pkh1 and Pkh2. (A) Pkh1 differentially activates Ypk1. Top, wild-type (WT; W303-1B), pkh1Δ (AC301), and pkh2Δ (YPT67) cells overexpressing from the GAL1 promoter on plasmids either Ypk1 (pAM75) or Ypk1-myc (pAM76), in the absence or presence of co-overexpression from the GAL1 promoter on plasmids of either Pkh1 (pAM73) or Pkh2 (pAM79), as indicated, were grown to mid-exponential phase and induced with galactose for 3 h. Extracts were prepared and identical amounts of total protein (1 mg) were immunoprecipitated with anti-Myc mAb 9E10. The immune complexes were washed extensively with protein kinase assay buffer and then incubated with a specific peptide substrate (Cross-tide) and [γ-32P]ATP, and the resulting product measured, as described in MATERIALS AND METHODS. Bottom, samples of each immunoprecipitate were treated with phosphatase in the absence and presence of phosphatase inhibitors, and examined by SDS-PAGE, as described in MATERIALS AND METHODS. (B) Pkh2 differentially activates Ykr2. Wild-type (WT; W303-1B), pkh1Δ (AC301), and pkh2Δ (YPT67) cells overexpressing from the GAL1 promoter on plasmids either Ykr2 (pAM1) or Ykr2-Myc (pAM78), in the absence or presence of co-overexpression from the GAL1 promoter on plasmids of either Pkh1 (pAM73) or Pkh2 (pAM79), as indicated, were grown to mid-exponential phase and induced with galactose for 3 h. Extracts were prepared and activity was measured, as in A. Values shown in A and B represent the average of three independent experiments, each performed in duplicate, and the error bars represent the range of values observed.
Immune complexes from wild-type cells expressing wild-type Ykr2-myc showed approximately a 10-fold increase in phosphotransferase activity compared with both negative controls: immune complexes from wild-type cells expressing untagged Ykr2 (Figure 3B) and immune complexes from wild-type cells expressing catalytically inactive Ykr2-myc (Torrance, 2000). When Ykr2-myc was isolated from either pkh1Δ or pkh2Δ cells, however, the increase in activity was reproducibly lower than that observed in wild-type cells, suggesting that both Pkh1 and Pkh2 contribute to phosphorylation and activation of Ykr2. Total activity was stimulated approximately threefold when Ykr2-myc was recovered from cells co-overexpressing Pkh2, but only ∼1.5-fold when Pkh1 was co-overexpressed (Figure 2B), indicating that phosphorylation and activation of Ykr2 are more responsive to Pkh2 than to Pkh1.
Loss of PKH2 (but Not PKH1) Exacerbates Slow Growth of ypk1Δ Cells
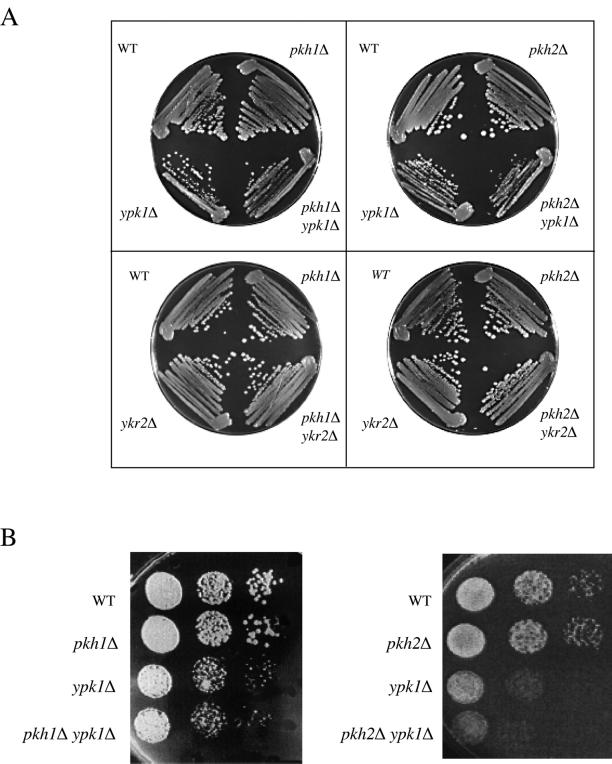
The biochemical assays discussed above indicated that Pkh1 preferentially activates Ypk1 and Pkh2 preferentially activates Ykr2. Given that both phk1Δ pkh2Δ and ypk1Δ ykr2Δ double mutants are inviable (Casamayor et al., 1999), if the discrimination observed in vitro with overexpressed proteins is even more stringent in vivo when these enzymes are expressed at their normal levels then it might be expected that certain mutant combinations might display genetic interaction. To construct all possible double mutant combinations between either ypk1Δ or ykr2Δ and either pkh1Δ or pkh2Δ, a ypk1Δ haploid was crossed to a pkh1Δ strain and to a pkh2Δ strain to create two diploid strains: PKH1/pkh1Δ::TRP1 YPK1/ypk1Δ::HIS3 and PKH2/pkh2Δ::HIS3 YPK1/ypk1Δ::TRP1. Likewise, a ykr2Δ haploid was crossed to a pkh1Δ strain and to a pkh2Δ strain to create two additional diploid strains: PKH1/pkh1Δ::TRP1 YKR2/ykr2Δ::HIS3 and PKH2/pkh2Δ::HIS3 YKR2/ykr2Δ::TRP1. The four doubly heterozygous diploid strains were sporulated. After tetrad dissection, viable Trp+ His+ haploid spores were readily recovered from all four diploids, indicating that all four double mutants (pkh1Δ ypk1Δ, pkh2Δ ypk1Δ, pkh1Δ ykr2Δ, and pkh2Δ ykr2Δ) are viable (Figure 4A). However, all of the pkh2Δ ypk1Δ spore clones grew significantly more slowly than any of the ypk1Δ spore clones or any of the pkh2Δ spore clones, when either streaked to single colonies on plates (Figure 4A) or examined by more definitive spot tests (Figure 4B). This finding provides genetic evidence in support of the biochemical results that the primary and physiologically relevant upstream activator of Ykr2 in vivo is Pkh2 (and not Pkh1). Nonetheless, a yeast cell can survive with either member of these two tiers of protein kinases. Hence, Pkh1 and Pkh2 must each be able to phosphorylate and activate either Ypk1 or Ykr2 to at least some significant degree.
Figure 4.
Slow growth of ypk1Δ cells is exacerbated by absence of Pkh2 (but not Pkh1). (A) Four spores of tetratype asci produced by doubly heterozyous diploids derived from the following four crosses were streaked to single colonies and tested for growth on YPGlc plates at 30°C: top left, a ypk1Δ strain (YFR107) against a pkh1Δ strain (YFR105); top right, a ypk1Δ strain (YFR107) against a pkh2Δ strain (YFR106); bottom left, a ykr2Δ strain (YFR119) against a pkh1Δ (YFR105) strain; and bottom right, a ykr2Δ strain (YFR64) against a pkh2Δ (YFR106) strain. (B) Serial dilutions of exponentially growing cultures of the indicated genotypes derived, respectively, from the crosses ypk1Δ (YFR107) against pkh1Δ (YFR105), left side, and ypk1Δ (YFR107) against pkh2Δ (YFR106), right side, were spotted on YPGlc plates and grown for 2 d at 30°C.
Loss of PKH1 (but Not PKH2) Alleviates Toxicity of Hyperactive Ypk1
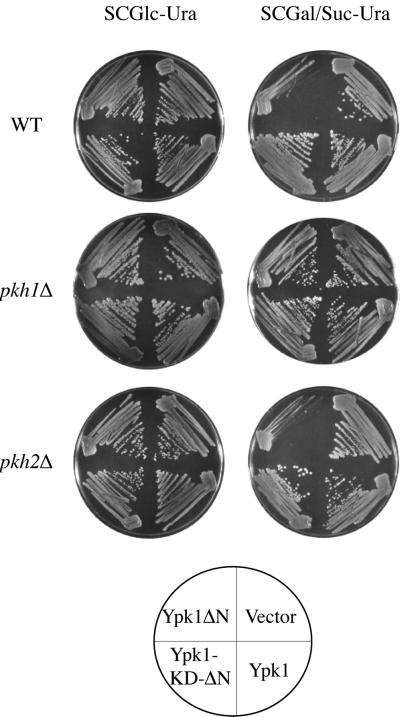
Ypk1 and Ykr2 share 88% identity within their 252-residue kinase domains and 75% identity within their downstream 75-residue C-terminal extensions. Ypk1 and Ykr2 also share considerable similarity within their 350–353-residue amino-terminal extensions: 22% identity within the first ∼100 residues and strikingly, 65% identity within the next 250 residues. To investigate what the large amino-terminal domain might contribute to the function of Ypk1, a truncation allele of Ypk1 was constructed in which the entire amino terminus (residues 2–336) was deleted. The YPK1(Δ2--336) allele, encoding Ypk1-ΔN, commences with Ser337; in Ypk1, the first Gly of the GxGxxG motif conserved in all protein kinases (Hanks and Hunter, 1995) lies at residue 354. To permit its conditional expression, YPK1(Δ2-336) was inserted in a multicopy vector under control of the GAL1 promoter. Induction of the resulting plasmid, pGAL-YPK1-ΔN (pAM101), in a wild-type strain (W303-1B) on galactose-containing medium was toxic as judged by the exceedingly slow growth of single colonies (Figure 5). Despite its toxicity, overexpression of Ypk1-ΔN was capable of restoring growth (albeit very slowly) to an otherwise inviable ypk1Δ ykr2Δ double mutant (Torrance, 2000). The observed toxicity required the catalytic activity of Ypk1-ΔN because a kinase-dead derivative, Ypk1(K376A)-ΔN, was not detectably growth inhibitory (Figure 4), even though immunoblotting indicated that, after induction, Ypk1-ΔN and Ypk1(K376A)-ΔN were expressed at equivalently high levels (Torrance, unpublished observations). Revealingly, the toxicity of Ypk1-ΔN was also alleviated in cells lacking Pkh1, but not in cells lacking Pkh2 (Figure 5). The fact that Pkh1 was required for the dominant toxicity of Ypk1-ΔN provides genetic evidence in support of the biochemical results that the primary and physiologically relevant upstream activator of Ypk1 in vivo is Pkh1 (and not Pkh2).
Figure 5.
Absence of Pkh1 (but not Pkh2) alleviates toxicity of constitutively active Ypk1. Wild-type (WT; W303-1B), pkh1Δ (AC301), or pkh2Δ (YPT67) strains were transformed with either an empty vector (YEp352GAL) or the same vector expressing from the GAL1 promoter either normal Ypk1 (pAM75), the C-terminal catalytic domain of Ypk1 (Ypk1-ΔN; pAM101), or a catalytically inactive derivative of the C-terminal catalytic domain of Ypk1 (Ypk1-KD-ΔN; pFR30). The resulting transformants were selected on glucose-containing medium and then representative isolates were streaked onto selective medium containing either Glc (left) or Gal (right) as the carbon source. Growth was assessed after 3 d at 30°C.
Differential Subcellular Localization of Pkh1, Pkh2, Ypk1, and Ykr2
One explanation for the observed preferential phosphorylation of Ypk1 by Pkh1, and of Ykr2 by Pkh2, is that the activating enzyme and its downstream target are confined to the same subcellular compartment. As an initial approach to examine localization, each of these four proteins was tagged at its N terminus with three tandem in-frame repeats of GFP and expressed from the GAL1 promoter in a multicopy plasmid. Both 3GFP-Pkh1 and 3GFP-Pkh2 were able to complement the temperature sensitivity of a pkh1ts pkh2Δ strain at restrictive temperature (37°C) on galactose-containing medium and even on glucose-containing medium (Roelants, unpublished observations), indicating that each construct was functional. Similarly, 3GFP-Ypk1 and 3GFP-Ykr2 retained their biological function (and transcriptional control was tighter) because each construct was able to complement the temperature sensitivity of ypk1-1ts ykr2Δ cells at the nonpermissive temperature (37°C) on galactose-containing medium (but not on glucose-containing medium) (Roelants, unpublished observations). In addition, as judged by immunoblotting with anti-GFP antibodies, each of the four tagged proteins was expressed intact and had the molecular weight expected for the full-length chimeric protein (Roelants, unpublished observations).
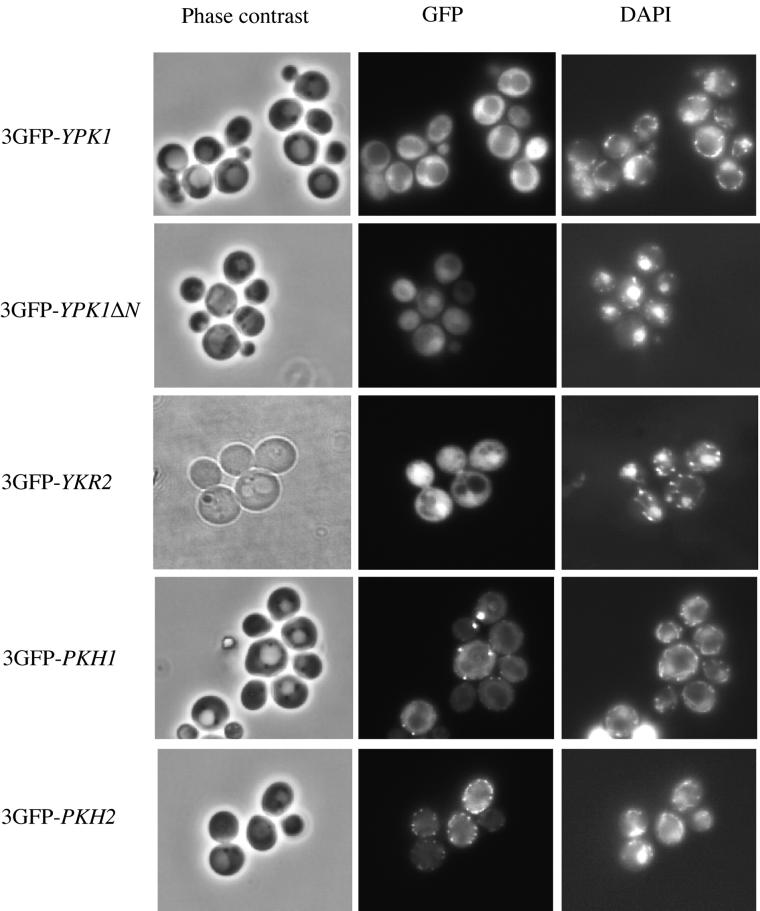
Live wild-type cells expressing each of the four fusions to 3GFP were examined under the fluorescence microscope (Figure 6). The 3GFP-Ypk1 chimera was found exclusively in the cytosol and was excluded from both the vacuole (whose position was observed by phase contrast microscopy of the same field) and the nucleus (whose position was revealed by growing the cells in the DNA-specific dye DAPI). In contrast, the 3GFP-Ykr2 chimera accumulated in the nucleus, congruent with the DAPI-stained DNA, although it was also readily detectable in the cytoplasm. Interestingly, when fused to 3GFP, the catalytic domain of Ypk1 (Ypk1ΔN), which by itself is toxic when overexpressed (see above), was located predominantly in the nucleus, unlike full-length 3GFP-Ypk1, suggesting that its toxicity may arise largely from its mislocalization. The same patterns of distribution for Ypk1 and Ykr2 were also observed if the cells were fixed, permeabilized, and stained, respectively, with polyclonal anti-Ypk1 and anti-Ykr2 antibodies (Torrance, unpublished observations). Likewise, identical patterns of distribution were observed when cells expressing Ypk1-myc or Ykr2-myc were examined by indirect immunofluorescence using anti-c-Myc mAb 9E10 (Roelants, unpublished observations).
Figure 6.
Subcellular localization of GFP-tagged Ypk1, Ykr2, Pkh1, and Pkh2. Wild-type (YPH499) cells were transformed with low copy number (CEN DNA-based) plasmids expressing from the GAL1 promoter either 3GFP-Ypk1 (pFR33), 3GFP-Ypk1-ΔN (pFR34), 3GFP-Ykr2 (pER2), 3GFP-Pkh1 (pFR37), or 3GFP-Pkh2 (pER3), as indicated. The transformants were grown to mid-exponential phase at 30°C in SCRaf/Suc-Leu, induced with galactose (2%) for 3 h, and samples of each culture were viewed directly under a fluorescence microscope. To permit visualization of the position of the nucleus, DAPI was added to the medium (1 μg/ml final concentration) during the last hour of induction.
As observed for 3GFP-Ypk1, 3GFP-Pkh1 was localized exclusively to the cytosol, and clearly excluded from both the vacuole and the nucleus (Figure 6). The most prominent feature of the 3GFP-Pkh1 staining was, however, bright puncta or larger patches situated at the cell cortex. These dots are not congruent with actin patches, as was revealed by costaining with rhodamine-labeled phalloidin (Roelants, unpublished observations). The same distribution pattern was observed if cells expressing Pkh1-(HA)3 were fixed, permeabilized, and examined by indirect immunofluorescence by using an anti-HA mAb and an appropriate fluorescently tagged secondary antibody (Roelants, unpublished observations). Unlike 3GFP-Pkh1, 3GFP-Pkh2 was not excluded from the nucleus, but like 3GFP-Pkh1, the most prominent feature of the staining was a large number of punctate bodies immediately subtending the plasma membrane (Figure 6), which were also distinct from actin patches (Roelants, unpublished observations).
Thus, taken together, these observations indicate that Pkh2 and Ykr2 are able to enter a compartment (the nucleus) from which Pkh1 and Ypk1 are normally excluded. Thus, these findings help to explain, at least in part, the greater dependence of Ypk1 activation on Pkh1 and the greater dependence of Ykr2 activation on Pkh2.
Genetic Analysis of Ypk1 and Ykr2 Function by Selection of Dosage Suppressors
To identify gene products that may be involved in processes both upstream and downstream of Ypk1 and Ykr2, we selected, first, for genes that when overexpressed from a URA3-marked multicopy vector, were able to restore growth to ypk1-1ts ykr2Δ cells at an otherwise nonpermissive temperature (35°C), as described in MATERIALS AND METHODS. From 20,000 Ura+ transformants, we recovered 18 plasmids that were able to support growth reproducibly at the restrictive temperature (Table 2). As expected, seven independent isolates of YKR2 and one isolate of YPK1 were obtained. Two of the other suppressor genes obtained, one encoding a putative chaperone (HLJ1) and the other encoding a component of RNA polymerase II holoenzyme (SRB4), may rescue because they stabilize or elevate expression of the temperature-sensitive Ypk1-1 enzyme, although this hypothesis was not tested directly. Another suppressor plasmid carried the YPC1 gene, which encodes an enzyme that can generate phytosphingosine from the corresponding phytoceramide (Mao et al., 2000) and hence presumably rescues by hyperstimulating Pkh1 and Pkh2. Indeed, we have shown previously that elevated Pkh1 can restore growth to ypk1-1ts ykr2Δ cells at otherwise restrictive temperature (Casamayor et al., 1999). Indeed, when excised from the original isolate and overexpressed from a completely different vector, YPC1 rescues the temperature sensitivity of ypk1-1ts ykr2Δ cells (Roelants, unpublished observations). In contrast, at least one other of the genes from the same insert, RPS6B, was not a suppressor on its own (Roelants, unpublished observations).
Table 2.
Dosage suppressors of the temperature-sensitive lethality of ypk1-1ts ykr2Δcells
| Complete open reading frame(s) in insert | Corresponding gene(s) | No. of independent isolates | Functiona |
|---|---|---|---|
| YMR104c | YKR2/YPK2 | 7 | Serine/threonine-specific protein kinase |
| YLR300w | EXG1 | 2 | Exo-β-1,3-glucanase |
| YBL104c | Uncharacterized | 2 | Unknown; 50% of null cells show shriveled cell surfaces and elongated buds |
| YKL126w | YPK1 | 1 | Serine/threonine-specific protein kinase |
| YMR008c | PLB1 | 1 | Phospholipase B |
| YMR161w | HLJ1 | 1 | Member of DnaJ family of putative protein chaperones |
| YER022w | SRB4 | 1 | Component of the “mediator” subcomplex of RNA polymerase II holoenzyme |
| YMR291w | Uncharacterized | 1 | Serine/threonine-specific protein kinase |
| YMR292w | GOT1 | Membrane protein required for ER-to-Golgi transport | |
| YBR181c | RPS6B | 1 | Ribosomal protein S6 isoform |
| YBR182c | SMP1 | Member of the MADS-box family of transcription factors | |
| YBR183w | YPC1 | Alkaline phytoceramidase | |
| YDR371w | CTS2 | 1 | Putative chitinase |
| YDR372c | Uncharacterized | Unknown; phosphoprotein | |
| YDR373w | FRQ1 | Frequenin; small Ca2+-binding regulatory protein | |
| YDR374c | Uncharacterized | Unknown |
Compiled from information available at the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces), and the Yeast Proteome Database maintained by Incyte, Inc (https://www.incyte.com/proteome/YPDsearch-quick.html).
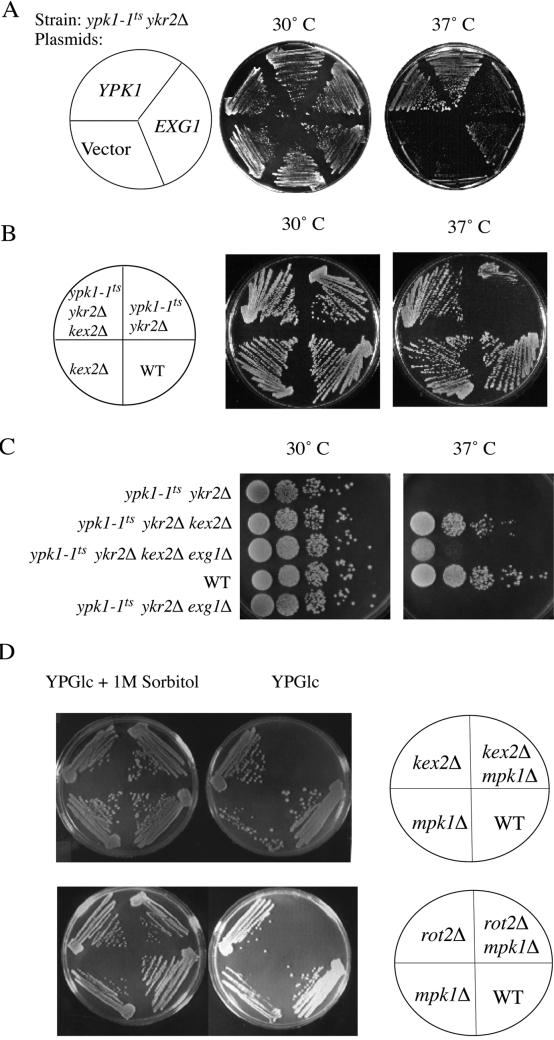
Revealingly, among the seven remaining dosage suppressors, two plasmids carried a single intact open reading frame corresponding to the EXG1 gene, which encodes the major exo-β(1,3)-glucanase involved in cell wall remodeling (Larriba et al., 1995). Indeed, when excised from the original isolate and expressed from a constitutive promoter (ADH1) in a completely different multicopy vector (see MATERIALS AND METHODS), elevated expression of EXG1 reproducibly suppressed, albeit weakly, the temperature-sensitive growth defect of ypk1-1ts ykr2Δ cells, even at 37°C (Figure 7A). Also obtained were two isolates of a locus (YBL104c) of unknown function, but which seems from the phenotype of a null allele to also have effects on cell wall structure (Obermaier et al., 1995). Another suppressor plasmid isolated carries multiple open reading frames, one of which is a candidate chitinase (Jacq et al., 1997), which may also influence cell wall structure. At least one of the other genes carried on this same plasmid, FRQ1 (Hendricks et al., 1999), is not responsible for the suppression and does not contribute to the suppression (Roelants, unpublished observations). The final two dosage suppressors encoded proteins that might act by enhancing the efficiency with which enzymes involved in cell wall biosynthesis or remodeling are delivered to their final destination and/or are activated there. One plasmid carried GOT1, which specifies a membrane protein thought to enhance the function of a t-SNARE heavy chain, Sed5, involved in vesicle-mediated protein transport from the endoplasmic reticulum (ER) to the Golgi (Conchon et al., 1999). The other plasmid carried only PLB1, which specifies the phospholipase B that is primarily responsible for the conversion of phosphatidylcholine and phosphatidylethanolamine in the exocellular leaflet of the plasma membrane to lysophosphatidylcholine (and glycerophosphocholine) and lyso-phosphatidylethanolamine (and glycerophosphoethanolamine), respectively (Lee et al., 1994). It is well documented that the activity of many classes of membrane-associated enzymes can be influenced dramatically (either stimulated or inhibited), depending on the nature of the phospholipids (or their derivatives) with which those enzymes associate (Dowhan, 1997).
Figure 7.
Relationship between dosage suppressor (EXG1) and extragenic suppressor (kex2Δ). (A) Overexpression of EXG1 suppresses the temperature sensitivity of ypk1-1ts ykr2Δ cells. A temperature-sensitive ypk1-1ts ykr2Δ strain (YPT40) was transformed with either an empty vector (pAD4M) or the same vector expressing from the ADH1 promoter either YPK1 (pADH-YPK1) or EXG1 (pAM88). Growth of the resulting transformants was assessed after 2 d at 30° and 37°C, as indicated. (B) Deletion of KEX2 suppresses the temperature sensitivity of ypk1-1ts ykr2Δ cells. A ypk1-1ts ykr2Δ strain (YAN2) was crossed to a kex2Δ strain (KRY24), and the resulting diploid cells (YFR66) were sporulated and dissected. The four spores of a tetratype ascus derived from this diploid were recovered and tested for growth on YPGlc at 30 and 37°C, as indicated. (C) Suppression by kex2Δ requires EXG1. A MATa ypk1-1ts ykr2Δ kex2Δ strain, derived as described in B, was crossed to an exg1Δ strain (YFR84) and the resulting diploid cells were sporulated and dissected. Serial dilutions of cultures of the indicated genotypes were spotted on YPGlc plates and growth was assessed after 3 d at 30 and 37°C, respectively. (D) Either kex2Δ cells or rot2Δ cells lacking Mpk1 are inviable. A kex2Δ strain (KRY24) was crossed to a mpk1Δ strain (YFR128), and the resulting diploid cells were sporulated and dissected on plates containing 1.2 M sorbitol. The four spores of a tetratype ascus derived from this diploid were recovered and tested for growth on YPGlc and YPGlc containing 1 M sorbitol at 30°C. The same procedure was applied to diploid cells resulting from crossing a rot2Δ strain (YFR129) and an mpk1Δ strain (YFR127).
Taken together, the nature of the dosage suppressors obtained, along with our observation that ypk1Δ cells showed a general increase in permeability to inhibitory drugs of several different chemical classes (see above), strongly suggested that the primary defect in Ypk1- and Ykr2-deficient cells involved some aspect of cell wall biosynthesis and/or structure.
Genetic Analysis of Ypk1 and Ykr2 Function by Selection of Suppressor Mutations
To gain further insight, and to corroborate the conclusion that absence of Ypk1 and Ykr2 compromises some aspect of cell wall structure, we also performed a selection for genes that, when interrupted by insertion of a transposon (Tn3::LacZ::LEU2), were able to restore the ability of ypk1-1ts ykr2Δ cells (strain YPT40) to grow at high temperature (35°C), as described in MATERIALS AND METHODS. From 80,000 Leu+ transformants, 16 haploid isolates were obtained that contained a transposon insertion and for which Leu+ segregated with the ability to grow at high temperature when the isolate was backcrossed to a ypk1-1ts ykr2Δ cell of opposite mating type (strain YAN2) (Table 3). One extragenic suppressor obtained inactivated a gene (SRN2) that interacts genetically with the machinery involved in nucleocytoplasmic transport. This mutation may simply enhance export of YPK1 mRNA and thus expression of the temperature-sensitive Ypk1-1 enzyme, although this hypothesis was not tested directly. Two other insertions disrupted uncharacterized loci of unknown function. Reassuringly, however, the 13 remaining extragenic suppressors fell in genes of known function, which were all involved in processes required for biosynthesis of normal cell wall glycoproteins.
Table 3.
Transposon insertions that suppress the temperature-sensitive lethality of ypk1-1ts ykr2 · cells
| Open reading frame disrupted | Corresponding gene | No. of independent isolates | Functiona |
|---|---|---|---|
| YPL227c | ALG5 | 4 | Dolichol-phosphate-β-glucosyltransferase |
| YBR229c | ROT2 | 3 | Catalytic subunit of glucosidase II |
| YNL238w | KEX2 | 2 | Golgi-localized precursor processing endoprotease |
| YMR162c | DNF3 | 2 | P-type ATPase involved in aminophospholipid transport |
| YBL082c | ALG3/RHK1 | 1 | α(1,3)-mannosyltransferase |
| YNL219c | ALG9 | 1 | α(1,2)- or α(1,6)-mannosyltransferase |
| YLR119w | SRN2 | 1 | Unknown; nonsense allele suppresses Ran-GAP mutation (rna1-1ts) |
| YLR350w | Uncharacterized | 1 | Unknown; has three predicted membrane-spanning helices; closely related to Orm1 |
| YLR404w | Uncharacterized | 1 | Unknown |
Compiled from information available at the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces), and the Yeast Proteome Database maintained by Incyte, Inc (https://www.incyte.com/proteome/YPDsearch-quick.html).
Four independent isolates represented transposon insertions in the ALG5 gene. Alg5 is an integral membrane enzyme that transfers glucose from UDP-Glc to the dolichol carrier that is used to attach the Glc residues to the immature Asn-linked (GlcNAc)2(Man)9(Glc)3 core oligosaccharide, which is added en bloc to cell wall mannoproteins and other secreted glycoproteins in the lumen of the ER (Runge et al., 1984). Three independent suppressors were insertions in a gene (ROT2) encoding the integral membrane enzyme that trims two α(1,3)-linked glucose residues from the (GlcNAc)2(Man)9(Glc)3 core during subsequent maturation of secreted glycoproteins (Herscovics, 1999). Two additional extragenic suppressors inactivated two other genes, ALG3/RHK1 (Aebi et al., 1996) and ALG9 (Burda et al., 1996), that encode mannosyltransferases involved in adding the sixth and seventh mannose residues, respectively, to the (GlcNAc)2(Man)9(Glc)3 core during its biosynthesis. Two other suppressor mutations fell in the gene (DNF3), which encodes an apparent transport ATPase for aminophospholipids (phosphatidylethanolamine and phosphatidylserine) (Catty et al., 1997), which may affect the composition of the ER membrane and thereby influence the activity of one or more of the enzymes involved in Asn-linked oligosaccharide biosynthesis mentioned above.
Strikingly, two more independent isolates (Table 3) corresponded to transposon insertions in the KEX2 gene, which encodes a Golgi-localized endoprotease that participates in maturation of secreted precursor glycoproteins by cleaving on the C-terminal side of pairs of basic residues (Rockwell et al., 1997). To verify that suppression was due to Kex2 loss of function, we crossed a ypk1-1ts ykr2Δ strain against a kex2-Δ2::LEU2 strain, in which the entire KEX2 open reading frame was deleted and replaced by the LEU2 gene, and examined the phenotype of the spores derived from resulting tetratype asci. Just like the original transposon insertions, a standard kex2 null allele also rescued the growth of ypk1-1ts ykr2Δ cells at the restrictive temperature (Figure 7B).
One possible explanation for this suppression is that, normally, Ypk1 and/or Ykr2 are negative regulators of Kex2 synthesis, function, or intracellular trafficking. In this regard, it was noteworthy that Kex2 is reportedly involved in processing of the precursor of the Exg1 exoglucanase (Basco et al., 1996). Thus, the fact that overexpression of Exg1 also rescued the temperature sensitivity of a ypk1-1ts ykr2Δ strain (Figure 7A) suggested that, perhaps, it is the unprocessed form of Exg1 that is responsible for the suppression, because this precursor form presumably accumulates when Kex2 is absent (due to mutation) or if Kex2 is limiting (when Exg1 is overproduced). To determine whether amelioration of the temperature sensitivity of ypk1-1ts ykr2Δ cells by loss of Kex2 involved Exg1, we deleted EXG1 in the ypk1-1ts ykr2Δ kex2Δ strain. Indeed, the absence of Exg1 greatly reduced the ability of the ypk1-1ts ykr2Δ kex2Δ cells to grow at nonpermissive temperature (Figure 7C), suggesting that an intact EXG1 gene is required for mediating, at least in part, the suppressive effect of loss of Kex2. The residual growth observed could be explained by the fact that the S. cerevisiae genome encodes 12 other demonstrated and presumptive glucanases (Cappellaro et al., 1998) whose precursors may also require Kex2-mediated processing.
However, there was another equally plausible explanation for the ability of kex2 mutations to suppress the temperature sensitivity of ypk1-1ts ykr2Δ cells that was consistent with all of the above-mentioned observations. Specifically, absence of Kex2 prevents processing of certain secreted cell wall mannoproteins, causing defects in the cell wall (Moukadiri et al., 1999). Moreover, various defects in the cell wall trigger activation of the Pkc1-dependent mitogen-activated protein (MAP) kinase Mpk1/Slt2 (de Nobel et al., 2000; de Groot et al., 2001) and induction of genes under its control (Jung and Levin, 1999), including EXG1 (Roberts et al., 2000). Indeed, in agreement with the hypothesis that the Pkc1-Mpk1 pathway is induced when cell wall structure is perturbed by a kex2Δ mutation and by at least one other of the extragenic suppressors (rot2Δ) we isolated, we found that these mutants are inviable when Mpk1 is absent but rescued on medium containing an osmotic support (Figure 7D). In fact, this synthetic lethality suggests that the only reason that kex2Δ and rot2Δ mutants are able to survive is that they induce the Pkc1–Mpk1 pathway, which up-regulates glucan synthases and many other enzymes necessary to repair, modify, and maintain the otherwise abnormal cell wall (Jung and Levin, 1999; de Nobel et al., 2000; Roberts et al., 2000).
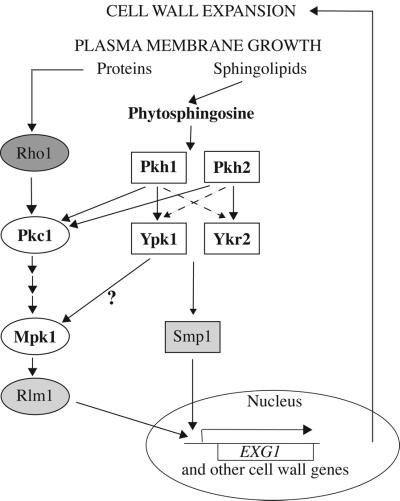
Thus, collectively, the above-mentioned findings suggested that Ypk1 and Ypk2 participate in a signaling pathway required for optimal cell wall integrity and that all of the dosage suppressors and extragenic suppressors rescue the lethality of ypk1-1ts ykr2Δ cells because they cause additional cell wall perturbations that induce the alternative Pkc1–Mpk1 cell wall integrity signaling pathway, and therefore bypass the need for efficient Ypk1- and Ykr2-dependent signaling.
Ypk1 and Ykr2 Are Involved in a Novel Cell Wall Integrity Signaling Pathway
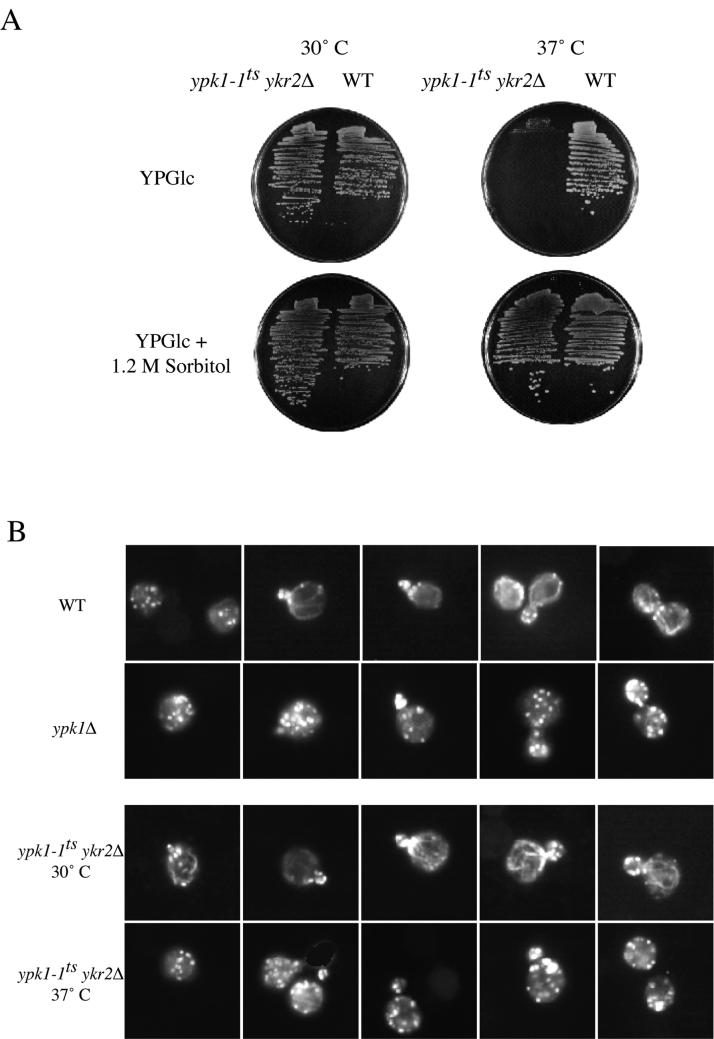
One diagnostic property of many of the conditional mutations whose primary defect is perturbation of cell wall structure is that the cells lose viability rapidly at the restrictive temperature because they undergo lysis (for review, see Cid et al., 1995). Indeed, we found that >50% of the population of ypk1-1ts ykr2Δ cells underwent lysis by 2 h after shift to nonpermissive temperature (37°C), and >90% of the cells were lysed by 4 h after the shift, as judged by staining with a commercial vital dye and by plating the cells for viable titer, whereas <2% of control cells were lysed under the same conditions (Torrance, 2000). A second hallmark of mutations that lead directly or indirectly to defects in cell wall structure is that inviability can be rescued in medium containing an osmotic support (Levin and Bartlett-Heubusch, 1992; Yoda et al., 2000). Again, consistent with a primary defect in cell wall integrity, the lysis phenotype of ypk1-1ts ykr2Δ cell could be completely prevented by the presence of an osmotic support (1.2 M sorbitol) in the growth medium (Figure 8A). Third, there is coupling between normal cell wall assembly and proper organization of the actin cytoskeleton (Helliwell et al., 1998; Delley and Hall, 1999). In normal cells, actin patches are confined to the bud and bundles of actin cables are found only in the mother cell (Pruyne and Bretscher, 2000). We found, first, that compared with isogenic wild-type cells, ypk1Δ cells showed a marked depolarization of the actin cytoskeleton even at 30°C, with pronounced actin patches in the mother cell and no detectable bundles of actin cables (Figure 8B). Likewise, at 30°C, ypk1-1ts ykr2Δ cells displayed normal actin polarization, whereas after shift to restrictive temperature (37°C) and before lysis, ypk1-1ts ykr2Δ displayed a pronounced defect in actin polarization, with numerous actin patches present in the mother cell at all stages of the cell cycle and no detectable bundles of actin cables (Figure 8B).
Figure 8.
Ypk1 and Ykr2 are required for maintenance of cell wall integrity. (A) Osmotic support permits growth of ypk1-1ts ykr2Δ cells at restrictive temperature. Either ypk1-1ts ykr2Δ cells (YPT40) or wild-type cells (WT; YPH499) were streaked onto YPGlc plates or YPGlc plates containing 1.2 M sorbitol and incubated for 2 d at 30 and 37°C, respectively. (B) Loss of Ypk1 causes defects in actin organization. Wild-type cells (WT; YPH499), top line, and ypk1Δ cells (YES3), second line, were grown to mid exponential phase at 30°C. Also, ypk1-1ts ykr2Δ cells (YPT40) were grown in YPGlc at 26°C for 18 h to an A600 nm = 0.5, third line, and then a portion of the same culture was shifted to 37°C for 2 h, bottom line. Samples of all four cultures were then fixed, stained with a reagent specific for visualizing F-actin (rhodamine-labeled phalloidin), and examined under the fluorescence microscope. (C) Hyperactivation of the Pkc1-dependent Mpk1 MAP kinase pathway rescues the temperature sensitivity of a ypk1-1ts ykr2Δ cells. Strain YPT40 (ypk1-1ts ykr2Δ) was transformed with an empty 2-μm DNA-based vector (YEp351) or with the same vector expressing from their native promoters either YPK1 (pAM21), PKC1 (pFR32), or BCK1-20 (pRS315-BCK1-20), or a different 2-μm DNA-based vector expressing EXG1 from the ADH1 promoter (pAM88). The resulting transformants were streaked onto SCGlc-Leu and growth was assessed after 3 d at the indicated temperatures (26, 35, and 36°C). (D) ypk1-1ts ykr2Δ cells lacking Mpk1 are inviable. A heterozygous ypk1-1ts/YPK1 ykr2Δ/YKR2 kex2Δ/KEX2 diploid strain (YFR66) was deleted for MPK1, and the resulting diploid strain was sporulated and dissected on YPGlc containing 1.2 M sorbitol. The indicated strains were streaked onto YPGlc plates and growth was assessed after 3 d at 26 and 30°C.
All of the above-mentioned phenotypes are also displayed by mutations that compromise the Pkc1-dependent cell wall integrity signaling pathway (for review, see Heinisch et al., 1999). In this pathway, Pkc1 (a protein kinase C-like enzyme) activates a MAP kinase cascade composed of a mitogen-activated protein kinase kinase kinase (Bck1), two redundant mitogen-activated protein kinase kinases (Mkk1 and Mkk2), and the Mpk1/Slt2 MAP kinase. Therefore, we tested directly the hypothesis that induction of the Pkc1-Mpk1 signaling pathway can bypass the defect in cells deficient in Ypk1- and Ykr2-mediated signaling. Consistent with this idea, we found that overexpression of PKC1 from its own promoter on a multicopy (2 μm DNA) plasmid was sufficient to rescue the temperature sensitivity of ypk1-1ts ykr2Δ cells (strain YPT40) and did so more efficiently than any of the dosage suppressors we selected directly, including EXG1 (Figure 8C). Similarly, PKC1 overexpression, like expression of YKR2 (as a control), was able to rescue the actin polarization defects of ypk1-1ts ykr2Δ cells at nonpermissive temperature (Roelants, unpublished observations). Likewise, expression of a constitutively active allele of BCK1 (BCK1-20) (Lee and Levin, 1992) was also able to restore growth at high temperature, although somewhat less efficaciously than PKC1 (Figure 8C).
Thus, in some senses, Pkc1 and the Mpk1 MAP kinase pathway act downstream of Ypk1 and Ykr2. On the other hand, several observations indicated that Ypk1 and Ykr2 do not act upstream of Pkc1, but rather in a parallel pathway. First, none of the dosage suppressors, none of the extragenic suppressors, and neither osmotic support nor overexpression of PKC1 or BCK1–20 was able to rescue the inviability of ypk1Δ ykr2Δ cells (Roelants and Torrance, unpublished observations). Thus, it seems that ypk1-1ts ykr2Δ cells retain some low level of throughput at the restrictive temperature and that elevation of Pkc1-induced processes can act in conjunction with that small signal but cannot substitute completely for it. Second, just as overexpression of PKC1 was unable to rescue ypk1Δ ykr2Δ cells, overexpression of YPK1 or YKR2 could not suppress the temperature sensitivity of the pkc1-2ts mutation (Roelants, unpublished observations). Third, and most revealingly, we found that a ypk1-1ts ykr2Δ mpk1Δ triple mutant was inviable on YPGlc medium at 30°C, conditions under which otherwise congenic ypk1-1ts ykr2Δ mutants, ykr2Δ mutants, mpk1Δ mutants, and ykr2Δ mpk1Δ mutants all grow well (Figure 8D). This synthetic lethality indicates that efficient Ypk1- and Ykr2-dependent signaling and signaling via the MAP kinase pathway downstream of Pkc1 are both required for viability. Hence, Ypk1 and Ykr2 most likely function in a pathway that acts in parallel to the Pkc1 pathway (or perhaps through Pkc1 effectors distinct from Mpk1).
DISCUSSION
Relative Roles of Ypk1 and Ykr2 Protein Kinases
Although either Ypk1 or Ykr2 is able to perform their shared, essential function at normal growth temperature (30°C), several lines of evidence support the conclusion that Ypk1 is the primary enzyme. First, although both ypk1Δ and ykr2Δ single mutants are alive (Chen et al., 1993; Schnieders, 1996), we found that ypk1Δ cells grow slowly at 30°C, are cold sensitive, and are hypersensitive to antibiotics and caffeine, whereas ykr2Δ cells display none of these phenotypes. The dramatic cold sensitivity of ypk1Δ cells is probably explained by the fact that recent analysis of global transcription patterns by using DNA microarrays has clearly shown that YKR2 (but not YPK1) is a gene strongly induced by heat stress (Gasch et al., 2000). Inspection of the 5′-flanking region of the YKR2 locus yields three matches to the consensus nucleotide sequence TTC(N)2–3GAA for the binding of the heat shock transcription factor (Hsf1) at −497, −404, and −104 from the ATG initiator codon. Thus, at lower temperatures, ypk1Δ cells presumably express YKR2 poorly and behave, therefore, like cells deficient in both enzymes, which are inviable. Other data also support the conclusion that, under normal growth conditions, Ypk1 is the enzyme most important for carrying out the function(s) essential for cell viability. Overexpression of catalytically inactive Ypk1 (Ypk1-KD) had detrimental effects on cell growth, whereas an equivalent level of overexpression of the analogous kinase-dead allele of Ykr2 did not. These detrimental effects were significantly exacerbated when there was no wild-type Ypk1 present to compete with the mutant protein. In other words, the presence of wild-type Ykr2 (in a ypk1Δ cell) was insufficient to overcome the toxicity imposed by Ypk1-KD, whereas the presence of wild-type Ypk1 (in a ykr2Δ cell) was able to ameliorate this toxicity to some extent. Furthermore, at steady state, Ypk1 was located exclusively in the cytosol, whereas Ykr2 was largely sequestered in the nucleus, suggesting that cytosolic targets of these enzymes are more important for normal growth than nuclear targets. Indeed, overexpression of the catalytic domain of Ypk1 was toxic, and this toxicity may be due to mislocalization because, when tagged with 3GFP, this constitutively active fragment accumulated in the nucleus. Nonetheless, overexpression of Ykr2 can overcome all of the phenotypes of ypk1Δ cells, providing evidence that both enzymes are able to perform the same essential function(s). Thus, the phenotypic differences between ypk1Δ and ykr2Δ mutants presumably arise primarily from differential expression of the corresponding genes, as discussed above, and from the differential subcellular localization of these proteins, rather than from differences in the intrinsic specificity of these enzymes for their substrates.
Division of Labor between Upstream Protein Kinases Pkh1 and Pkh2
Pkh1 and Pkh2 are responsible for the activation of several downstream protein kinases, and we have shown that either enzyme can fulfill this function (Casamayor et al. 1999). This role is sufficient to explain why Pkh1 and Pkh2 are essential gene products because the targets of Pkh1 and Pkh2 include protein kinases that are themselves known to be required for cell viability, such as Pkc1 (Inagaki et al., 1999) and, together, Ypk1 and Ykr2 (Casamayor et al., 1999). Our findings suggest, however, that Pkh1 and Pkh2 have differential roles with regard to phosphorylation and activation of Ypk1 and Ykr2. First, as measured by immune-complex kinase assays, absence of Pkh1 substantially reduced Ypk1 activity in cell extracts, whereas absence of Pkh2 did not; in contrast, absence of Pkh2 did reduce Ykr2 activity. Conversely, overproduction of Pkh1 increased Ypk1 activity in cell extracts much more than overproduction of Pkh2, whereas overproduction of Pkh2 increased Ykr2 activity more than overproduction of Pkh1. Second, as expected if Pkh1 is the primary activator of Ypk1, absence of Pkh1 (but not Pkh2) suppressed the toxicity resulting from overexpression of a constitutively active, carboxy-terminal fragment containing the catalytic domain of Ypk1. This result is in accord with our previous observations that overexpression of Pkh1 (but not Pkh2) was able to suppress the temperature sensitivity of ypk1-1ts ykr2Δ cells (Casamayor et al., 1999) and that absence of Pkh1 (but not Pkh2) resulted in a detectable decrease in incorporation of [32PO43-] into Ypk1 that was immunoprecipitated from extracts of metabolically labeled cells (Casamayor et al., 1999). Third, as expected if Pkh2 is the primary activator of Ykr2, the slow growth and drug sensitivity of ypk1Δ cells were exacerbated by absence of Pkh2 (but not Pkh1). Fourth, at steady state, Pkh1 and Ypk1 are confined to the cytosol and excluded from the nucleus, whereas Pkh2 and Ykr2 are not excluded from the nucleus. Taken together, these findings support the conclusion that, in the cell, Pkh1 preferentially activates Ypk1 and Pkh2 preferentially activates Ykr2. However, several other findings indicate that this separation is by no means complete. First, when overexpressed, either Pkh1 or Pkh2 was able to stimulate either Ypk1 or Ykr2 activity over that observed in the controls. Second, in the case of Ykr2, absence of either Pkh1 or Pkh2 caused a similar reduction in Ykr2 activity. Finally, and most convincingly, all four double mutant combinations (pkh1Δ ypk1Δ, pkh1Δ ykr2Δ, pkh2Δ ypk1Δ, and pkh2Δ ykr2Δ) were recovered as viable haploids at the expected frequency after tetrad dissection of the appropriate doubly heterozygous diploid strains. Therefore, despite the apparent Pkh1-Ypk1 and Pkh2-Ykr2 dichotomy, the capacity exists for significant cross talk between these enzyme-substrate pairs.
Physiological Function of Pkh1-Ypk1 and Pkh2-Ykr2 Cascades
There is compelling evidence in both yeast (Bagnat et al., 2000) and mammalian cells (Ikonen, 2001) that membrane microdomains enriched in sphingolipids and sterols, referred to as rafts, are involved in the biosynthetic delivery of certain proteins to the cell surface. Likewise, there is substantial evidence both in yeast (Zanolari et al., 2000) and in animal cells (Nichols and Lippincott-Schwartz, 2001) that sphingolipid rafts also play a role in a clathrin-independent route of endocytosis. It has been reported recently (Friant et al., 2001) that the sphingoid base requirement for the internalization step of endocytosis may be to activate Pkh1 and Pkh2, and that Pkc1, a known Pkh1 and Pkh2 target (Inagaki et al., 1999), acts as a downstream effector in this signaling pathway. In this regard, it is noteworthy that we found GFP-tagged Pkh1 and Pkh2 both localized primarily to prominent cortical puncta, distinct from actin patches, that seem to be membrane-associated, as judged by three-dimensional reconstruction of images taken using deconvolution fluorescence microscopy (Roelants, unpublished observations). We are currently exploring whether these structures represent microdomains enriched in sphingolipids, although there are, to our knowledge, no reliable cytological markers for such structures currently available. While our studies were in progress, it was also reported that the growth inhibitory effect of an antibiotic myriocyn (also known as ISP-1) that causes sphingolipid depletion can be overcome by overexpression of Ypk1 (Sun et al., 2000), suggesting that Ypk1 may also be a downstream target in response to phytosphingosine-dependent activation of Pkh1 and Pkh2, in agreement with our previous findings demonstrating that Pkh1 and Pkh2 act upstream of phosphorylate and activate Ypk1 and Ykr2 (Casamayor et al., 1999). Even more recently, it has been claimed that Ypk1 (but not Ykr2) is involved directly in endocytosis (deHart et al., 2002). However, proper actin assembly is critical for endocytosis in yeast (Munn, 2001; Shaw et al., 2001), and we found that loss of Ypk1 alone had profound effects on actin organization. Thus, the apparent role of Ypk1 in endocytosis may have been inferred from a rather indirect effect that is secondary to its primary function.
Several properties of ypk1-1ts ykr2Δ cells, including rapid lysis at restrictive temperature, rescue of the lysis phenotype by osmotic support, altered actin organization, and increased sensitivity to many different toxic agents, were all consistent with a cell wall defect. To gain further insight about the physiological role of Ypk1 and Ykr2, we selected both dosage suppressors and transposon insertions that restored viability to ypk1-1ts ykr2Δ cells at an otherwise nonpermissive temperature. The majority of both classes of suppressors provided additional evidence for a direct connection between Ypk1 and Ykr2 and cell wall biosynthesis. In most cases, suppression could be attributed to imposition of further cell wall damage sufficient to trigger the Pkc1-mediated activation of the Mpk1 MAP kinase-dependent cell wall integrity pathway, suggesting that Ypk1 and Ykr2 themselves are components of a novel pathway also responsible for activation of the transcription of genes involved in cell wall maintenance and remodeling. Indeed, while our article was in preparation, a study was published that also linked Ypk1 and Ykr2 to cell integrity signaling on the basis of a completely independent approach (Schmelzle et al., 2002). Because the rate of phytosphingosine generation will depend on the phytoceramide concentration in the plasma membrane, the Pkh1-Ypk1 and Pkh2-Ykr2 cascades could represent a feedback control mechanism whereby membrane growth via insertion of sphingolipid-enriched vesicles is monitored and coordinately coupled to appropriate expansion of the cell wall (Figure 9). This signaling route seems to be distinct from, but work in parallel with, the Pkc1 pathway for cell wall maintenance, in which plasma membrane proteins (such as Wsc1/Slg1 and Mid2) serve as sensors of cell turgor pressure and function by stimulating guanine nucleotide exchange factors for the small GTPase Rho1, a known activator of Pkc1 (for review, see Heinisch et al., 1999).
Figure 9.
Pkh1-Ypk1 and Pkh2-Ykr2 cascades and cell wall integrity signaling. For proper cell enlargement, plasma membrane growth needs to be accommodated by expansion of the cell wall. This coordinated coupling is achieved through at least two pathways. As shown previously, one route for monitoring plasma membrane growth is via the insertion of transmembrane proteins that affect the state of activation of the small GTPase, Rho1. Rho1, in turn, stimulates the protein kinase, Pkc1, and the downstream MAP kinase kinase cascade (Bck1 → Mkk1 and Mkk2 → Mpk1). The Mpk1 MAP kinase kinase activates transcription factors, including Rlm1, that regulate genes for cell wall enzymes. As shown in this study, a two-tiered cascade of functionally redundant protein kinases (Pkh1-Ypk1 and Pkh2-Ypk2) appears to monitor growth of the plasma membrane via a distinct mechanism. In this novel pathway, a product (phytosphingosine) derived from the vesicle-mediated insertion of sphinolipids into the plasma membrane stimulates Pkh1 and Pkh2. These enzymes, in turn, phosphorylate and activate Ypk1 and Ykr2, primarily in the cytosol and the nucleus, respectively. Genetic evidence indicates that Ypk1 and Ykr2 may act through the transcription factor, Smp1, which shares DNA-binding specificity with Rlm1. Nuclear entry is only one potential level at which the activity of these transcription factors might be regulated by these signaling pathways. Cross talk between these pathways probably occurs at two levels: full activation of Pkc1 requires phosphorylation by Pkh1 and/or Pkh2 (Inagaki et al., 1999) and Ypk1 (and perhaps Ykr2) may contribute (directly or indirectly) to Mpk1 activation (Schmelzle et al., 2002). See DISCUSSION for additional details.
One possibility is that Ypk1 and Ykr2 feed into the known cell integrity signaling pathway by providing a Pkc1-independent route to activate Mpk1 itself. Consistent with this idea is our observation that ypk1-1ts ykr2Δ cells are inviable when Mpk1 is absent and with the fact that Mpk1 phosphorylation induced by heat stress is reduced in a ypk1Δ mutant (Schmelzle et al., 2002). Moreover, the strongest suppressors we obtained were loss-of-function mutations in the Kex2-processing enzyme, which is known to cause abnormalities in the cell wall (Basco et al., 1996; Moukadiri et al., 1999), and such abnormalities evoke Pkc1- and Mpk1-dependent signaling (de Nobel et al., 2000). Not all Pkc1-dependent responses are achieved via the Mpk1 MAP kinase pathway (Delley and Hall, 1999; Li et al., 2000; Nanduri and Tartakoff, 2001). However, we found that kex2Δ mutants are inviable when Mpk1 is absent, suggesting that Kex2-deficient cells can only survive because genes under Mpk1 control are activated. Likewise, it has been observed by others that kex2Δ mutants seem to have cell wall defects (Fuller, personal communication) and are inviable when they are defective in components of a signaling pathway that monitors extracellular Ca2+, including calmodulin (Cmd1) and calmodulin-activated phosphoprotein phosphatase 2B/calcineurin (Cna1, Cna2, and Cnb1) (Davis, personal communication), or the calcineurin-activated C2H2-type zinc finger transcription factor (Crz1/Tcn1) (Cunningham, personal communication), or are exposed to known calcineurin inhibitors, cyclosporin A, or FK506 (Fuller, personal communication). Strikingly, it has been shown previously that Crz1 provides an independent means to regulate many of the same genes that are under the control of the Pkc1- and Mpk1-dependent pathway (Zhao et al., 1998; Yoshimoto et al., 2002). Additional evidence that absence of Kex2 leads to up-regulation of the transcriptional initiation of genes essential for viability (including, presumably, cell wall synthesis) is provided by two previously obscure findings. First, it was observed that kex2Δ mutations bypass temperature-sensitive mutations in the largest subunit of RNA polymerase II (Rpo21) and also temperature-sensitive mutations in other RNA polymerase II subunits (Martin and Young, 1989). Second, and conversely, kex2Δ mutations are synthetically lethal when combined with otherwise viable null mutations (ppr2Δ) in the gene encoding a factor (TFIIS) needed for efficient transcriptional elongation by RNA polymerase II (Davie and Kane, 2000).
However, for several reasons, we currently favor the idea that Ypk1 and Ykr2 activate genes for cell wall remodeling independently of the Pkc1-activated Mpk1 MAP kinase kinase pathway. One of the transcription factors under Mpk1 control is Rlm1 (676 residues), a member of the Mcm1, Agamous, Deficiens, Serum Response Factor-box family of transactivators (Watanabe et al., 1995; Dodou and Treisman, 1997). In this regard, it seems more than a coincidence that one of the dosage suppressors of the temperature-sensitive lethality of ypk1-1ts ykr2Δ cells that we isolated carries the SMP1 gene, which encodes another Mcm1, Agamous, Deficiens, Serum Response Factor-box transcription factor related to Rlm1. Smp1 (452 residues) shares near identity to Rlm1 in its ∼60 residue, N-terminal, DNA-binding domain (but bears little similarity to Rlm1 beyond that), recognizes the same (but a somewhat more extended) sequence motif as Rlm1, and is even able to form heterodimers with Rlm1 (Dodou and Treisman, 1997). Indeed, when excised from the original isolate and expressed from a completely different multi-copy vector, elevated SMP1 expression reproducibly suppressed the temperature-sensitive growth defect of ypk1-1ts ykr2Δ cells and did so better than YPC1 and as well as EXG1 (Roelants, unpublished observations). Moreover, Smp1 contains three consensus Ypk1 phosphorylation sites (-R-x-R-x-x-S/T-Hyd-, where Hyd indicates any bulky hydrophobic residue; Casamayor et al., 1999), and in preliminary experiments, Smp1 can be phosphorylated by Ypk1 in vitro (Roelants, unpublished observations). Hence, currently we favor the idea that Ypk1 and Ykr2 do not act by leading to Mpk1 activation per se, but rather by providing an independent input through Smp1 that acts in parallel to or in conjunction with Rlm1 to activate genes involved in cell wall metabolism, including EXG1 (Figure 9). Experiments to test the above-mentioned ideas, including analysis of global transcription profiles with DNA microarrays, are underway.
ACKNOWLEDGMENTS
We thank Antonio Casamayor and Dario Alessi for hospitality and expert advice; Satoru Uzawa and W. Zacheus Cande for assistance with deconvolution fluorescence microscopy and three-dimensional image reconstruction; Gabriel Schlenstedt, Michael Snyder, and David E. Levin for the gift of plasmids and/or strains; Robert S. Fuller, Trisha N. Davis, and Kyle W. Cunningham for the communication of unpublished results; Amar Nijagal and Ellyn Rosenthal for technical assistance; Brian Krechman for comments on the manuscript; and Elisabeth A. Schnieders, Matthias L.A. Versele, Nathan C. Rockwell, and other members of the Thorner laboratory for constructive criticism during the course of these studies. This work was supported by National Cancer Institute Postdoctoral Traineeship CA-09041 (to F.M.R.), by National Institutes of Health Predoctoral Traineeship GM-07232 and a National Science Foundation Predoctoral Fellowship (to P.D.T.), by funds from the undergraduate Work-Study Program of the University of California at Berkeley (to N.B.), and by National Institutes of Health Research Grant GM-21841 and facilities provided by the Cancer Research Laboratory at the University of California at Berkeley (to J.T.)
Note added in proof.
Inspection of ypk1Δ mutants, and ypk1-1ts ykr2Δ mutants at nonpermissive temperature, stained with Alexa-564-phalloidin using deconvolution fluorescence microscoy (performed by Isabelle Sagot and David Pellman, Dana Farber Cancer Research Institute, Boston, MA) revealed that these cells do contain actin cables, which were obscured when the cells were viewed by standard epifluorescence microscopy due to the increased number, brightness, and delocalization of the actin patches in these mutants.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–04–0201. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–04–0201.
REFERENCES
- Aebi M, Gassenhuber J, Domdey H, te Heesen S. Cloning and characterization of the ALG3 gene of Saccharomyces cerevisiae. Glycobiology. 1996;6:439–444. doi: 10.1093/glycob/6.4.439. [DOI] [PubMed] [Google Scholar]
- Alessi DR, Cohen P, Ashworth A, Cowley S, Leevers SJ, Marshall CJ. Assay and expression of mitogen-activated protein kinase, MAP kinase kinase, and Raf. Methods Enzymol. 1995;255:279–290. doi: 10.1016/s0076-6879(95)55031-3. [DOI] [PubMed] [Google Scholar]
- Alessi DR, Kozlowski MT, Weng Q-P, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- Bagnat M, Keranen S, Shevchenko A, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basco RD, Cueva R, Andaluz E, Larriba G. In vivo processing of the precursor of the major exoglucanase by Kex2 endoprotease in the Saccharomyces cerevisiae secretory pathway. Biochim Biophys. 1996;1310:110–118. doi: 10.1016/0167-4889(95)00156-5. [DOI] [PubMed] [Google Scholar]
- Benton BM, Zang J-H, Thorner J. A novel FK506- and rapamycin-binding protein (FPR3 gene product) in the yeast Sacharomyces cerevisiae is a proline rotomase localized to the nucleus. J Cell Biol. 1994;127:623–639. doi: 10.1083/jcb.127.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Lacroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genetics. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brazil DP, Hemmings BA. Ten years of protein kinase B signaling. a hard Akt to follow. Trends Biochem Sci. 2001;26:657–664. doi: 10.1016/s0968-0004(01)01958-2. [DOI] [PubMed] [Google Scholar]
- Burda P, te Heesen S, Brachat A, Wach A, Dusterhoft A, Aebi M. Stepwise assembly of the lipid-linked oligosaccharide in the endoplasmic reticulum of Saccharomyces cerevisiae: identification of the ALG9 gene encoding a putative mannosyl transferase. Proc Natl Acad Sci USA. 1996;93:7160–7165. doi: 10.1073/pnas.93.14.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellaro C, Mrsa V, Tanner W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J Bacteriol. 1998;180:5030–5037. doi: 10.1128/jb.180.19.5030-5037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A, Torrance PD, Kobayashi T, Thorner J, Alessi DR. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol. 1999;9:186–197. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- Catty P, de Kerchove d'Exaerde A, Goffeau A. The complete inventory of the yeast Saccharomyces cerevisiae P-type transport ATPases. FEBS Lett. 1997;409:325–332. doi: 10.1016/s0014-5793(97)00446-8. [DOI] [PubMed] [Google Scholar]
- Chen P, Lee KS, Levin DE. A pair of putative protein kinase genes (YPK1 and YPK2) is required for cell growth in Saccharomyces cerevisiae. Mol Gen Genet. 1993;236:443–447. doi: 10.1007/BF00277146. [DOI] [PubMed] [Google Scholar]
- Chou MM, Hou W, Johnson J, Graham LK, Lee MH, Chen CS, Newton AC, Schaffhausen BS, Toker A. Regulation of protein kinase C zeta by PI 3-kinase and PDK-1. Curr Biol. 1998;8:1069–1077. doi: 10.1016/s0960-9822(98)70444-0. [DOI] [PubMed] [Google Scholar]
- Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- Cid VJ, Duran A, del Rey F, Snyder MP, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conchon S, Cao X, Barlowe C, Pelham HR. Got1p and Sft2p: membrane proteins involved in traffic to the Golgi complex. EMBO J. 1999;18:3934–3946. doi: 10.1093/emboj/18.14.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR. ‘Marker swap’plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Currie RA, Walker KS, Gray A, Deak M, Casamayor A, Downes CP, Cohen P, Alessi DR, Lucocq J. Role of phosphatidylinositol 3,4,5-trisphosphate in regulating the activity and localization of 3-phosphoinositide-dependent protein kinase-1. Biochem J. 1999;337:575–583. [PMC free article] [PubMed] [Google Scholar]
- Davie JK, Kane CM. Genetic interactions between TFIIS, and the Swi-Snf chromatin-remodeling complex. Mol Cell Biol. 2000;20:5960–5973. doi: 10.1128/mcb.20.16.5960-5973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Emr SD, McPherson PS, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- de Groot PWJ, et al. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp Funct Genomics. 2001;2:124–142. doi: 10.1002/cfg.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHart AK, Schnell JD, Allen DA, Hicke L. The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J Cell Biol. 2002;156:241–248. doi: 10.1083/jcb.200107135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delley PA, Hall MN. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J Cell Biol. 1999;147:163–174. doi: 10.1083/jcb.147.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nobel H, Ruiz C, Martin H, Morris W, Brul S, Molina M, Klis FM. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase, and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance, and thermotolerance. Microbiology. 2000;146:2121–2132. doi: 10.1099/00221287-146-9-2121. [DOI] [PubMed] [Google Scholar]
- Dodou E, Treisman R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:1848–1859. doi: 10.1128/mcb.17.4.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- Evans GI, Lewis GK, Ramsey G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields FO. Biochemical and molecular genetic analysis of protein kinase C function in the yeast Saccharomyces cerevisiae. Ph.D. Thesis. Berkeley, CA: University of California, Berkeley; 1991. [Google Scholar]
- Fields FO, Thorner J. Genetic suppression analysis of the function of a protein kinase C (PKC1 gene product) in Saccharomyces cerevisiae cell cycle progression: the SKCd mutations. Cold Spring Harb Symp Quant Biol. 1991;56:51–60. doi: 10.1101/sqb.1991.056.01.008. [DOI] [PubMed] [Google Scholar]
- Friant S, Lombardi R, Schmelzle T, Hall MN, Riezman H. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 2001;20:6783–6792. doi: 10.1093/emboj/20.23.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman DA, Rameh LE, Cantley LC. Phosphoinositide binding domains: embracing 3-phosphate. Cell. 1999;97:817–820. doi: 10.1016/s0092-8674(00)80792-8. [DOI] [PubMed] [Google Scholar]
- Fuller RS, Brake A, Thorner J. Yeast prohormone processing enzyme (Kex2 gene-product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci USA. 1989;86:1434–1438. doi: 10.1073/pnas.86.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1988. [Google Scholar]
- Hawkins PT, Stephens LR, Piggott JR. Analysis of inositol metabolites produced by Saccharomyces cerevisiae in response to glucose stimulation. J Biol Chem. 1993;268:3374–3383. [PubMed] [Google Scholar]
- Heinisch JJ, Lorberg A, Schmitz HP, Jacoby JJ. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol Microbiol. 1999;32:671–680. doi: 10.1046/j.1365-2958.1999.01375.x. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Schmidt A, Ohya Y, Hall MN. The Rho1 effector Pkc1, but not Bni1 mediates signaling from Tor2 to the actin cytoskeleton. Curr Biol. 1998;8:1211–1214. doi: 10.1016/s0960-9822(07)00511-8. [DOI] [PubMed] [Google Scholar]
- Hendricks KB, Wang BQ, Schnieders EA, Thorner J. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat Cell Biol. 1999;1:234–241. doi: 10.1038/12058. [DOI] [PubMed] [Google Scholar]
- Herscovics A. Processing glycosidases of Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1426:275–285. doi: 10.1016/s0304-4165(98)00129-9. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Ikonen E. Roles of lipid rafts in membrane transport. Curr Opin Cell Biol. 2001;13:470–477. doi: 10.1016/s0955-0674(00)00238-6. [DOI] [PubMed] [Google Scholar]
- Inagaki M, Schmelzle T, Yamaguchi K, Irie K, Hall MN, Matsumoto K. PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast. Mol Cell Biol. 1999;19:8344–8352. doi: 10.1128/mcb.19.12.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq C, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome IV. Nature. 1997;387:75–78. [PubMed] [Google Scholar]
- Jung US, Levin DE. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signaling pathway. Mol Microbiol. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- King CC, Zenke FT, Dawson PE, Dutil EM, Newton AC, Hemmings BA, Bokoch GM. Sphingosine is a novel activator of 3-phosphoinositide-dependent kinase 1. J Biol Chem. 2000;275:18108–18113. doi: 10.1074/jbc.M909663199. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Deak M, Morrice N, Cohen P. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem J. 1999;344:189–197. [PMC free article] [PubMed] [Google Scholar]
- Kozma SC, Thomas G. Regulation of cell size in growth, development, and human disease PI3K, PKB and S6K. Bioessays. 2002;24:65–71. doi: 10.1002/bies.10031. [DOI] [PubMed] [Google Scholar]
- Kubo K, Ohno S, Matsumoto S, Yahara I, Suzuki K. A novel yeast gene coding for a putative protein kinase. Gene. 1989;76:177–180. doi: 10.1016/0378-1119(89)90021-8. [DOI] [PubMed] [Google Scholar]
- Kuo CJ, Chung J, Fiorentino DF, Flanagan WM, Blenis J, Crabtree GR. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature. 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larriba G, Andaluz E, Cueva R, Basco RD. Molecular biology of yeast exoglucanases. FEMS Microbiol Lett. 1995;125:121–126. doi: 10.1111/j.1574-6968.1995.tb07347.x. [DOI] [PubMed] [Google Scholar]
- Lawlor MA, Alessi DR. PKB/Akt. a key mediator of cell proliferation, survival and insulin responses? J Cell Sci. 2001;114:2903–2910. doi: 10.1242/jcs.114.16.2903. [DOI] [PubMed] [Google Scholar]
- Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- Lee KS, Levin DE. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Patton JL, Fido M, Hines LK, Kohlwein SD, Paltauf F, Henry SA, Levin DE. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J Biol Chem. 1994;269:19725–19730. [PubMed] [Google Scholar]
- Levin DE, Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE, Fields FO, Kunisawa R, Bishop JM, Thorner J. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell. 1990;62:213–224. doi: 10.1016/0092-8674(90)90360-q. [DOI] [PubMed] [Google Scholar]
- Li Y, Moir RD, Sethy-Coraci IK, Warner JR, Willis IM. Repression of ribosome, and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol Cell Biol. 2000;20:3843–3851. doi: 10.1128/mcb.20.11.3843-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM. Cloning, and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J Biol Chem. 2000;275:31369–31378. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- Martin GA, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Martin C, Young RA. KEX2 mutations suppress RNA polymerase II mutants and alter the temperature range of yeast cell growth. Mol Cell Biol. 1989;9:2341–2349. doi: 10.1128/mcb.9.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer P, Redd M, Solsbacher J, Bischoff FR, Greiner M, Podtelejnikov AV, Mann M, Stade K, Weis K, Schlenstedt G. The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates, and the yeast Ran-binding-protein 1 (Yrb1p) Mol Biol Cell. 2001;12:539–549. doi: 10.1091/mbc.12.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer RA. Isolation of a yeast protein kinase gene by screening with a mammalian protein kinase cDNA. DNA. 1988;7:469–474. doi: 10.1089/dna.1.1988.7.469. [DOI] [PubMed] [Google Scholar]
- Moukadiri I, Jaafar L, Zueco J. Identification of two mannoproteins released from cell walls of a Saccharomyces cerevisiae mnn1 mnn9 double mutant by reducing agents. J Bacteriol. 1999;181:4741–4745. doi: 10.1128/jb.181.16.4741-4745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL. Molecular requirements for the internalization step of endocytosis. insights from yeast. Biochim Biophys Acta. 2001;1535:236–257. doi: 10.1016/s0925-4439(01)00028-x. [DOI] [PubMed] [Google Scholar]
- Nanduri J, Tartakoff AM. The arrest of secretion response in yeast. signaling from the secretory path to the nucleus via Wsc proteins and Pkc1p. Mol Cell. 2001;8:281–289. doi: 10.1016/s1097-2765(01)00312-4. [DOI] [PubMed] [Google Scholar]
- Nichols BJ, Lippincott-Schwartz J. Endocytosis without clathrin coats. Trends Cell Biol. 2001;11:406–412. doi: 10.1016/s0962-8924(01)02107-9. [DOI] [PubMed] [Google Scholar]
- Obermaier B, Gassenhuber J, Piravandi E, Domdey H. Sequence analysis of a 78.6 kb segment of the left end of Saccharomyces cerevisiae chromosome II. Yeast. 1995;11:1103–1112. doi: 10.1002/yea.320111112. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Reneke JE, Blumer KJ, Courchesne WE, Thorner J. The carboxy-terminal segment of the yeast α-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Wang GT, Krafft GA, Fuller RS. Internally consistent libraries of fluorogenic substrates demonstrate that Kex2 protease specificity is generated by multiple mechanisms. Biochemistry. 1997;36:912–917. doi: 10.1021/bi961779l. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Sheehan A, Friddle C, Roeder GS, Snyder M. Transposon mutagenesis for the analysis of protein production, function, and localization. Methods Enzymol. 1999;303:512–532. doi: 10.1016/s0076-6879(99)03031-1. [DOI] [PubMed] [Google Scholar]
- Runge KW, Huffaker TC, Robbins PW. Two yeast mutations in glucosylation steps of the asparagine glycosylation pathway. J Biol Chem. 1984;259:412–417. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. ed. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Helliwell SB, Hall MN. Yeast protein kinases, and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol Cell Biol. 2002;22:1329–1339. doi: 10.1128/mcb.22.5.1329-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnieders EA. Biochemical and genetic analysis of a phosphatidylinositol 4-kinase (PIK1 gene product) in the yeast Saccharomyces cerevisiae, Ph.D. Thesis, Appendix, YPK1 and YKR2, 158–185. Berkeley, CA: University of California; 1996. [Google Scholar]
- Shaw JD, Cummings KB, Huyer G, Michaelis S, Wendland B. Yeast as a model system for studying endocytosis. Exp Cell Res. 2001;271:1–9. doi: 10.1006/excr.2001.5373. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L, et al. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B [see comments] Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- Sun Y, Taniguchi R, Tanoue D, Yamaji T, Takematsu H, Mori K, Fujita T, Kawasaki T, Kozutsumi Y. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol Cell Biol. 2000;20:4411–4419. doi: 10.1128/mcb.20.12.4411-4419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. p70s6k/p85s6k: mechanism of activation, effects of rapamycin and role in mitogenesis. Biochem Soc Trans. 1993;21:901–904. doi: 10.1042/bst0210901. [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Wigler M. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 1988;2:517–527. doi: 10.1101/gad.2.5.517. [DOI] [PubMed] [Google Scholar]
- Toker A, Newton AC. Cellular signaling. pivoting around PDK-1. Cell. 2000;103:185–188. doi: 10.1016/s0092-8674(00)00110-0. [DOI] [PubMed] [Google Scholar]
- Torrance PD. Regulation, and function of the protein kinases, Ypk1, and Ykr2, in the yeast Saccharomyces cerevisiae. Ph.D. Thesis. Berkeley, CA: University of California, Berkeley; 2000. [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection. more than just a road to PKB. Biochem J. 2000;346:561–576. [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Irie K, Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Kawada T, Kaibara C, Fujie A, Abe M, Hashimoto H, Shimizu J, Tomishige N, Noda Y, Yamasaki M. Defect in cell wall integrity of the yeast Saccharomyces cerevisiae caused by a mutation of the GDP-mannose pyrophosphorylase gene, VIG9. Biosci Biotechnol Biochem. 2000;64:1937–1941. doi: 10.1271/bbb.64.1937. [DOI] [PubMed] [Google Scholar]
- Yon J, Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, H., Saltsman, K., Gasch, A.P., Li, H.X., Ogawa, N., Botstein, D., Brown, P.O., Cyert, M.S. (2002). Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. in press. [DOI] [PubMed]
- Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson BJ, Riezman H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000;19:2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Jung US, Garrett-Engele P, Roe T, Cyert MS, Levin DE. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol Cell Biol. 1998;18:1013–1022. doi: 10.1128/mcb.18.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]