Abstract
The spindle checkpoint plays a central role in the fidelity of chromosome transmission by ensuring that anaphase is initiated only after kinetochore-microtubule associations of all sister chromatid pairs are complete. In this study, we find that known spindle checkpoint proteins do not contribute equally to chromosome segregation fidelity in Saccharomyces cerevisiae. Loss of Bub1 or Bub3 protein elicits the largest effect. Analysis of Bub1p reveals the presence of two molecular functions. An N-terminal 608-amino acid (nonkinase) portion of the protein supports robust checkpoint activity, and, as expected, contributes to chromosome segregation. A C-terminal kinase-encoding segment independently contributes to chromosome segregation through an unknown mechanism. Both molecular functions depend on association with Bub3p. A 156-amino acid fragment of Bub1p functions in Bub3p binding and in kinetochore localization by one-hybrid assay. An adjacent segment is required for Mad1p binding, detected by deletion analysis and coimmunoprecipitation. Finally, overexpression of wild-type BUB1 or MAD3 genes leads to chromosome instability. Analysis of this activity indicates that the Bub3p-binding domain of Bub1p contributes to this phenotype through disruption of checkpoint activity as well as through introduction of kinetochore or spindle damage.
INTRODUCTION
Protein components of the spindle checkpoint were first defined genetically through studies in the budding yeast Saccharomyces cerevisiae by analysis of mutants that lack the ability to arrest in the presence of spindle damage introduced by antimicrotubule drug exposure or by manipulation of temperature conditional spindle proteins (Hoyt et al., 1991; Li and Murray, 1991; Weiss and Winey, 1996). The spindle checkpoint thus defined has been shown to control at least two functionally distinct steps within mitosis. First, at metaphase, the checkpoint acts to detect a lack of bipolar attachment or tension for any sister chromatid pair. This condition delays anaphase in the presence of even a single unattached kinetochore or a lack of tension on a single chromatid pair (Spencer and Hieter, 1992; Rieder et al., 1994; Li and Nicklas, 1995). Second, entry into G1 (mitotic exit) is prevented in cells that have suffered spindle damage sufficient to preclude the delivery of a daughter nucleus into the bud (for review, see Taylor, 1999; Gardner and Burke, 2000).
Metaphase arrest due to activation of the spindle checkpoint depends upon a well-conserved pathway that regulates the degradation of the anaphase inhibitor protein Securin (budding yeast Pds1p; for review, see Amon, 1999; Zachariae and Nasmyth, 1999). Anaphase is normally initiated as Separin (Esp1p) is liberated from its binding partner Securin (Pds1p) after Securin is targeted for degradation by the Cdc20-associated form of the anaphase promoting complex. Cdc20p is a target of the metaphase checkpoint arrest pathway and physically interacts with other checkpoint proteins during metaphase arrest (for review, see Shah and Cleveland, 2000; Hoyt, 2001; Sorger, 2001). Maintenance of the arrest induced by kinetochore damage also requires arrest of the mitotic exit pathway (Krishnan et al., 2000), indicating a functional connection between the distinct control pathways that operate at anaphase initiation and mitotic exit. In budding yeast, both of these steps are inhibited by the presence of Pds1 protein (Cohen-Fix and Koshland, 1999; Tinker-Kulberg and Morgan, 1999), and thus anaphase and exit control may be related to one another by a key role played by Pds1p or Esp1p at both cell cycle positions (Fraschini et al., 2001; Sullivan et al., 2001; Jensen et al., 2001; Stegmeier et al., 2002).
Initiation of metaphase arrest in response to a lack of bipolar attachment requires at least six proteins in S. cerevisiae: Mad1p, Mad2p, Mad3p, Bub1p, Bub3p, and Mps1p (Hoyt et al., 1991; Li and Murray, 1991; Wang and Burke, 1995; Pangilinan and Spencer, 1996; Hardwick et al., 1999). These proteins function together in kinetochore surveillance, activating a checkpoint-governed arrest in response to microtubule defects, kinetochore protein defects, or centromere DNA mutations (Hoyt et al., 1991; Li and Murray, 1991; Wang and Burke, 1995; Pangilinan and Spencer, 1996; Hardwick et al., 1999). The proteins involved in kinetochore surveillance are remarkably conserved in eukaryotes, and homologs have been found in fission yeast, flies, maize, frog, mouse, and human (for review, see Amon, 1999). In systems with robust cytology, homologus of Mad1p, Mad2p, Bub1p, Bub3p, and Mps1p have been observed to concentrate at unattached kinetochores in prometaphase. Thus, these proteins behave as expected components of a molecular structure that broadcasts an inhibitory signal that will be extinguished upon achievement of bipolar attachment and/or associated tension from spindle forces exerted in opposite directions (for review, see Gillett and Sorger, 2001; Hoyt, 2001; Nasmyth, 2001).
Physical association studies have shown that the metaphase arrest proteins reside in several complexes that contain overlapping components, and that these complexes exhibit alterations in a cell cycle-regulated manner. In budding yeast, Mad1p/Mad2p, Bub1p/Bub3p, and Mad3p/Bub3p complexes are detected in interphase, whereas cells in damage-induced metaphase arrest contain a Bub1p/Bub3p/Mad1p complex, as well as a Cdc20p/Mad2p/Mad3p/Bub3p complex (Hardwick and Murray, 1995; Farr and Hoyt, 1998; Brady and Hardwick, 2000; Hardwick et al., 2000). Moreover, at metaphase arrest, both Bub1p and Mad1p exhibit shifts in gel migration consistent with hyperphosphorylated states (Hardwick and Murray, 1995; Farr and Hoyt, 1998; Brady and Hardwick, 2000). Movement of constituents among protein complexes may represent the spatial communication from an activated (unattached) kinetochore to site(s) where the Cdc20-associated form of the anaphase promoting complex is poised to initiate the degradation of Pds1p. Although biochemical characterization of protein complexes has provided insight into features of checkpoint activation, the nature of the spatial regulation imposed at metaphase by the presence of unattached kinetochores has not been precisely elucidated. Indeed, it is possible that different kinetochore states, such as kinetochore-microtubule attachment or the presence of tension, may be handled at metaphase by either overlapping or distinct signaling pathways (Waters et al., 1998; Skoufias et al., 2001; Stern and Murray, 2001).
Chromosome missegregation associated with loss of kinetochore surveillance by the spindle checkpoint has been observed. In budding yeast, Li and Murray (1991) observed an increase in chromosome missegregation in mad1, mad2, and mad3 mutants upon recovery from aberrant mitoses induced by exposure to the antimicrotubule drug nocodazole. In the absence of intentional spindle damage, chromosome missegregation has been detected in budding yeast bub1 and mad2 mutants (Pangilinan and Spencer, 1996) as well as in Drosophila melanogaster bub1 (Basu et al., 1999), Schizosaccharomyces pombe Δbub1 (Bernard et al., 1998), and Caenorhabditis elegans mdf-1 and mdf-2 mutants (Kitagawa and Rose, 1999). The chromosome missegregation phenotypes observed suggest that the spindle checkpoint plays a role in many cell cycles (even in the absence of induced damage), or that Bub1 and Mad2 checkpoint proteins have additional roles in kinetochore function.
In this report, we present a quantitative survey of the segregation roles of five nonessential metaphase checkpoint proteins that govern kinetochore surveillance (Mad1, Mad2, Mad3, Bub1, and Bub3) in cells without additional spindle damage. We find that these spindle checkpoint proteins differ in their contributions, and that the absence of Bub1p or Bub3p has the greatest impact on segregation. Further analysis of the role of Bub1p leads to a model in which Bub1 protein provides chromosome stability through two separate mechanisms.
MATERIALS AND METHODS
Yeast Media
All yeast media are as described in Rose et al. (1990).
Yeast Strains
Strains used in this study are listed in Table 1. Except where noted, experiments were conducted in an S288c laboratory background and are related by DNA-mediated transformation or isogenic mating and sporulation. Figure 1B, C, and E show data for strains that are derivatives of W303-1a. The one-hybrid assay was carried out in YJL128 (Ortiz et al., 1999) and transformants derived from it.
Table 1.
Yeast strains used in this study
| Name | Genotype | Source or Reference |
|---|---|---|
| S288c background | ||
| YPH278 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1+CFIII (CEN3.L.YPH278) URA3 SUP11 | Spencer et al. 1990 |
| YFP2 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 bub1Δ::LEU2+CFIII (CEN3.L.YPH278) URA3 SUP11 | Pangilinan and Spencer, 1996 |
| YCD165 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 bub2Δ::LEU2+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YFS1100 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 bub3Δ::LEU2+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YFS1120 | MATa ura3–52 lys2–801 ade2–101 his3Δ200 trp1Δ leu2Δ1 mad1Δ::kanMX+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YCD173 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 mad2Δ::HIS3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YFS1205 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 mad3Δ::kanMX+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YCD279 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 BUB1::HIS3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YCD280 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 bub1-1::HIS3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YCD281 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 bub1K733R::HIS3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YML101 | MATa ura3–52 lys2–801 ade2–101 his3Δ200 trp1Δ63 leu2Δ1 MPS1::GAL1-Nmyc-MPS1-URA3 | This study |
| YCD362 | MATa ura3–52 lys2–801 ade2–101 his3Δ200 trp1Δ63 leu2Δ1 bub1::natMx MPS1::GAL1-Nmyc-MPS1-URA3 | This study |
| YCD371 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 bub1[1–367]::HIS3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YCD358 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 bub1[1–608]::HIS3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YCD407 | MATα ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 bub1[211–1021]::HIS3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YFS377 | MATa ura3–52 lys2–801 ade2–101 his3Δ200 trp1Δ63 leu2Δ1 ctf18Δ::LEU2 | This study |
| YJH18.3 | MATa ura3–52 lys2–801 ade2–101 his3Δ200 trp1Δ63 leu2Δ1 ctf18Δ::LEU2 mad2Δ::HIS3 | Hanna et al., 2001 |
| YCD251 | MATa/MATα ura3–52/ura3–52 lys2–801/lys2–801 ade2–101/ade2–101 HIS3/his3Δ200 trp1Δ63/TRP1 leu2Δ1/leu2Δ1+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| W303-1a background | ||
| YKH231 | MATa ura3-1 leu2,3–112 his3–11 trp1-1 ade2-1+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YRJ112 | MATa ura3-1 leu2,3–112 his3–11 trp1-1 ade2-1 bub1Δ::HIS3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YRJ113 | MATa ura3-1 leu2,3–112 his3–11 trp1-1 ade2-1 bub2Δ::TRP1+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YRJ114 | MATa ura3-1 leu2,3–112 his3–11 trp1-1 ade2-1 bub3Δ::TRP1+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YMB111 | MATa ura3-1 leu2,3–112 his3–11 trp1-1 ade2-1 mad1Δ::URA3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YMB113 | MATa ura3-1 leu2,3–112 his3–11 trp1-1 ade2-1 mad2Δ::LEU2+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YRJ111 | MATa ura3-1 leu2,3–112 his3–11 trp1-1 ade2-1 mad3Δ::URA3+CFIII (CEN3.L.YPH278) URA3 SUP11 | This study |
| YKH300 | MATa bub1Δ::URA3 BUB3-(Myc)13::G418 ura3-1 leu2,3–112 his3–11 trp1-1 ade2-1 | This study |
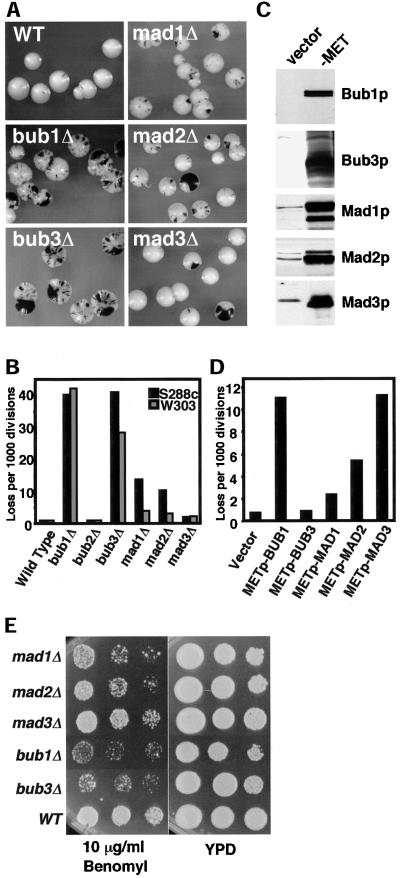
Figure 1.
Spindle checkpoint mutants exhibit different rates of chromosome loss. (A) Null mutant sectoring phenotypes. The strains shown are wild type (YPH278), bub1Δ (YFP2), bub3Δ (YFS1100), mad1Δ (YFS1120), mad2Δ (YCD173), and mad3Δ (YFS1205). (B) Chromosome loss rates in null mutants determined by half-sector analysis. Wild type: 49 half-sectored colonies/61,276 total colonies (YPH278); 8/9,305 (YKH231). bub1Δ: 192/4,784 (YFP2); 89/2,121 (YRJ112). bub2Δ: 17/22,101 (YCD165); 3/3,440 (YRJ113). bub3Δ: 137/3,362 (YFS1100); 63/2,237 (YRJ114). mad1Δ: 139/12,394 (YFS1120); 32/8,552 (YMB111). mad2Δ: 87/10,153 (YCD173); 9/3,106 (YMB113). mad3Δ: 57/29,364 (YFS1205); 29/13,186 (YRJ111). (C) Immunoblots showing overexpression from a MET25 promoter. Extracts were taken after 2 h of inductionin media lacking methionine and were analyzed by Western blot using antibody specific for each protein. The left lane (vector, p423MET) shows the endogenous Bub1p expression level where detected; the right lane (–MET) shows protein expressed from the MET25 promoter. All strains were generated from YKH231 by introduction of p423MET-derived plasmids containing full-length open reading frames cloned adjacent to the MET25 promoter. (D) Chromosome loss associated with overexpression of checkpoint genes. Half-sector analysis was performed after plating the strains in C on plates lacking methionine. Vector: 14/19,030. METpBUB1: 195/17,640. METpBUB3: 17/17,875. METpMAD1: 28/11,920. METpMAD2: 92/17,065. METpMAD3: 137/12,115. Two or more additional independent transformants tested for each construct showed the same chromosome instability phenotype by colony sectoring assay. (E) Benomyl sensitivity of checkpoint null mutants. Log phase cultures were spotted in a 10-fold dilution series on rich medium (YPD) or rich medium plus Benomyl. Strains were mad1Δ (YMB111), mad2Δ (YMB113), mad3Δ (YRJ111), bub1Δ (YRJ112), and bub3Δ (YRJ114).
Chromosome Loss Rate
This assay was performed as previously described (Hieter et al., 1985b; Spencer et al., 1990). Strains containing a nonessential SUP11-marked test chromosome and plasmids were grown in selective media and were plated at a density of ∼200 colonies per plate on minimal (SD) medium, including 20 μg/ml uracil, 40 μg/ml l-lysine, 6 μg/ml adenine sulfate, 20 μg/ml l-histidine, 30 μg/ml l-tryptophan, and 220 μg/ml l-leucine when required to cover auxotrophies. The limiting adenine supplementation was used to facilitate red pigment development in ade2-101 cells. Chromosome loss events during the first cell division were visualized as colonies that were at least one-half red. The loss rate for the SUP11-marked chromosome is expressed as loss per chromosome per cell division, and is calculated by dividing the number of half-sectored colonies by the total number of colonies scored.
Determination of the bub1-1 Mutation
The bub1-1 allele was captured on a yeast-bacterial shuttle vector by gap repair from MAY1726 (Roberts et al., 1994), and the entire open reading frame was sequenced from two independent transformants. The sole change observed was G to A at position 997, which substitutes a conserved glutamic acid with lysine in the putative Bub3p binding region. Therefore, bub1-1 is referred to as bub1-E333K.
Introduction of bub1 Mutations at the Genomic Locus
Plasmid-borne mutant alleles were created adjacent to a HIS3 marker, amplified in a single DNA fragment with the selectable marker by high-fidelity polymerase chain reaction (PCR), and integrated by homologous recombination into the native BUB1 locus. The resulting genomic structure contained the native BUB1 promoter, a mutant bub1 allele, an HIS3 downstream marker gene, and finally, natural BUB1 3′-flanking sequence. Details of the constructions are available upon request.
One-Hybrid Assay
The one-hybrid assay was performed essentially as described (Ortiz et al., 1999). YJL128 was transformed with activation domain fusion constructs, and multiple independent transformants were plated on SD-LEU supplemented with 5 mM 3-amino-triazole (3-AT). Plates were incubated at 30°C for up to 2 wk.
MPS1 Overexpression
A GAL-MPS1 allele (Hardwick et al. 1996) was created by integration of pAFS120 at the MPS1 locus of YFS589 yielding yeast strain YML101. BUB1 overexpression plasmids were introduced into YML101, and two independent transformants were picked. Cultures were grown overnight in selective media lacking histidine and uracil supplemented with 2% raffinose, diluted into selective media lacking histidine, uracil, and methionine (to derepress the MET25 promoter) supplemented with 2% raffinose and were grown to early log phase. To induce MPS1 overexpression, galactose was added to a final concentration of 3%. Samples taken at t = 0 and t = 4 h were fixed in1 M sorbitol, 50 mM KPO4, pH 7.5, and 3.7% formaldehyde, 4,6-diamidino-2-phenylindole (DAPI) stained, and scored for bud and nuclear morphology. A minimum of 200 cells was scored for each sample.
Plasmids
All overexpression constructs were made in either p423MET (2μ HIS3) or p415MET (CEN/ARS/LEU2) vectors containing the methionine-repressible MET25 promoter and the CYC1 terminator sequence flanking the multiple cloning site (Mumberg et al., 1994). For one-hybrid analysis, GAL4-AD fusions were constructed by cloning each PCR-generated open reading frame into pGADT7 (Clontech, Palo Alto, CA). All plasmids generated by PCR were verified by sequence analysis. Details are available upon request.
Immunoblotting and Coimmunoprecipitation
Immunoblotting and coimmunoprecipitation were carried out as described previously (Hardwick and Murray, 1995; Brady and Hardwick, 2000). The lysis buffer for coimmunoprecipitation was 50 mM HEPES, pH 7.6, 75 mM KCl, 50 mM NaF, 1 mM Na vanadate, 1 mM MgCl2, 1 mM EGTA, 0.1% Na Deoxycholate, 1 mM phenyl methyl sulfoxide, “complete EDTA-free protease inhibitor cocktail” (Roche, Indianapolis, IN), and 1 mM dithiothreitol. Rabbit α-Mad1, Mad2, Mad3, Bub1, and Bub3 antibodies have been previously described (Hardwick and Murray, 1995; Brady and Hardwick, 2000; Hardwick et al., 2000).
RESULTS
Spindle Checkpoint Mutants Exhibit Different Rates of Chromosome Loss
In previous work, it was apparent that chromosome loss of bub1 and mad2 mutants differed from one another (Pangilinan and Spencer, 1996). To determine the requirement for spindle checkpoint proteins in accurate chromosome segregation during normal unperturbed mitosis, null mutants of six checkpoint genes (BUB1, BUB2, BUB3, MAD1, MAD2, and MAD3) were generated in an otherwise isogenic background. To follow chromosome segregation fidelity, the loss of a nonessential test chromosome (Spencer et al., 1990) was ascertained by a colony color assay (Hieter et al., 1985a). In this assay, haploid colonies containing the test chromosome bearing a SUP11 (ochre-suppressing tRNA) gene are white, whereas cells that have lost the test chromosome accumulate a red pigment due to the host ade2-101 (ochre) mutation. Thus, loss events give rise to red sectors during colony growth.
The checkpoint mutant strains were plated on color indicator plates and chromosome loss rates were evaluated visually by colony sectoring morphology and by half-sector analysis (Hieter et al., 1985a). In half-sector analysis, the rate of first division missegregation events is directly measured by observing the number of colonies that are at least one-half red, and dividing by the total number of colonies that were established by cells with a test chromosome. In an S288c background, bub1Δ and bub3Δ cells exhibited the highest rates of chromosome loss, 50-fold higher than the wild-type rate of 0.8 loss events per 1000 divisions (Figure 1, A and B). mad1Δ and mad2Δ strains also showed an increased chromosome loss rate, but at a level two- to threefold lower than bub1Δ and bub3Δ. mad3Δ exhibited a slight increase above wild type, whereas bub2Δ was indistinguishable from control. To test the generality of this result, the null mutants were characterized in a different laboratory strain background, W303-1a. The strong phenotypes for bub1Δ and bub3Δ were again observed, but the smaller differences among the mad null mutants were less apparent in W303-1a strains. At a minimum, the chromosome loss phenotypes indicate that Bub1 and Bub3 proteins have an additional role that is important to chromosome segregation during culture in the absence of intentional spindle damage.
Kinetochore surveillance checkpoint proteins perform their functions in the context of multiprotein complexes. To test whether cells are sensitive to protein dosage, each full-length open reading frame was placed under the control of the MET25 promoter (MET25p), whose transcriptional strength is controlled by altering the environmental methionine concentration (Mumberg et al., 1994). MET25p-controlled expression of the five checkpoint proteins led to steady-state protein levels in excess of wild type (Figure 1C).
The MET25p-controlled alleles were introduced into a wild-type yeast strain containing the test chromosome for monitoring chromosome segregation. Cultures grown in the presence of methionine were diluted in water and plated at ∼200 cells/plate on media lacking methionine. Half-sector analysis indicated that overexpression of Bub1p and Mad3p led to a 15-fold increase in test chromosome missegregation over the wild-type rate (0.7 loss events per 1000 divisions; Figure 1D). High-level expression of Mad1p and Mad2p caused a smaller increase in chromosome missegregation (three- and sevenfold), whereas high level expression of Bub3p had no effect.
Commonly used assays for the presence of checkpoint deficiency measure cell survival in the presence of antimicrotubule drugs such as Benomyl. The Benomyl sensitivity elicited by the absence of each kinetochore surveillance checkpoint protein was determined using the panel of null alleles. Strain viability was tested in the presence of a concentration of drug that delays but does not arrest wild-type cell growth. Cells containing bub1 and bub3 mutations were more Benomyl sensitive than mad1, mad2, or mad3 mutants by an order of magnitude (Figure 1E). Thus, checkpoint proteins differ in their contribution to the maintenance of cell viability in response to mild spindle damage. Note that the order of Benomyl sensitivity correlates with the relative intrinsic chromosome loss rates observed (Figure 1A). In principle, Benomyl sensitivity of mutants in this assay may reflect a sum of defective mechanisms contributing to cell death, including drug-induced hindrance of microtubule dynamics, null mutant kinetochore structural defects, and inappropriate cell cycle progression.
BUB1 and BUB3 Cooperate in a Chromosome Segregation Role
To determine whether the overexpression phenotype of Bub1 was due to discrete domain(s), a series of BUB1 truncation alleles was constructed capable of expressing the N-terminal 210, 367, or 608 amino acids as well as amino acid segments 211-1021 and 211–367 (Figure 2A). Western analysis using a Bub1p-specific antibody raised to the N-terminal 216 amino acids indicated that MET25-promoted expression led to significant protein accumulation in cells grown in the absence of methionine (Figure 2B). A serial dilution analysis indicated that the full-length overexpression product reached ∼50-fold that of wild type.
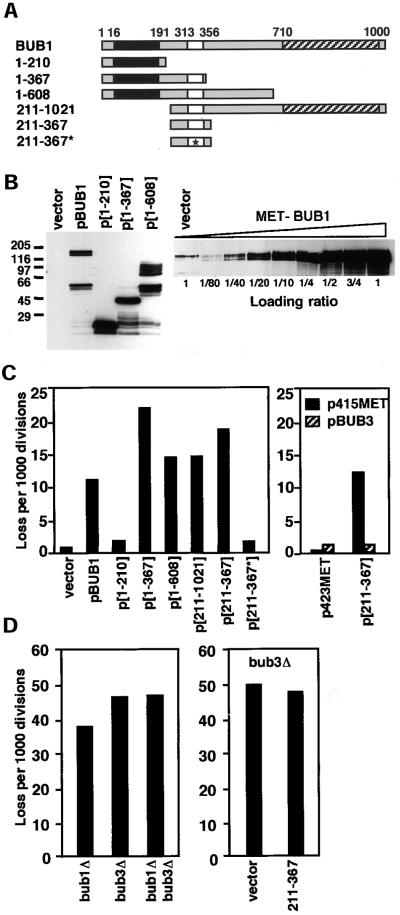
Figure 2.
The N terminus of Bub1p contributes to the overexpression phenotype and is counterbalanced by additional BUB3. (A) Diagram of BUB1 protein and protein fragments. The boxes indicate positions of conserved regions of BUB1p. Black: Mad3 like. White: Bub3 binding. Hatched: kinase domain. Star: E333K mutation. (B) Western blot detection of BUB1 overexpression alleles. Left: Wild-type cells (YPH278) containing MET25-promoted Bub1 alleles in p423MET were grown in the absence of methionine. Western blot analysis using an antibody raised to the N-terminal 216 amino acids of Bub1p (Brady and Hardwick, 2000) detects protein bands with migrations consistent with eachconstruct, in addition to faster migrating degradation products. Endogenous Bub1p is not detected at this exposure (vector lane). Right: A dilution series Western blot indicates that the overexpression level for full-length BUB1 is ∼50-fold. (C) Chromosome missegregation induced by BUB1 overexpression alleles. Left: Full-length and partial BUB1 alleles expressed from the MET25 promoter of p423MET were introduced into a wild-type strain (YPH278). Chromosome loss was determined by half-sector analysis after plating to methionine-free medium. Vector (no insert) and pBUB1 data are from D. p[1-210]: 25/14,087. p[1-367]: 127/5,741. p[1-608]: 96/6,592. p[211-1021]: 157/10,697. p[211-367]: 247/13,781. p[211-367*]: 9/5,117. Right: Chromosome loss was determined in YPH278 containing plasmid pairs as shown. p423MET + p415MET: 1/2,616. p423MET + pBUB3: 7/4,998. p[211-367] + p415MET: 37/3,015. p[211-367] + pBUB3: 6/4,510. At least two additional independent transformants of each construct were tested by visual sectoring assay and showed the same chromosome instability phenotype. (D) Chromosome loss in bub1 and bub3 null mutants. Left: Half sector analysis was used to compare chromosome loss rates of bub1Δ, bub3Δ, and bub1Δ bub3Δ mutant strains derived from sporulation of a wild-type diploid (YCD251) into which heterozygous bub1Δ::natMX and bub3Δ::kanMX alleles were introduced by transformation. bub1Δ: 186/5019 (one spore); bub3Δ: 262/5406 (one spore); bub1Δ bub3Δ: 601/12566 (four spores). Right: Chromosome missegration in a bub3Δ strain (YFS1100) containing the vector p423MET (284/5,675) or overexpression plasmid p[211-367] (289/6,069).
Each of the truncation constructs was introduced into wild-type cells on high-methionine medium, where expression is suppressed. Chromosome loss was quantitated for several independent transformants by half-sector analysis after plating on low-methionine medium (Figure 2C, left). Under these conditions, the full-length construct exhibits a 15-fold increase in loss (from Figure 1D). Overexpression of the N-terminal 367 or 608 amino acids of Bub1p from plasmids (p[1–367] and p[1–608], respectively) caused a 30- and 20-fold increase in chromosome loss. Overexpression of the N-terminal 210 amino acids had little effect (2.4-fold, Figure 2C). The segment common to p[1–367] and p[1–608], but absent from p[1–210], contains a well-conserved homology box predicted to mediate association between Bub1p and Bub3p (Taylor et al., 1998). This result suggested that overexpression of a Bub3-binding region of Bub1p might cause the chromosome loss. To test this hypothesis, a construct expressing only amino acids 211–367 under the control of the MET25 promoter was created. It too was found to induce chromosome loss at high expression levels (26-fold greater than the vector control).
Several additional lines of in vivo evidence now strongly support the interpretation that the overexpression phenotype is mediated through disruption of a Bub1p/Bub3p interaction. First, the bub1-1 point mutation (Hoyt et al., 1991), which is suppressed by a low-level increase in BUB3 gene dosage, was cloned and identified as E333K (see “Materials and Methods”). This mutation is located within the 211–367 segment. Second, when the E333K mutation was introduced into the p[211–367] plasmid, this allele failed to induce chromosome instability (Figure 2C, left panel). Third, expression of additional BUB3 from a MET25-controlled allele (from plasmid pBUB3) on a centromere vector (p415MET) reversed the chromosome instability phenotype of p[211–367] (Figure 2C, right). Fourth, a BUB1/BUB3 cooperative role in chromosome segregation implied by this interpretation was tested by analyzing the chromosome loss rate of a bub1Δ bub3Δ mutant (Figure 2D, left). The rate observed in the double mutant (48 events in 1000 divisions) is consistent with a shared role for Bub1p and Bub3p. Finally, if the presence of excessive bub1[211–367]p interferes with a Bub1p-Bub3p association, then this protein fragment should not elicit additional missegregation in the absence of the complex. Indeed, its overexpression does not augment chromosome loss in a bub3Δ null mutant (Figure 2D, right). We conclude that an interaction between Bub1 and Bub3 proteins is likely to be mediated by amino acids 211–367 of Bub1p in vivo, and disruption of this interaction contributes significantly to the overexpression phenotype associated with excessive Bub1p.
Yeast Bub1p Can Associate with Kinetochores in a One-Hybrid Assay
Previous experiments have demonstrated kinetochore localization of Bub1 protein in experimental systems where these structures are cytologically visible (Taylor and McKeon, 1997; Bernard et al., 1998; Jablonski et al., 1998; Basu et al., 1999; Sharp-Baker and Chen, 2001). To date, localization of checkpoint proteins to budding yeast kinetochore structures has not been achieved. In a one-hybrid assay (Ortiz et al., 1999), kinetochore protein components can activate a HIS3 reporter gene located immediately adjacent to the centromere of chromosome III. This reporter system depends on the presence of an active centromere, reveals association of known kinetochore components, and has been successfully used to identify new kinetochore proteins (Ortiz et al., 1999). We used this assay to ask whether kinetochore association of full-length Bub1p or truncation alleles can be detected in budding yeast.
Independent transformants containing Gal4-activation domain fusions of Bub1 and Bub1 fragments were spotted onto minimal medium lacking leucine and histidine supplemented with 5 mM 3-AT. Figure 3 shows that fusions of BUB1[1–367], BUB1[1–608], and BUB1[211–367] can activate transcription of the centromere reporter, indicating that these proteins can localize to kinetochores. Reporter activation with the full-length BUB1 fusion protein is not observed, likely due to a lower level of fusion protein accumulation or the presence of a nonfunctional conformation.
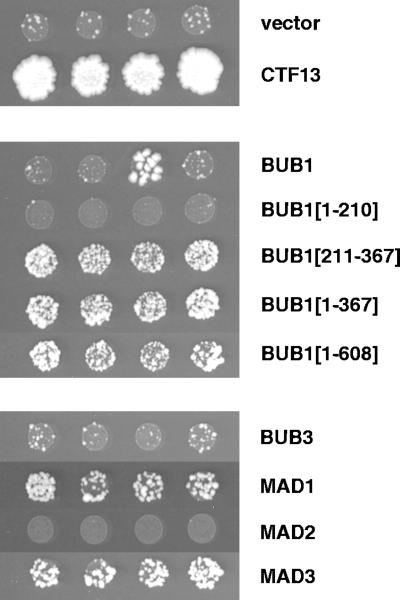
Figure 3.
Localization of activation domain fusions to kinetochores in a one-hybrid assay. In the one-hybrid assay, fusion of the GAL4-activation domain to a kinetochore-binding protein induces transcription of a centromere-adjacent HIS3 reporter allele (Ortiz et al., 1999). GAL4AD fusion constructs were introduced into strain YJL128. The fusion moiety is indicated to the right; GAL4AD-CTF13 (top) served as a positive control. Four independent transformants were grown to saturation in SD-LEU, diluted to 1.5 × 107 cells/ml, and spotted (3 μl) on SD-HIS, LEU + 5 mM 3-AT. The spots shown were incubated at 30°C for 14 d. The large papillae that appear occasionally in the GAL4AD-BUB1 transformants (observed in ∼25% of transformants) may reflect the occurrence of truncating mutations. All constructs were similarly tested in YJL148, a strain containing a mutant centromere sequence adjacent to the HIS3 reporter (Ortiz et al., 1999). No growth above vector background was observed in these controls. All fusion constructs shown were functional in a two-hybrid assay.
Activation domain fusions of other kinetochore surveillance proteins were also tested. MAD1 and MAD3 fusions activated the reporter (Figure 3), whereas BUB3 and MAD2 fusions did not. CTF13-AD is shown as a control: it activates the HIS3 reporter and supports growth on 3-AT within 3–4 d at 30°C, whereas the checkpoint fusion proteins tested require up to 14 d to show evidence of activation over vector background. This weak signal is consistent with a transient association, as would be expected for proteins that associate with a subset of kinetochores for a subset of the cell cycle. An unequal transcriptional activation efficiency for different fusion proteins may also be a contributing factor. We conclude from these experiments that the budding yeast kinetochore surveillance proteins Bub1, Mad1, and Mad3 can associate with yeast kinetochores, as is predicted from localizations of their studied orthologs.
The BUB1 Overexpression Phenotype Includes Disruption of Both Checkpoint and Segregation Functions
High-level expression of Bub1p (and fragments of this protein) may disrupt kinetochore checkpoint signaling, a segregation function, or both. To address whether checkpoint signaling was disrupted, strains overexpressing full-length Bub1p or protein fragments were tested for checkpoint competence in two different assays.
The first took advantage of the spindle checkpoint-dependent delay exhibited by ctf18Δ cells, which is associated with a partial defect in sister chromatid cohesion (Hanna et al., 2001). This delay is detected as an accumulation of G2/M phase cells during early log phase using flow cytometry (Figure 4A, left column). MET25-controlled alleles were introduced into ctf18Δ cells and transformants were selected on high-methionine medium. Four independent transformants were then grown in medium without methionine for 18–24 h (O.D. ∼0.4) and were analyzed for DNA content using flow cytometry (Figure 4A). Diminution of the G2/M phase peak indicated that the delay can be disrupted by overexpression of full-length Bub1p, Bub1[1–367]p, Bub1[1–608]p, and Bub1p[211–367]p. The G2/M reduction is consistent with an observed decrease in the proportion of budded cells (Figure 4A), as well as a reduction in viability determined by growth on solid medium with or without methionine (Figure 4B). The degree of delay diminution and reduced viability correlates with the amount of chromosome loss induced by the overexpression alleles (see Figure 2B). We observed similar loss of delay and viability in cells lacking CTF19 (C.D. Warren, unpublished data), a gene that encodes a nonessential kinetochore protein (Hyland et al., 1999). We conclude from these experiments that overexpression of Bub1p, and Bub1p fragments that cause chromosome loss, does have the capacity to disrupt a spindle checkpoint-dependent delay.
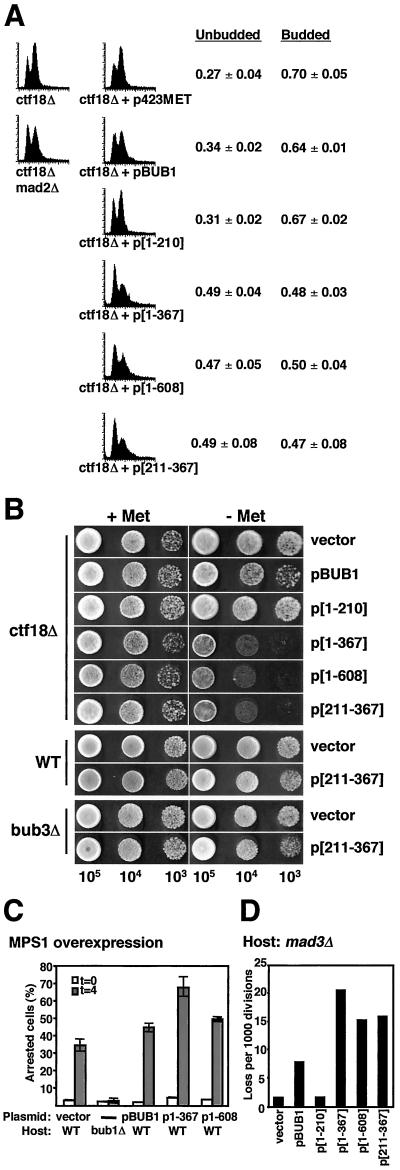
Figure 4.
Overexpression of Bub1p or Bub1p fragments both disrupts the spindle checkpoint and causes damage. (A) Disruption of a ctf18Δ-induced checkpoint delay. ctf18Δ strains containing p423MET plasmids expressing no (vector), full-length (BUB1), or partial (1–210, 1–367, 211–367, 1–608) alleles of BUB1 were created by transformation of YFS377. Cells were grown to early log phase in the absence of methionine for 18–24 h, and were prepared for flow cytometry (as in Hanna et al., 2001). A representative histogram is given for each genotype; four independent transformants were analyzed. The fractions of budded and unbudded cells were determined and are shown as mean ± SD (3 d.f.). (B) Disruption of the ctf18Δ delay results in decreased viability. Two independent isolates from each of the ctf18Δ strains described in A were grown to early log phase in media containing methionine. Cells were spotted onto solid medium without methionine in a 10-fold serial dilution series. Overexpression of bub1-[211-367]p in wild-type or bub3Δ cells did not result in a significant reduction in mortality (bottom). (C) Competence to arrest in response to MPS1 overexpression. Log phase cells grown in raffinose media lacking methionine (to induce expression of the BUB1 alleles) were treated with 3% galactose to induce MPS1 expression. Samples were taken at t = 0 and t = 4 h, formaldehyde fixed, DAPI stained, and scored for bud and nuclear morphology. The graph shows the percentage arrested (large-budded uninucleate) cells at each time point. Two independent transformants were analyzed (average ± range indicated). All strains were derived from YML101 (GAL-MPS1). All plasmids overexpressing BUB1 alleles in this strain were derived from p423MET (vector). YCD362 (bub1Δ) was included as a control for the assay. (D) Chromosome missegregation in a mad3Δ host. Chromosome loss rates were determined by half-sector analysis on plates lacking methionine. Vector: 5/3,327. pBUB1: 18/2,295. p[1–210]: 4/2,408. p[1–367]: 51/2,500. p[1–608]: 42/2,766. p[211–367]: 37/2,349. Data for a representative transformant are shown; at least two independent transformants were analyzed for each construct. The strains were YFS1205 derivatives created by introduction of p423MET (vector) and related BUB1 allele overexpression plasmids.
In a second system, we obtained evidence that the checkpoint arrest pathway is not completely dysfunctional. In this experiment, checkpoint activation by overexpression of the MPS1 protein kinase was used to cause cell cycle arrest (Hardwick et al., 1996; Weiss and Winey, 1996). MPS1-induced arrest is dependent on each of the known BUB and MAD checkpoint genes (Hardwick et al., 1996). If overexpression of Bub1p or Bub1p fragments completely disrupts the checkpoint pathway, similar to a null mutant, then MPS1 overexpression will not cause cell cycle arrest. Wild-type strains containing integrated GAL1-MPS1 as well as MET-controlled BUB1 overexpression plasmids were grown in medium lacking methionine to induce high-level expression of the BUB1 truncation alleles. Samples were taken before and 4 h after addition of galactose (for overexpression of MPS1). Formaldehyde-fixed cells were stained with DAPI and were scored for morphological evidence of metaphase arrest. Strains overexpressing the full-length Bub1p, Bub1[1–367]p, or Bub1[1–608]p arrested in response to MPS1 overexpression, whereas control bub1Δ cells did not (Figure 4C). The presence of an MPS1-induced arrest indicates that the overexpression of Bub1 full-length protein or the truncation alleles does not fully abrogate spindle checkpoint function. We speculate that even a single remaining active kinetochore may be sufficient to arrest cells in response to MPS1 overexpression.
Both of the experiments above address the checkpoint competence of strains containing extra Bub1p or Bub1p fragments. Neither addresses whether disruption of checkpoint control is responsible for the chromosome loss introduced by overexpression of Bub1p, or whether a separate mechanism causes missegregation (e.g., competition for a kinetochore structural component). Note that mad3Δ null cells have a quite modest chromosome instability phenotype (Figure 1, A and B), although they are markedly defective in preanaphase arrest (Straight et al., 1996; Hardwick et al., 2000; our unpublished data). To explore the cause of chromosome loss, overexpression interference of BUB1 alleles was tested in a mad3Δ yeast host (Figure 4D). The chromosome loss rates observed closely parallel those induced by the Bub1 overexpression alleles in wild-type cells. This result indicates that the mechanism responsible for chromosome loss incorporates a defect distinct from loss of a functional checkpoint pathway.
Two Domains of Bub1p Play Distinct Roles in Chromosome Segregation
Because the molecular defects engendered by overexpression may be complex, genomic loss-of-function alleles have also been characterized. Chromosome loss rates of two bub1 missense mutants were measured by half-sector analysis (Figure 5, A and B). The bub1-1 allele (i.e., bub1-E333K) has apparent partial function because its checkpoint defect is suppressible by additional copies of the BUB3 gene on a centromere plasmid, whereas that of a bub1 null mutation is not (Hoyt et al., 1991; Roberts et al., 1994). However, the chromosome missegregation rate measured for bub1-1 (Figure 5, A and B) is similar to that of bub1Δ (Figure 1, A and B). Like the bub1-1 checkpoint defect, the bub1-1 chromosome missegregation phenotype is suppressible by extra copies of the BUB3 gene introduced on a centromere plasmid (Figure 5B).
Figure 5.
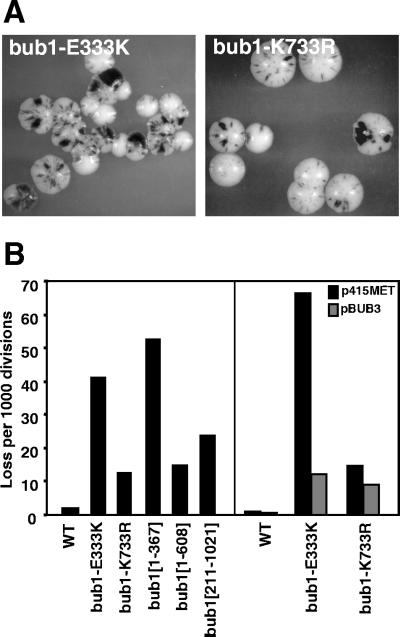
Chromosome missegregation associated with genomic alleles of BUB1. (A) Colony sectoring phenotypes of bub1-E333K (bub1-1) and bub1-K733R. Strains were YCD280 and YCD281. (B) Chromosome loss caused by genomic alleles. Left: Half sector analysis was used to analyze the mutants as shown. WT: 8/8,503. bub1-E333K: 248/5,964. bub1-K733R: 83/6,576. bub1[1-210]: 354/6,769. bub1[1-367]: 477/9,019. bub1[1-608]: 222/14,105. bub1[211-1021]: 355/14,837. Strains were YCD279, YCD280, YCD281, YCD371, YCD358, and YCD407. Right: A centromere plasmid containing a MET25-inducible BUB3 gene (pBUB3) or vector alone (p415MET) was introduced into wild-type (WT, YPH278), bub1-E333K (YCD280), or bub1-K733R (YCD281) strains. Half sector analysis was performed after plating on methionine-free media. WT + p415MET: 1/3,728. WT + pBUB3: 3/4,925. bub1-E333K + p415MET: 104/1,567. bub1-E333K + pBUB3: 48/3,941. bub1-K733R + p415MET: 53/3,654. bub1-K733R + pBUB3: 36/4,102.
The bub1-K733R allele, which alters a conserved lysine residue in the Bub1 protein kinase domain, has been previously characterized as deficient in checkpoint competence and protein kinase activity (Roberts et al., 1994). The chromosome stability defect of bub1-K733R is less severe than that of bub1-E333K (Figure 5, A and B). The addition of extra BUB3 gene copies to bub1-K733R does not markedly alter its chromosome segregation phenotype.
BUB1 truncation alleles were tested at single copy in the native genomic locus under the control of the BUB1 promoter in haploid cells. Although the genomic bub1[1-367] allele exhibits a phenotype similar to the null mutant, the bub1[1-608] allele supports a chromosome loss rate that is intermediate (16 events per 1000 divisions, Figure 5B). This rate is similar to that observed for the kinase region missense mutant bub1-K733R (13 events per 1000). Thus, an intermediate level of chromosome stability is observed for two alleles of BUB1, with defective or absent protein kinase activity, indicating a role for the N-terminal portion of Bub1p in segregation.
Note that the chromosome segregation competence conferred by the N-terminus of Bub1p does not account for the very high fidelity of segregation in wild-type cells. To test if chromosome stability can be provided by a Bub1p C-terminal protein fragment, a deletion allele expressing amino acids 211-1021 was constructed at the genomic locus. This mutant has a chromosome stability phenotype that is between wild-type and bub1Δ, at 24 events per 1000 (Figure 5B). We conclude that the C-terminal portion of Bub1p also contributes to chromosome stability.
In summary, the bub1-E333K allele appears to be virtually null for both checkpoint and chromosome segregation activities, although it encodes a protein whose functions are rescued by additional expression of its binding partner Bub3p. This argues that an association with Bub3p is required for both checkpoint and segregation activities. Separate N-terminal (bub1[1-608]) and C-terminal (bub1[211-1021]) protein fragments contribute to chromosome stability, each providing an intermediate level of segregation fidelity. We note that the loss rates of these two partial protein alleles sum to a value that is the same as the loss rate observed in the null mutant (40 per 1000, Figure 1B).
An N-Terminal Segment of Bub1p Is Necessary and Sufficient for Its Checkpoint Function
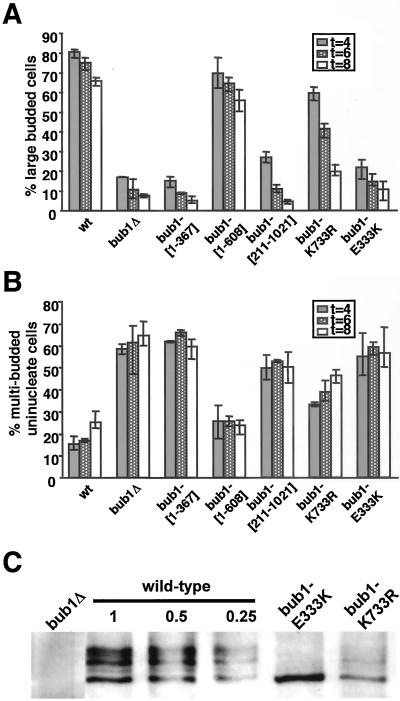
Genomic loss-of-function alleles were tested for checkpoint competence by evaluating arrest after spindle damage. An arresting concentration of the antimicrotubule drug nocodazole (15 μg/ml) was added to asynchronous cultures grown in rich medium at t = 0. Samples at 4, 6, and 8 h were fixed in formaldehyde, DAPI stained, and scored for the frequency of uninuclear large-budded (arrested) or multibudded (inappropriately progressing) cellular phenotypes (Figure 6, A and B). The bub1[1-367] and bub1-E333K mutants behaved like bub1Δ, consistent with their null chromosome instability phenotypes. However, bub1[1-608] gave results similar to wild type for both arrest and inappropriate progression tests. This indicates that the kinase domain can be deleted without loss of the checkpoint arrest function of Bub1p.
Figure 6.
Checkpoint competence of BUB1 genomic alleles. (A) Cell cycle arrest. Logarithmically growing cultures of strains containing integrated alleles were transferred to YPD + 15 μg/ml nocodazole. At t = 0, 4, 6, and 8 h after shift into nocodazole, aliquots were formaldehyde fixed and stained with DAPI. Two hundred cells from each were scored for the arrested fraction (large-budded uninucleate cells), and the mean ± standard deviation for three independent integrants is shown. The strains were YPH278, YFP2, YCD371, YCD358, YCD281, and YCD280. (B) Failure of cell cycle arrest. The same samples were scored for the fraction of cells that exhibited multibudded uninucleate cells, an indication mitotic exit in the absence of nuclear division. (C) Anti-Bub1p immunoprecipitates from the genotypes indicated were analyzed by Western blot for the presence and abundance of Bub1 protein (as described in Brady and Hardwick, 2000). Immunoprecipitation product from the wild-type cells was loaded in a dilution series (1, 0.5, and 0.25) for comparison with lanes containing immunoprecipitations from mutant extracts. The strains were YFP2, YPH278, YCD280, and YCD281.
The previous report that bub1-K733R is checkpoint deficient (Roberts et al., 1994) led to the hypothesis that the kinase-encoding portion of BUB1 is the checkpoint-functional moiety of the protein. We observe that although bub1-K733R cells are indeed checkpoint deficient, the timing of arrest failure indicates the presence of partial function (Figure 6A). Moreover, the bub1-[211-1021] allele failed to exhibit a checkpoint arrest in nocodazole. These results, taken together with the arrest competence of bub1-[1-608], indicate that the protein kinase activity of Bub1p is not responsible for nocodazole-induced arrest.
The proteins encoded by bub1-E333K and bub1-K733R alleles were further investigated. Western blot analysis of anti-Bub1p immunoprecipitates reveals the presence of a stable protein pool in bub1-E333K mutant cells (Figure 6C). The bub1-E333K protein is significantly underphosphorylated, strongly suggesting that function of the wild-type Bub1 protein depends upon its phosphorylation. In contrast, bub1-K733Rp appears to be less abundant, and modified forms are readily detected (Figure 6C). The low steady-state abundance of bub1-K733Rp may reflect a high protein turnover rate. This prediction suggests an hypothesis in which the inability of bub1-K733Rp to maintain a checkpoint arrest is in part due to a gradual loss of the mutant protein in arrested cells.
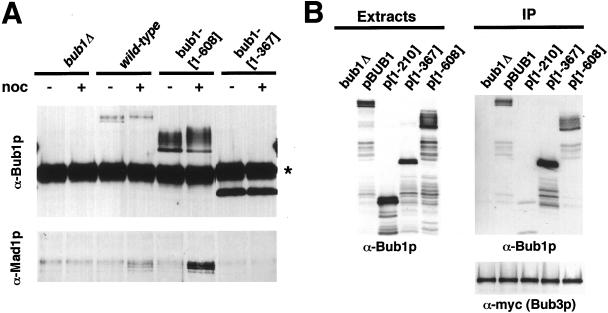
Formation of a Mad1p-Bub1p-Bub3p complex is crucial for spindle checkpoint function (Brady and Hardwick, 2000). Therefore, we tested whether the Bub1 protein fragment alleles could form such a complex by assaying for coimmunoprecipitation with Mad1p or myc-tagged Bub3p (Figure 7). First, immunoprecipitates prepared with an α-Bub1p antibody were characterized for the presence of Bub1p and Mad1p (Figure 7A). Full-length Bub1p and bub1[1-608]p expressed from the genomic locus were found to coprecipitate Mad1p in nocodazole-arrested cells, whereas bub1[1-367]p did not. Second, immunoprecipitation was carried out to test for association between a genomic myc-tagged BUB3 allele and Bub1 truncation proteins expressed from the MET25 promoter on a 2-μm plasmid (Figure 7B). Anti-myc precipitates containing equivalent amounts of Bub3-myc protein (Figure 7B, bottom) also contained appreciable amounts of full-length Bub1p, bub1[1-367]p, and bub1[1-608]p, but not bub1[1-210]p.
Figure 7.
Bub1-[1-608]p associates with Mad1p and Bub3p. (A) Coimmunoprecipitation of full-length Bub1p and bub1-[1-608]p with Mad1p. The strains shown, containing integrated Bub1 alleles expressed from the wild-type BUB1 promoter, were grown to log phase and were incubated with ± 15 μg/ml nocodazole for 2 h at 24°C. Immunoprecipitates were prepared using an α-Bub1p antibody, separated by SDS-PAGE, and transferred to nitrocellulose. The immunoblots were then probed with α-Bub1p and α-Mad1p rabbit antibodies as indicated. The strong band labeled (*) in the Bub1 blot is IgG heavy chain from the immunoprecipitation. Strains shown are YPH278, YFP2, YCD358, and YCD371. (B) Coimmunoprecipitation of full-length Bub1p, bub1-[1-367]p, and bub1-[1-608]p with Bub3p. All strains contained a BUB3-myc allele in the genome. The experimental strains contained a wild-type BUB1 gene in addition to episomal MET25-promoted alleles as indicated. A bub1Δ strain served as control. Left: Bub1p Western blot using a rabbit α-Bub1p antibody. Right: Immunoprecipitation with an α-myc antibody recovered an equivalent amount of myc-tagged Bub3 protein (bottom). The immunoprecipitates were probed with rabbit α-Bub1p antibody (top). The strains were YKH300 (bub1Δ) or YKH238 with pBUB1, p[1-210], p[1-367], or p[1-608].
In summary, bub1[1-608]p exhibits biochemical characteristics of a functional Bub1 protein capable of coprecipitation with both Mad1p and Bub3p. Moreover, bub1[1-608]p is heavily phosphorylated in all of our Western blots (Figures 2B and 7; confirmed by lambda protein phosphatase treatment; K.G. Hardwick, unpublished data), whereas bub1[1-367]p and bub1[1-210]p are not. In a functional assay, a genomic allele of bub1[1-608] supports a robust checkpoint arrest in the presence of nocodazole. Thus, we conclude that the bub1[1-608] protein is sufficient for BUB1 checkpoint arrest function, and exhibits biochemical properties expected for this activity.
DISCUSSION
Nonessential spindle checkpoint proteins from budding yeast differ in their importance to chromosome stability in cells where spindle assembly dynamics are not challenged by intentional introduction of damage. The disparity in chromosome loss rates observed among the checkpoint null mutants indicates the presence of functional differentiation. bub2Δ cells exhibit a wild-type chromosome loss rate, in agreement with BUB2's primary role in mitotic exit rather than in kinetochore surveillance at metaphase. Each of the other mutants conferred a chromosome loss rate higher than wild type, indicating one or more roles important for high-fidelity chromosome transmission. BUB1 and BUB3 genes in particular appear to influence chromosome segregation more strongly than MAD1, MAD2, and MAD3 genes. We speculate that differential roles among these genes may include distinct kinetochore structural contributions that influence segregation, detection of different types of kinetochore status in the context of checkpoint signaling (e.g., tension vs. attachment), or communication of checkpoint signaling to diverse target molecules that mediate different aspects of checkpoint delay or recovery. It was recently argued that although mammalian Mad2p responds to the lack of microtubule attachment, the Bub proteins respond to both microtubule attachment and a lack of tension (Waters et al., 1998; Skoufias et al., 2001). However, evidence from budding yeast suggests that the spindle checkpoint in this organism responds to the lack of tension in a mitotic spindle, and that this checkpoint-associated delay is Mad2 dependent (Stern and Murray, 2001). Further work is needed to clarify roles of the checkpoint proteins.
In this work, we have endeavored to explain the relatively high rate of loss exhibited by bub1Δ cells and to find evidence for the presence of two distinct contributions to chromosome segregation. One is encoded within the first 608 amino acids in a protein segment that is both necessary and sufficient for a nocodazole-induced checkpoint arrest. The other is encoded in the kinase domain, which is not required for checkpoint arrest and whose function is unknown. Previous work in budding yeast has indicated that a missense allele predicted to disrupt kinase activity (bub1-K733R) was also defective in checkpoint arrest (Roberts et al., 1994). In apparent contradiction, an in vitro experiment using a Xenopus extract system has provided evidence that a kinase-defective missense allele can support an active checkpoint (Sharp-Baker and Chen, 2001). Here, we find that the genomic bub1-[1-608] allele, entirely lacking the conserved kinase domain, exhibits checkpoint competence after spindle disruption, whereas bub1-K733R exhibits a transient arrest that decays rapidly. Examination of the steady-state abundance of bub1-K733R encoded protein indicates a decreased accumulation. Taken together, these studies indicate that the checkpoint defect associated with bub1-K733R is more likely due to insufficient gene product than to a dysfunctional kinase activity.
The protein encoded by bub1-[1-608] exhibits several interesting properties relevant to its checkpoint function. The immunoprecipitation experiments reveal association of this truncation product with both Bub3p and Mad1p. The BUB1 partial protein allele series indicates the involvement of specific amino acid segments of Bub1p in complex formation. The segment from amino acid 211 to 367 is required for complex formation with Bub3p. Similarly, the segment from 367 to 608 is required for Mad1p association. In our analysis of the partial protein alleles, the presence of both Bub1p and Mad1p binding correlates with the accumulation of phosphorylated forms of Bub1p, as well as the presence of checkpoint arrest competence. These observations strongly support the current model that a Bub1-Bub3-Mad1 protein complex is required for checkpoint arrest, and they suggest that the phosphorylation in the N-terminal one-half of Bub1p may also be a requirement.
The 211–367-amino acid segment can localize a GAL4 transcriptional activation domain to the yeast kinetochore in the one-hybrid assay. This activity, as well as Bub3p binding, is consistent with previous work on murine Bub1p, which defined a conserved homology (Taylor et al., 1998) with similar functions in an overexpression assay. In general, our overexpression results in budding yeast parallel studies in mammalian cells where overexpression of Bub1p mutant alleles from an ectopic promoter leads to disruption of checkpoint function (Taylor and McKeon, 1997; Cahill et al., 1998). However, the mammalian studies have been controversial (see Tighe et al., 2001) due to differing outcomes from similar experiments. In budding yeast, under partial induction of the checkpoint (e.g., in ctf18Δ or ctf19Δ cycling populations), overexpression of Bub1p or fragments was sufficient to “silence” checkpoint signaling. We assume that in ctf18Δ or ctf19Δ mutants, many cells experience a delay due to the failure of one kinetochore (or a few) to achieve stable bipolar attachment with normal timing. In contrast, under the same Bub1-overexpression conditions, checkpoint activation by extra Mps1p was sufficient for cell cycle arrest. We speculate that because the checkpoint is strongly induced with overexpression of Mps1p, even a single remaining active checkpoint-signaling complex may cause cell cycle arrest. Comparison of the results from these two tests for checkpoint function in yeast highlights a cautionary note where partial induction or disruption of checkpoint activity is involved. For example, in vertebrate cell culture systems, seemingly subtle variation (e.g., in genotype or culture conditions) may contribute to quantitative aspects of checkpoint competence and may affect the outcome.
In the overexpression survey of checkpoint proteins, extra Mad3p caused a chromosome loss rate similar to that conferred by extra Bub1p. Although Mad3p exhibits similarity in protein alignment to the Bub1p N-terminal segment, each gene is independently required for checkpoint activity and, therefore, they are not functionally equivalent. The mad3 null chromosome loss rate is notably subtle in comparison with the MAD3 overexpression phenotype, indicating that compromise of Mad3p's overexpression binding partners is more important to segregation than Mad3 protein itself. Because the interaction between Bub1p and Bub3p contributes to the BUB1 overexpression phenotype, and because Mad3p associates with Bub3p (Hardwick et al., 2000), it is likely that interference with Bub3p function is causal for the chromosome missegregation induced by Mad3p overexpression. Interestingly, the amounts of Bub3p at a single human kinetochore have been estimated to be around 1000 copies (Martinez-Exposito et al., 1999), an abundance that is suggestive of its having a structural role as well as a signaling one.
In conclusion, a quantitative study of the roles played by spindle checkpoint genes in chromosome segregation indicates the presence of functional differentiation beyond their essential contributions to the spindle checkpoint. Further studies of loss-of-function alleles that define distinct functional contributions, and overexpression alleles that disrupt in vivo relationships, hold promise for elucidating the in vivo importance of biochemical properties of checkpoint components.
ACKNOWLEDGMENTS
We thank A. Hoyt and C. Dougherty for helpful discussions as well as for sharing the original bub1-1 strain and bub1-K733R plasmid; M. Winey for the GAL-MPS1 construct; J. Lechner for one-hybrid yeast strains and constructs; and M. Lee and F. Pangilinan for plasmids and strains. We also thank A. Hoyt, J. Boeke, F. Pangilinan, D. Warren, and our reviewers for comments on the manuscript. This work was supported by GM50842 from the National Institutes of Health to F.S.; the Program in Human Genetics and Molecular Biology (to C.D.W.); The Wellcome Trust (to K.G.H. and D.M.B.), of which K.G.H. is a Wellcome Trust Senior Research Fellow; and the Medical Research Council (to R.C.J.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–04–0203. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–04–0203.
REFERENCES
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Basu J, Bousbaa H, Logarinho E, Li Z, Williams BC, Lopes C, Sunkel CE, Goldberg ML. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Hardwick K, Javerzat JP. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J Cell Biol. 1998;143:1775–1787. doi: 10.1083/jcb.143.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Koshland D. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr KA, Hoyt MA. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Beretta A, Lucchini G, Piatti S. Role of the kinetochore protein Ndc10 in mitotic checkpoint activation in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;266:115–125. doi: 10.1007/s004380100533. [DOI] [PubMed] [Google Scholar]
- Gardner RD, Burke DJ. The spindle checkpoint: two transitions, two pathways. Trends Cell Biol. 2000;10:154–158. doi: 10.1016/s0962-8924(00)01727-x. [DOI] [PubMed] [Google Scholar]
- Gillett ES, Sorger PK. Tracing the pathway of spindle assembly checkpoint signaling. Dev Cell. 2001;1:162–164. doi: 10.1016/s1534-5807(01)00032-6. [DOI] [PubMed] [Google Scholar]
- Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Li R, Mistrot C, Chen RH, Dann P, Rudner A, Murray AW. Lesions in many different spindle components activate the spindle checkpoint in the budding yeast Saccharomyces cerevisiae. Genetics. 1999;152:509–518. doi: 10.1093/genetics/152.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hieter P, Mann C, Snyder M, Davis RW. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985a;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hieter P, Pridmore D, Hegemann JH, Thomas M, Davis RW, Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985b;42:913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Hoyt MA. A new view of the spindle checkpoint. J Cell Biol. 2001;154:909–911. doi: 10.1083/jcb.200108010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hyland KM, Kingsbury J, Koshland D, Hieter P. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- Jensen S, Segal M, Clarke DJ, Reed SI. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J Cell Biol. 2001;152:27–40. doi: 10.1083/jcb.152.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Rose AM. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat Cell Biol. 1999;1:514–521. doi: 10.1038/70309. [DOI] [PubMed] [Google Scholar]
- Krishnan R, Pangilinan F, Lee C, Spencer F. Saccharomyces cerevisiae BUB2 prevents mitotic exit in response to both spindle and kinetochore damage. Genetics. 2000;156:489–500. doi: 10.1093/genetics/156.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Martinez-Exposito MJ, Kaplan KB, Copeland J, Sorger PK. Retention of the BUB3 checkpoint protein on lagging chromosomes. Proc Natl Acad Sci USA. 1999;96:8493–8498. doi: 10.1073/pnas.96.15.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BT, Farr KA, Hoyt MA. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Shah JV, Cleveland DW. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H, Chen RH. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, Andreassen PR, Lacroix FB, Wilson L, Margolis RL. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc Natl Acad Sci USA. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger, P. (2001). Tracing the pathway of spindle assembly checkpoint signaling. Dev. Cell, 162–164,. [DOI] [PubMed]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Hieter P. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:8908–8912. doi: 10.1073/pnas.89.19.8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Stern BM, Murray AW. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Lehane C, Uhlmann F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS. Chromosome segregation: dual control ensures fidelity. Curr Biol. 1999;9:R562–564. doi: 10.1016/s0960-9822(99)80355-8. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Albertella M, Taylor SS. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker-Kulberg RL, Morgan DO. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Burke DJ. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]