Abstract
Microtubule assembly is initiated by the γ-tubulin ring complex (γ-TuRC). In yeast, the microtubule is nucleated from γ-TuRC anchored to the amino-terminus of the spindle pole body component Spc110p, which interacts with calmodulin (Cmd1p) at the carboxy-terminus. However, mammalian protein that anchors γ-TuRC remains to be elucidated. A giant coiled-coil protein, CG-NAP (centrosome and Golgi localized PKN-associated protein), was localized to the centrosome via the carboxyl-terminal region. This region was found to interact with calmodulin by yeast two-hybrid screening, and it shares high homology with the carboxyl-terminal region of another centrosomal coiled-coil protein, kendrin. The amino-terminal region of either CG-NAP or kendrin indirectly associated with γ-tubulin through binding with γ-tubulin complex protein 2 (GCP2) and/or GCP3. Furthermore, endogenous CG-NAP and kendrin were coimmunoprecipitated with each other and with endogenous GCP2 and γ-tubulin, suggesting that CG-NAP and kendrin form complexes and interact with γ-TuRC in vivo. These proteins were localized to the center of microtubule asters nucleated from isolated centrosomes. Pretreatment of the centrosomes by antibody to CG-NAP or kendrin moderately inhibited the microtubule nucleation; moreover, the combination of these antibodies resulted in stronger inhibition. These results imply that CG-NAP and kendrin provide sites for microtubule nucleation in the mammalian centrosome by anchoring γ-TuRC.
INTRODUCTION
The microtubule fulfills essential functions in chromosome segregation in mitosis and in determining organelle positioning and cell polarity. Nucleation of microtubules is initiated by the γ-tubulin ring complex (γ-TuRC) at microtubule organizing centers, such as the centrosome of mammalian cells or the spindle pole body (SPB) of yeast. γ-TuRC is a ring-shaped multiprotein complex containing γ-tubulin, which is found in the cytoplasm as well as in the centrosome (Zheng et al., 1995; Schiebel, 2000). The cytoplasmic pool of γ-TuRC may be a source of nucleating complexes that are recruited to the centrosome when increased microtubule nucleation is required (Khodjakov and Rieder, 1999). Budding yeasts have a simple γ-TuRC consisting of three proteins, Spc97p, Spc98p, and Tub4p (Knop et al., 1997), which are conserved among other organisms (Martin et al., 1998; Murphy et al., 1998; Tassin et al., 1998). In mammalian cells, at least 6 proteins have been identified in γ-TuRC, including GCP2, GCP3 (also named HsSpc98), and γ-tubulin, which are the orthologues of Spc97p, Spc98p, and Tub4p, respectively (Murphy et al., 1998, 2001; Tassin et al., 1998).
Little is known about how γ-TuRCs are anchored at the centrosome in mammalian cells. Mammalian centrosome is composed of a pair of centrioles surrounded by an electron-dense cloud of pericentriolar material (PCM) (Kellog et al., 1994). PCM seems to be an interconnected meshwork of proteins that forms a matrix or lattice composed of a high proportion of coiled-coil proteins. The microtubule nucleating capacity of the centrosome is localized to the PCM (Gould and Borisy, 1977). In budding yeast, Spc72p in outer plaque and Spc110p in inner plaque have been identified as mediating microtubule nucleation by anchoring γ-TuRC (Knop and Schiebel, 1997, 1998). Spc110p is a coiled-coil protein and anchors γ-TuRC through interaction with Spc97p and Spc98p at the amino-terminal region. The carboxyl-terminal region associates with Cmd1p (calmodulin) that is integrated into the SPB near the nuclear envelope (Sundberg et al., 1996). Several groups have attempted to identify anchoring proteins for γ-TuRC in other organisms. Human and Xenopus laevis Spc110p-related proteins were found by cross-reactivity of monoclonal antibodies to Spc110p (Tassin et al., 1997). Mouse pericentrin and γ-tubulin are found in a protein complex and organized into a lattice at the centrosome (Dictenberg et al., 1998). Recently, a larger isoform of pericentrin, human kendrin (or pericentrin B) (Li et al., 2001) was found to share homology with the Cmd1p-binding domain of the yeast Spc110p and thus seems to be the mammalian orthologue of Spc110p (Flory et al., 2000). However, it is not known whether these candidates can mediate microtubule nucleation or bind to the γ-TuRC.
We have reported that a giant coiled-coil protein, CG-NAP (centrosome and Golgi localized PKN-associated protein), is localized to the centrosome throughout the cell cycle and the Golgi apparatus at interphase (Takahashi et al., 1999). CG-NAP shares partial homology with pericentrin (Takahashi et al., 1999). Its localization to the centrosome is independent of microtubules, whereas that to the Golgi apparatus is disrupted by microtubule depolymerization. Furthermore, CG-NAP associates with protein kinases (PKN, PKA, and PKCε) and protein phosphatases (PP1 and PP2A) (Takahashi et al., 1999, 2000). CG-NAP may thus coordinate the location and activity of these enzymes to regulate the phosphorylation states of specific substrates at the centrosome and the Golgi apparatus.
In this study, we demonstrate that CG-NAP (also named AKAP350 [Schmidt et al., 1999] and AKAP450 [Witczak et al., 1999]) and kendrin anchor γ-TuRC through binding with GCP2 and/or GCP3 at their amino-terminal regions. CG-NAP and kendrin are localized to the centrosome via their carboxyl-terminal regions, which interact with calmodulin. Furthermore, endogenous CG-NAP and kendrin form complexes together with GCP2 and γ-tubulin in vivo. Microtubule aster formation from isolated centrosomes is suppressed by pretreatment with antibodies to CG-NAP and/or kendrin. These observations imply that CG-NAP and kendrin provide microtubule nucleation sites by anchoring γ-TuRC in mammalian centrosome.
MATERIALS AND METHODS
Cell Culture and Transfection
COS7, HeLa S3, and HEK293T cells were grown in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin at 37°C in a humidified 5% CO2 atmosphere. For Chinese hamster ovary (CHO)-K1 cells, Ham's F12 medium was used as a basal medium. COS7 cells were transfected with expression plasmid(s) by electroporation using GenePulser II (Bio-Rad, Hercules, CA). HEK293T cells were transfected by use of LipofectAMINE PLUS Reagent (Invitrogen, Carlsbad, CA).
Preparation of Full-Length cDNA of Human Kendrin, GCP2, GCP3, and γ-Tubulin
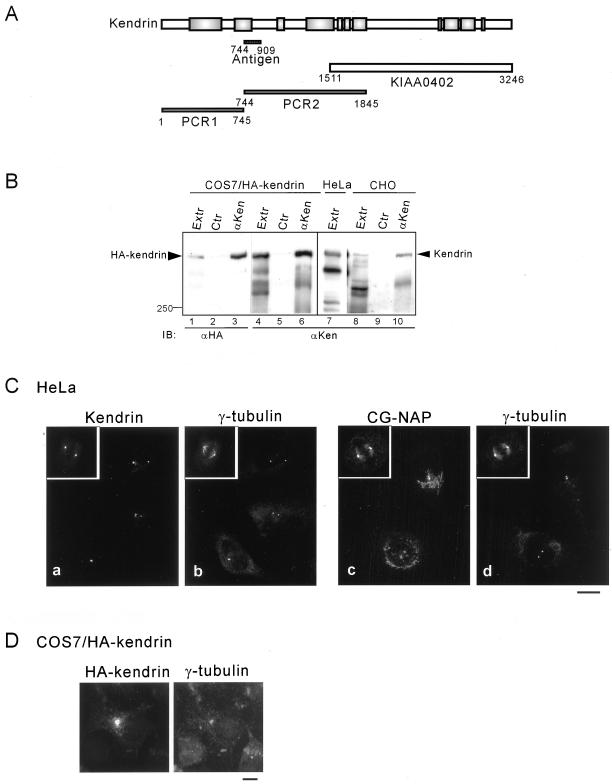
The full-length cDNA sequence of kendrin was found in GenBank with accession number U52962. KIAA0402 encoding the carboxyl-terminal half of kendrin (amino acids 1511 to the stop codon) was obtained from Kazusa DNA Research Institute Foundation (Kazusa, Japan). The amino acid sequence encoded by KIAA0402 was slightly different from that of kendrin (U52962): it lacks amino acids 2828–2906, and the carboxyl-terminal 13 amino acids were replaced with 16 amino acids. The rest of the kendrin cDNA was obtained by PCR using the HeLa Marathon-ready cDNA library (Clontech, Palo Alto, CA) with appropriate oligonucleotide primer sets for the cDNA fragments encoding amino acids 1–744 and 745-1845 (see Figure 5A). These fragments were assembled into pBluescript to create the full-length cDNA of kendrin encoding 3246 amino acids. Expression plasmids for the full-length and deletion mutants of kendrin were constructed by subcloning the corresponding fragments into pTB701-HA (hemagglutinin) or pTB701-FLAG (Takahashi et al., 1999).
Figure 5.
Centrosomal localization of endogenous and recombinant full-length kendrin. (A) Preparation of full-length cDNA of kendrin. Schematic representation of kendrin is shown with the fragments used to construct full-length cDNA as described in MATERIALS AND METHODS. Positions are shown with the corresponding amino acid residues. KIAA0402 was obtained from Kazusa DNA Research Institute. Position of the fragment to generate specific antibody is shown as Antigen. (B) Specificity of anti-kendrin (αKen) antibody. Cell extracts of COS7 cells expressing HA-tagged kendrin, HeLa, or CHO cells were immunoprecipitated with αKen followed by immunoblotting with anti-HA (lanes 1–3) or αKen (lanes 4–10). (C) Centrosomal localization of endogenous kendrin. HeLa cells were fixed with MeOH, and then double-stained with αKen and anti-γ-tubulin (a, b) or anti-CG-NAP (αEE) and anti-γ-tubulin (c, d). (D) Centrosomal localization of recombinant kendrin. COS7 cells expressing HA-tagged kendrin were fixed with MeOH, and then double-stained with anti-HA and anti-γ-tubulin. Bars, 10 μm.
Full-length cDNAs for human GCP2 and GCP3 were obtained by PCR using the HeLa Marathon-ready cDNA library (Clontech) with appropriate oligonucleotide primer sets. Then the cDNA fragments were cloned into pTB701-HA or pTB701-FLAG. γ-Tubulin cDNA was obtained by PCR using the same library with a primer set designed to create the XhoI site in place of the stop codon, and then cloned into pcDNA3.1-MycHis (Invitrogen) to express γ-tubulin carboxyl-terminally tagged with Myc and His6.
Antibodies
Polyclonal antibodies to human CG-NAP (αEE and αBH) were described previously (Takahashi et al., 1999). Immunoprecipitation of endogenous CG-NAP was performed by combination of αEE and αBH. Immunostaining and immunoblotting were done with αEE. Polyclonal antibodies to rat CG-NAP (αrXN), human kendrin (αKen), and human GCP2 (αGCP2) were generated by immunizing rabbits with bacterially expressed glutathione S-transferase (GST)-tagged antigens prepared as follows. A partial rat CG-NAP cDNA fragment (1.34 kb) was cloned by hybridization screening of rat brain cDNA library (Ono et al., 1988) by using the human CG-NAP cDNA fragment #2–43 (Takahashi et al., 1999) as a probe. The cDNA sequence was submitted to DDBJ/GenBank/EMBL Data Bank with accession number AB071391. The nucleotide number 757-1341 was subcloned into pGEX4T (Amersham Biosciences, Piscataway, NJ) to express GST-tagged antigen. The cDNA fragments encoding amino acids 744–909 of kendrin and 762–902 of GCP2 were cloned into pGEX4T. GST-tagged antigens were expressed in Escherichia coli and purified by use of glutathione-Sepharose beads (Amersham Biosciences) as described previously (Takahashi et al., 1999). Antibodies were affinity-purified by use of antigen-coupled Sepharose beads according to the manufacturer's instruction (Amersham Biosciences).
The following antibodies were purchased: anti-γ-tubulin GTU88, anti-α-tubulin DM1A, and anti-FLAG M2 (Sigma, St. Louis, MO); rat anti-HA 3F10 (Roche Diagnostics, Basel, Switzerland); anti-His6 antibody RGS-His (Qiagen, Hilden, Germany); rhodamine-conjugated anti-rabbit IgG and FITC-conjugated anti-mouse IgG (Chemicon International, Temecula, CA); and horseradish peroxidase (HRP)-conjugated anti-Myc and HRP-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunofluorescence Microscopy
Cells grown on cover glasses were fixed with cold methanol for 5 min with or without prior extraction by 0.1% Triton X-100 in 80 mM 1,4-piperazinediethanesulfonic acid at pH 6.9, 5 mM EDTA, and 1 mM MgCl2 at room temperature for 2 min. The fixed cells were blocked with 5% normal donkey serum in phosphate-buffered saline-containing Triton X-100 (PBST) (20 mM phosphate buffer at pH 7.5, 150 mM NaCl, and 0.03% Triton X-100) and then incubated with the relevant antibody for 1 h at room temperature. After the cells had been washed with PBST, the primary antibody was visualized by subsequent incubation with the appropriate secondary antibody conjugated with either rhodamine or FITC. The fluorescence of rhodamine and FITC was observed under a fluorescence microscope (Zeiss, Oberkochen, Germany) equipped with a CCD camera (Hamamatsu Photonics, Hamamatsu, Japan).
Yeast Two-Hybrid Screening and Interaction Analysis
The cDNA fragment encoding CG-NAP3510–3828 was fused to the Gal4 DNA binding domain (Gal4bd) by subcloning into pGCKT7 (Clontech). The resultant plasmid was transformed to the yeast reporter strain AH109 together with a HeLa cDNA library fused to the Gal4 activation domain (Gal4ad) in pGAD GH (Clontech). Screening was done as described previously (Mukai et al., 1996).
GST Pull-down Assay
Bacterial expression plasmids for GST-tagged CG-NAP3510–3829 and His6-tagged calmodulin 2 were constructed by inserting the corresponding cDNA fragments into pGEX4T and pRSET (Invitrogen), respectively. Tagged proteins were purified as described previously (Takahashi et al., 1999). The His6-tagged calmodulin 2 was incubated with the GST-tagged CG-NAP3510–3829 in a buffer containing 50 mM Tris-HCl at pH 7.5, 1% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 2 mM MgCl2, 10 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride (PMSF) at 4°C for 1 h. Then, glutathione-Sepharose 4B beads (Amersham Biosciences) were added and incubated for 30 min. After the resin had been washed with the same buffer, the bound proteins were analyzed by immunoblotting.
Immunoprecipitation and Immunoblotting
Cells were lysed with buffer containing 50 mM Tris-HCl at pH 7.5, 1% NP-40, 150 mM NaCl, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 1.5 mM MgCl2, 1 mM PMSF, 20 μg/ml aprotinin, and 10 μg/ml leupeptin. After centrifugation, cell extracts were saved and incubated with an appropriate antibody at 4°C for 2 h. Then protein A–Sepharose beads (Amersham Biosciences) were added and incubated for 30 min. The resin was washed with the same buffer, and the bound proteins were processed for immunoblotting.
Isolation of Centrosomes and In Vitro Microtubule Nucleation
Centrosomes were isolated from CHO cells according to the method of Moudjou and Bornens (1994) with minor modifications. In brief, CHO cells were incubated with 10 μg/ml nocodazole and 5 μg/ml cytochalacin B for 2 h, rinsed with isolation buffer (1 mM Tris-HCl at pH 8.0, 0.5 mM EDTA, and 0.1% β-mercaptoethanol), and then lysed by swaying the dishes in the isolation buffer containing 0.5% NP-40 at 4°C for 10 min. The lysates were spun at 2500 × g for 10 min to remove cell debris and nuclei. The resultant supernatant was subjected to the discontinuous sucrose density gradient with 60 and 40% (wt/wt) solutions from the bottom and spun at 120,000 × g for 1 h. Fractions were collected from the bottom and stored at −80°C. For immunoblotting, fractions were eightfold diluted with 10 mM PIPES at pH 6.9 followed by centrifugation at 18,500 × g for 10 min to precipitate centrosomes, and then the precipitates were dissolved in SDS sample buffer.
The microtubule nucleating activity of the isolated centrosomes was tested according to Mitchison and Kirschner (1984). The centrosome suspension was incubated with 2.5 mg/ml bovine tubulin (Cytoskeleton) and 1 mM GTP at 37°C for 4–8 min. After glutaraldehyde fixation and sedimentation on glass coverslips, the microtubules were visualized by use of an anti-α-tubulin antibody. To test the effect of antibodies on microtubule nucleation, the centrosome suspension was preincubated with affinity-purified antibodies for 30 min at 4°C. Then, a nucleation reaction was done for 4 min.
RESULTS
CG-NAP Is Localized to the Centrosome via the Carboxyl-Terminal Region
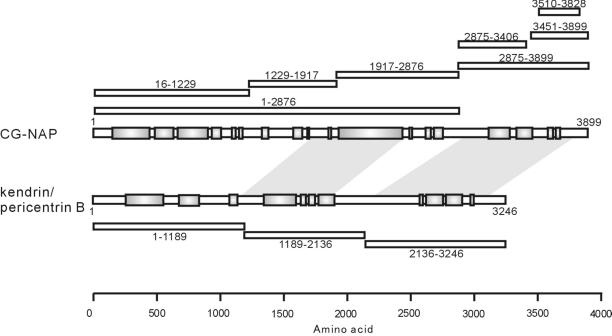
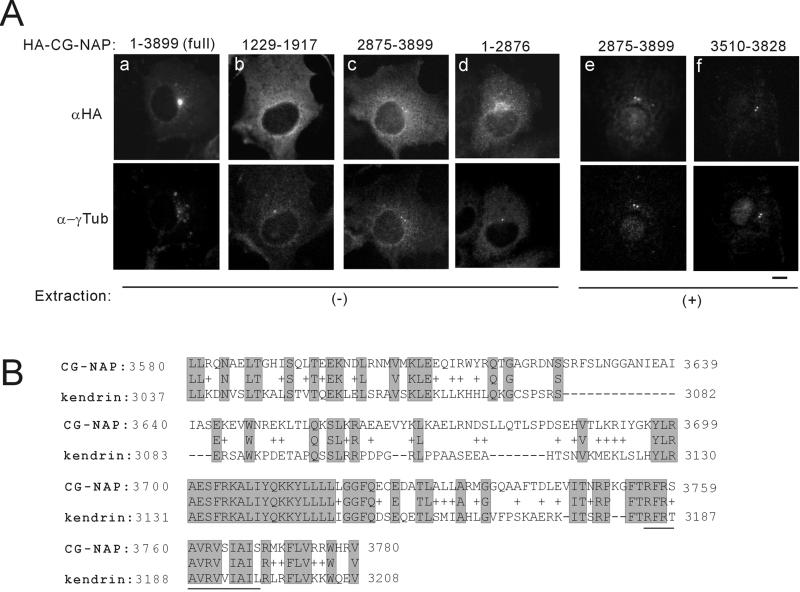
A schematic representation of the centrosomal proteins CG-NAP, kendrin, and their deletion mutants used in this study is shown in Figure 1. In the text, deletion mutants are designated as CG-NAPX-X or kendrinX-X, where X-X represents amino acid residues. We first analyzed subcellular localization of various deletion mutants of CG-NAP expressed in COS7 cells. Most of the deletions were distributed in the cytosol (for instance, Figure 2Ab). On the other hand, the carboxyl-terminal fragment CG-NAP2875–3899 and a further deletion CG-NAP3510–3828 were well colocalized with γ-tubulin at the centrosomes (Figure 2A, c, e, and f, respectively). Moreover, CG-NAP1–2876 lacking the carboxyl-terminal region distributed diffusely at perinuclear area that was not colocalized with γ-tubulin (Figure 2Ad). These results indicate that the carboxyl-terminal region containing the amino acid residues 3510–3810 is responsible for the centrosomal localization of CG-NAP. BLAST search of CG-NAP3510–3828 revealed that this region shares high homology with the carboxyl-terminal region of another centrosomal coiled-coil protein kendrin (Figure 2B). CG-NAP and kendrin have three coiled-coil regions flanked by noncoiled regions (Figure 1). BLAST search using full-length CG-NAP yielded kendrin at two regions with relatively high homology (Figure 1, shaded areas), and the carboxyl-terminal part contained the sequence shown in Figure 2B.
Figure 1.
Schematic representation of CG-NAP, kendrin, and their deletion mutants. Schematic structure of CG-NAP and kendrin are shown with predicted coiled-coil regions in shaded boxes. Positions of the deletion mutants of CG-NAP and kendrin are shown with amino acid residues on the upper and lower sides, respectively. Shaded areas between CG-NAP and kendrin represent the regions sharing homology found by BLAST search. Total amino acid residue of kendrin used in this study was 3246, as described in MATERIALS AND METHODS, which is shorter than that (3321) deposited to GenBank with accession number U52962.
Figure 2.
Centrosomal localization of the carboxyl-terminal region of CG-NAP. (A) Subcellular localization of deletion mutants of CG-NAP. HA-tagged full-length and deletion mutants of CG-NAP were transiently expressed in COS7 cells, and then the cells were fixed with methanol directly (a–d) or after brief extraction with detergent to visualize proteins associated with intracellular structures (e, f). Then, the cells were double-stained with anti-HA and anti-γ-tubulin (α-γTub). Bar, 10 μm. (B) Sequence homology of the centrosomal-localization region of CG-NAP with kendrin. Aligned sequences are the result of BLAST search with CG-NAP3510–3828. The amino acid residues of kendrin shown are of U52962 and are different from those of our kendrin construct. Possible calmodulin binding sequence of kendrin homologous to that of yeast Spc110p (Flory et al., 2000) is underlined.
The Carboxyl-Terminal Region of CG-NAP Associates with Calmodulin
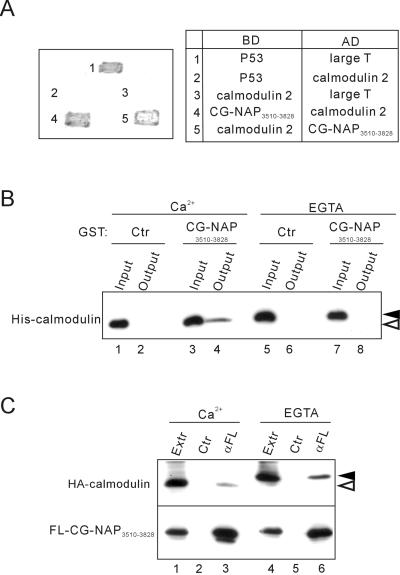
To search for the proteins interacting with the centrosomal-localization region of CG-NAP, we used yeast two-hybrid screening of a HeLa cDNA library using CG-NAP3510–3828 as bait. Among ∼1500 clones obtained, most of the clones carried calmodulin 2 and calmodulin 3 cDNAs. Calmodulin 2 and 3 have identical amino acid sequences, although they are coded by distinct genes (Berchtold et al., 1993). These clones contained varying lengths of 5′- and 3′-noncoding sequences, indicating that they were independent clones. We confirmed the interaction between CG-NAP3510–3828 and calmodulin by different combinations in a yeast two-hybrid system (Figure 3A). Furthermore, GST pull-down assay revealed that CG-NAP3510–3828 interacts directly with calmodulin in a Ca2+-dependent manner (Figure 3B). However, exogenously expressed CG-NAP3510–3828 in COS7 cells was coimmunoprecipitated with calmodulin in either the presence or absence of Ca2+ (Figure 3C). A mobility shift of calmodulin on electrophoresis confirmed that the Ca2+-dependent conformational change (Rhyner et al., 1992) occurred under these experimental conditions.
Figure 3.
Association of the centrosomal-localization region CG-NAP3510–3828 with calmodulin. (A) Yeast two-hybrid analysis of interaction between CG-NAP3510–3828 and calmodulin 2. Interaction of the proteins fused to the Gal4 DNA binding domain (BD) and activation domain (AD) was assessed by growth and development of blue color of the transfected yeasts. Combinations of BD and AD constructs are shown on the right. p53 and SV40 large T antigen were used as controls. (B) Direct and Ca2+-dependent binding of CG-NAP3510–3828 with calmodulin. Bacterially expressed His6-tagged calmodulin 2 and GST-tagged CG-NAP3510–3828 were mixed and incubated in the presence of 2 mM CaCl2 or EGTA. After removal of aliquots (Input), glutathione-Sepharose beads were added to the mixture and incubated further, and then the proteins bound to the beads were collected (Output) and immunoblotted with anti-His. Black and white arrowheads indicate the positions of Ca2+-unbound and -bound forms of calmodulin, respectively. (C) Ca2+-independent coimmunoprecipitation of calmodulin with CG-NAP3510–3828. HA-tagged calmodulin 2 and FLAG-tagged CG-NAP3510–3828 were coexpressed in COS7 cells, and then cell extracts (Extr) were prepared in the presence of 2 mM CaCl2 or EGTA. The extracts were immunoprecipitated with anti-FLAG (αFL) or control (Ctr) mouse IgG followed by immunoblot with anti-HA (top) or anti-FLAG (bottom). Black and white arrowheads indicate the positions of calcium-unbound and -bound forms of calmodulin, respectively.
The Amino-Terminal Region of CG-NAP Associates with γ-TuRC through Interaction with GCP2 and/or GCP3
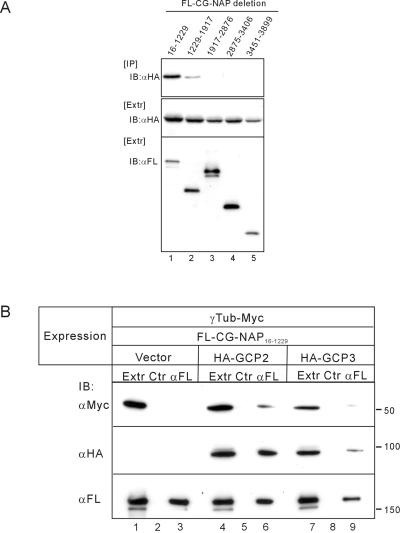
During the course of this study, kendrin was proposed to be a human orthologue of yeast Spc110p, because kendrin has a calmodulin-binding sequence homologous to that of Spc110p (Flory et al., 2000). A similar sequence was also found in CG-NAP3510–3828 (Figure 2B), which indeed interacted with calmodulin (Figure 3). Therefore, CG-NAP may function as a mammalian orthologue of Spc110p and anchor γ-TuRC. To test this possibility, we first carried out an immunoprecipitation study of endogenous proteins. Endogenous γ-tubulin was coimmunoprecipitated with CG-NAP by anti-CG-NAP antibody from HeLa cell extracts. However, γ-tubulin was not coimmunoprecipitated with any of the deletion mutants of CG-NAP expressed in COS7 cells (our unpublished results). Spc110p indirectly associates with γ-tubulin through binding with Spc97p and Spc98p, the components of γ-TuRC (Knop and Schiebel, 1997). Mammalian orthologues of Spc97p and Spc98p were identified as GCP2 and GCP3 (also named HsSpc98p), respectively (Murphy et al., 1998; Tassin et al., 1998). We thus examined whether CG-NAP binds with these proteins. GCP2 was efficiently coimmunoprecipitated with the amino-terminal region of CG-NAP, CG-NAP16–1229, and weakly with CG-NAP1229–1917 (Figure 4A). The binding property of GCP3 to CG-NAP was similar to that of GCP2 (our unpublished results). We next examined whether CG-NAP16–1229 interacts with γ-tubulin through binding with GCP2 or GCP3 (Figure 4B). As expected, γ-tubulin was coimmunoprecipitated with CG-NAP16–1229 only when GCP2 or GCP3 was coexpressed (Figure 4B, lanes 3, 6, and 9). The binding affinity of GCP3 to CG-NAP16–1229 seemed to be lower than that of GCP2. GCP3 might indirectly associate with CG-NAP16–1229 through interaction with GCP2, because GCP3 was coimmunoprecipitated with GCP2 (our unpublished results). These results indicate that CG-NAP anchors γ-TuRC through binding with GCP2 and/or GCP3.
Figure 4.
Association of CG-NAP16–1229 with γ-tubulin through binding with GCP2/GCP3. (A) Association of GCP2 with deletion mutants of CG-NAP. FLAG-tagged (FL) deletion mutants of CG-NAP were expressed in COS7 cells together with HA-tagged GCP2. Then the cell extracts were immunoprecipitated (IP) with anti-FLAG followed by immunoblot (IB) with anti-HA. (B) Indirect association of γ-tubulin (γTub) with CG-NAP16–1229 through binding with GCP2 or GCP3. COS7 cells were transfected with various combinations of expression plasmids as shown at the top. Then the cell extracts were immunoprecipitated with anti-FLAG or control mouse IgG followed by immunoblot with anti-Myc for γ-tubulin (top), anti-HA for GCP2 or GCP3 (middle), or anti-FLAG for CG-NAP16–1229 (bottom).
The Amino-Terminal Region of Kendrin Also Associates with γ-TuRC through Interaction with GCP2
Although kendrin was proposed to be a mammalian orthologue of Spc110p, its interaction with γ-TuRC has not been presented. We thus asked whether kendrin also interacts with γ-TuRC in a manner similar to CG-NAP. To do this, we prepared full-length cDNA of kendrin and specific antibody to kendrin (αKen) as described in MATERIALS AND METHODS (Figure 5A). HA-tagged kendrin expressed in COS7 cells and endogenous kendrin in HeLa or CHO cells were well recognized by αKen in immunoblotting and immunoprecipitation (Figure 5B). The mobility of these bands may agree with the deduced molecular mass of ∼370 kDa. We next examined the subcellular localization of kendrin. αKen gave centrosomal staining in HeLa cells (Figure 5C, a and b) and other cells such as CHO (our unpublished results). HA-tagged full-length kendrin was also localized to the centrosome (Figure 5D). The carboxyl-terminal fragment of kendrin, kendrin2136–3246, was localized to the centrosome and associated with calmodulin in a manner similar to CG-NAP2875–3899 (our unpublished results).
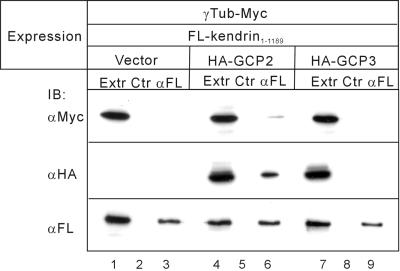
The amino-terminal fragment of kendrin, kendrin1–1189, associated with GCP2, and with γ-tubulin when GCP2 was coexpressed (Figure 6, lanes 3 and 6). However, association of kendrin1–1189 with GCP3 was very weak, and coexpression of GCP3 had little effect on the association with γ-tubulin (Figure 6, lane 9). These results indicate that kendrin also associates with γ-TuRC through binding with GCP2, although it remains unclear whether GCP3 is involved in this association.
Figure 6.
Association of kendrin with γ-tubulin (γTub) through binding with GCP2. COS7 cells were transfected with various combinations of expression plasmids as shown at the top. Then the cell extracts (Extr) were immunoprecipitated with anti-FLAG (αFL) or control (Ctr) mouse IgG followed by immunoblot with anti-Myc for γ-tubulin (top), anti-HA for GCP2 or GCP3 (middle), or anti-FLAG for kendrin1–1189 (bottom).
Endogenous CG-NAP and Kendrin Form Complexes and Associate with GCP2 and γ-Tubulin
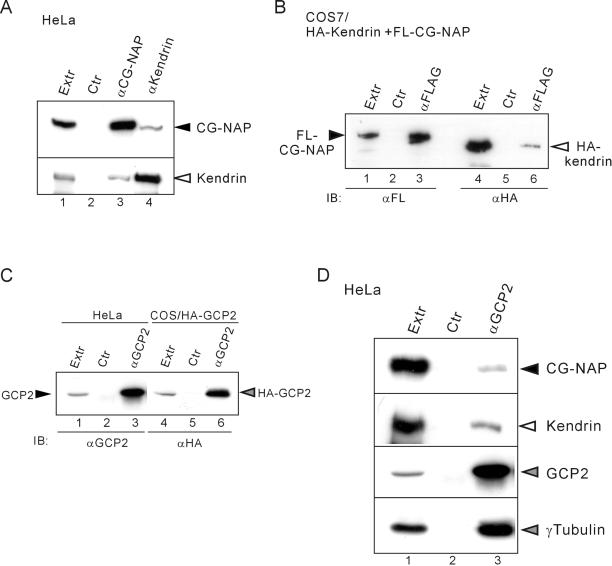
CG-NAP and kendrin were both localized to the centrosome (Figure 5C) and have coiled-coil regions that are thought to mediate protein-protein interaction. We thus examined whether CG-NAP interacts with kendrin. Endogenous kendrin was coimmunoprecipitated with endogenous CG-NAP from HeLa cell extracts by anti-CG-NAP antibody (Figure 7A, lane 3), and vice versa (Figure 7A, lane 4). The interaction was also observed between exogenously expressed CG-NAP and kendrin (Figure 7B). These results indicate that CG-NAP and kendrin are associated in vivo at the centrosome.
Figure 7.
Association of endogenous kendrin, CG-NAP, γ-tubulin, and GCP2. (A) Association of endogenous CG-NAP and kendrin. HeLa cell extracts (Extr) were immunoprecipitated with anti-CG-NAP (αEE+αBH) or anti-kendrin (αKen) or control (Ctr) rabbit IgG followed by immunoblot with anti-CG-NAP (αEE) (top) or αKen (bottom). (B) Association of recombinant kendrin with CG-NAP. 293T cells were cotransfected with FLAG-tagged (FL) CG-NAP and HA-tagged kendrin. Then the cell extracts were immunoprecipitated with anti-FLAG or control mouse IgG followed by immunoblot with anti-FLAG (lanes 1–3) or anti-HA (lanes 4–6). (C) Specificity of anti-GCP2 antibody. The extracts of COS7 cells transfected with HA-tagged GCP2 or HeLa cells were immunoprecipitated with αGCP2 or control rabbit IgG followed by immunoblot with αGCP2 (lanes 1–3) or anti-HA (lanes 4–6). (D) Association of GCP2 with CG-NAP and kendrin as well as with γ-tubulin. HeLa cell extracts were immunoprecipitated with αGCP2 or control rabbit IgG followed by immunoblot with anti-CG-NAP (αEE), αKen, αGCP2, or anti-γ-tubulin.
To examine whether CG-NAP and kendrin form complexes with γ-TuRC in vivo, we performed an immunoprecipitation study using anti-GCP2 antibody (αGCP2). αGCP2 was prepared as described in MATERIALS AND METHODS and was confirmed to recognize and immunoprecipitate endogenous GCP2 (Figure 7C). Endogenous CG-NAP and kendrin were significantly coimmunoprecipitated with GCP2 together with γ-tubulin (Figure 7D). These data suggest that CG-NAP and kendrin form complexes and associate with γ-TuRC in vivo.
Antibodies to CG-NAP and Kendrin Inhibit Microtubule Nucleation from Isolated Centrosomes
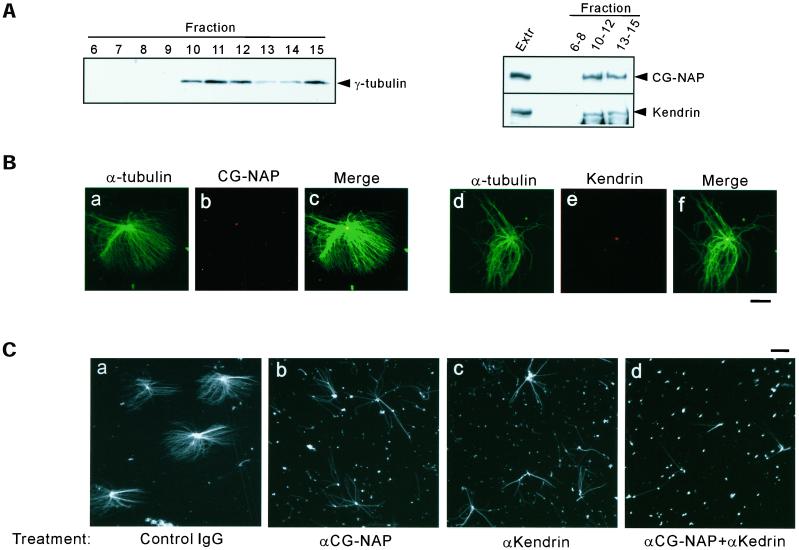
To examine whether CG-NAP and kendrin were present in isolated centrosomes, we fractionated the lysates of nocodazole/cytochalacin B–treated CHO cells by sucrose density gradient centrifugation. Fractions were subjected to immunoblotting and assayed for microtubule nucleation in vitro. γ-Tubulin was enriched in the fractions 10–12 corresponding to the sucrose densities of between 40 and 60% (Figure 8A). CG-NAP and kendrin were cosedimented with γ-tubulin in these fractions (Figure 8A). Microtubule asters were efficiently assembled in vitro by using the fractions 10 and 11, and CG-NAP and kendrin were detected in the center of microtubule asters (Figure 8B).
Figure 8.
Inhibition of microtubule nucleation from isolated centrosomes by antibodies to CG-NAP and kendrin. (A) Presence of CG-NAP and kendrin in the fractions enriched with centrosomes. CHO cell lysates were fractionated by sucrose density gradient as described in MATERIALS AND METHODS. Then the fractions were examined for the presence of γ-tubulin (left), CG-NAP, and kendrin (right) by immunoblotting. Fractions 10–12 corresponding to the interphase between 40 and 60% sucrose were enriched with centrosomes. (B) Localization of CG-NAP and kendrin at the center of microtubule asters formed in vitro. Centrosome fractions were incubated with bovine tubulin in the presence of 1 mM GTP for 8 min at 37°C as described in MATERIALS AND METHODS. The resultant microtubule asters were fixed with glutaraldehyde and spun down on coverslips. Then the coverslips were processed for double-staining with anti-CG-NAP (αEE) and anti-α-tubulin (a–c), or αKen and anti-α-tubulin (d–f). Bar, 10 μm. (C) Inhibition of microtubule nucleation by pretreatment of the centrosomes with antibodies to CG-NAP and kendrin. Centrosome fractions were pretreated with antibodies to CG-NAP (αrXN) or kendrin (αKen) at a concentration of 1.2 mg/ml or with the combination of αrXN and αKen (1.2 mg/ml each), or control rabbit IgG (2.4 mg/ml) for 30 min on ice. Then microtubule nucleation was performed by incubation with bovine tubulin for 4 min. Microtubule asters were visualized by immunostaining with anti-α-tubulin. Bars, 10 μm. The results shown are representative of five different experiments.
Next, we examined the effect of antibodies to CG-NAP and kendrin on the microtubule nucleation. The centrosome fractions were preincubated with αCG-NAP and/or αKen or control IgG and then assayed for the microtubule nucleation in vitro. Normal rabbit IgG had no effect on asters formation (Figure 8Ca), whereas αCG-NAP and αKen moderately reduced the number of microtubules nucleated from the centrosomes (Figure 8C, b and c, respectively). Furthermore, combination of the two antibodies inhibited the microtubule nucleation more efficiently (Figure 8Cd). It is of interest that microtubules escaping from this inhibition had almost the same length as those in the control. These results suggest that antibodies to CG-NAP and kendrin inhibit the initiation of nucleation but not the elongation process of microtubules.
DISCUSSION
Several candidates have been proposed as mammalian anchoring proteins for γ-TuRC at the centrosome, such as Ctr100, pericentrin, and kendrin (Tassin et al., 1997; Dictenberg et al., 1998; Flory et al., 2000). However, none of them have been demonstrated to interact with components of γ-TuRC or to mediate microtubule nucleation, although pericentrin was found in the complex containing γ-tubulin (Dictenberg et al., 1998). In this study, we have shown that the amino-terminal regions of CG-NAP and kendrin indirectly associated with γ-tubulin through interaction with other components of γ-TuRC, GCP2/GCP3 and GCP2, respectively. Interaction among endogenous proteins was also detected by immunoprecipitation study (Figure 7), although it remains to be established that CG-NAP and kendrin constitute primary anchoring sites for γ-TuRC in vivo. Furthermore, pretreatment of isolated centrosomes by antibodies to CG-NAP and/or kendrin suppressed the initiation of microtubule nucleation (Figure 8C), suggesting the involvement of CG-NAP and kendrin in microtubule nucleation from the centrosome. The antigens used to generate these antibodies are located in the amino-terminal region that interacted with γ-TuRC; thus, the epitopes might be close to or overlap with the binding sites for γ-TuRC. It was revealed that the amount of γ-tubulin was decreased in the centrosomes treated with antibodies to CG-NAP and/or kendrin by immunoblotting analysis (our unpublished results). Therefore, these antibodies suppressed microtubule nucleation, probably by displacement of γ-TuRC from CG-NAP and/or kendrin in the centrosome. The majority of microtubule nucleation appears to be limited to the centrosome despite the presence of substantial amounts of cytoplasmic γ-TuRC. Therefore, on recruitment to CG-NAP and/or kendrin in the centrosome, γ-TuRC might represent the active form, or activation of γ-TuRC might be prerequisite for the recruitment. Additional studies will be necessary to address each of these possibilities.
CG-NAP and kendrin were localized to the centrosome via their carboxyl-terminal regions, which were found to associate with calmodulin. The role of calmodulin in the targeting of CG-NAP and kendrin to the centrosome remains unclear. It was reported that green fluorescent protein–tagged calmodulin is localized to the centrosome only at mitotic phase in HeLa cells (Li et al., 1999). We could also detect only a small amount of calmodulin in the centrosomal fractions and almost no specific staining of endogenous calmodulin at the centrosomes (our unpublished results). Centrosomal targeting of CG-NAP and kendrin might be mediated by calmodulin at mitotic phase and by some undefined protein(s) at interphase. Another possibility is that calmodulin may serve to chaperone the carboxyl-terminal region of CG-NAP and kendrin, similar to the role of Cmd1p in the proper assembly of Spc110p at SPB (Sundberg et al., 1996), and be released after centrosomal targeting, as discussed by Gillingham and Munro (2000). It is also unclear whether calmodulin binding to these proteins is regulated by Ca2+: it was Ca2+-dependent in vitro (Figure 3B) but Ca2+-independent in the immunoprecipitation study (Figure 3C). The interaction between these proteins might be modified by the presence of some other protein(s) under cellular conditions. In addition to the calmodulin-binding region, the centrosomal-localization regions contain several conserved sequences between CG-NAP and kendrin (Figure 2B), which may provide interaction sites for other proteins. Our screening using CG-NAP3510–3828 as bait thus far has yielded only calmodulin clones, which might be attributed to the abundance of calmodulin mRNA in mammalian cells. Further screening by using different bait constructs may be one approach to assess these possibilities.
CG-NAP and kendrin have coiled-coil regions (Figure 1) that may form a filamentous complex. Indeed, endogenously and exogenously expressed CG-NAP and kendrin formed complexes (Figure 7, A and B). Moreover, certain combinations of CG-NAP and kendrin deletions were coimmunoprecipitated (our unpublished results), suggesting that the interaction between CG-NAP and kendrin is direct rather than indirect as components of a large protein complex. Heterodimers (or oligomers) of CG-NAP and kendrin may serve as components of PCM and provide anchoring sites for γ-TuRC. CG-NAP also forms a homodimer (or oligomer) (Takahashi et al., 1999); thus, CG-NAP homodimers (or oligomers) may also provide the anchoring sites. Do CG-NAP and kendrin play distinct roles in the centrosome? In yeast, γ-TuRC anchoring proteins Spc110p and Spc72p play independent roles by their different subcellular localization, inner plaque and outer plaque, respectively. Although CG-NAP and kendrin represent differences in terms of the additional localization of CG-NAP in the Golgi apparatus, they are localized to the centrosome in a very similar manner (Figure 5C). Some difference was observed in the efficiency of binding with γ-TuRC. CG-NAP16–1229 associated with GCP2 and γ-tubulin more efficiently than kendrin1–1189 (compare Figures 4B and 6B), which might be attributed to the relatively low homology between these regions (Figure 1). We may not be able to conclude that CG-NAP has a higher affinity to γ-TuRC, because these results were obtained with deletion constructs of CG-NAP and kendrin and from cells at random stages of the cell cycle. Cell cycle–dependent phosphorylation and regulation of yeast SPB proteins, such as Spc110p (Friedman et al., 2001) and Spc98p (Pereira et al., 1998), have been demonstrated. Similarly, centrosomal proteins may be regulated by phosphorylation, for instance, at mitotic phase. From interphase to metaphase, the amount of γ-tubulin at the centrosome increases at least threefold and decreases rapidly by late anaphase (Khodjakov and Rieder, 1999). The recruitment of γ-TuRC to the centrosome may contribute to the increase in microtubule number associated with mitotic versus interphase centrosomes (Kuriyama and Borisy, 1981). It is attractive to postulate that mitotic phosphorylation of CG-NAP or kendrin or GCP2/GCP3 alters affinity among these proteins or to the component(s) of PCM, which may result in increased recruitment of γ-TuRC to the centrosomes.
We have found that CG-NAP interacts with Rho-activated protein kinase PKN (Amano et al., 1996; Shibata et al., 1996; Watanabe et al., 1996), PKA, PKCε, and protein phosphatases PP1 and PP2A, and thus, CG-NAP may target them to the proximity of specific substrates at the centrosome and the Golgi apparatus (Takahashi et al., 1999, 2000). It is possible that the complex containing CG-NAP, kendrin, and γ-TuRC serves as a substrate for these enzymes and is regulated downstream of various signals such as Rho and cAMP. Kendrin also anchors PKA (Diviani et al., 2000). CG-NAP and kendrin may form matrix as integral components of PCM to provide anchoring sites for γ-TuRC as well as serving as targeting machinery for various signaling enzymes to the centrosome. Further studies will be necessary to elucidate the role of CG-NAP and kendrin in the regulation of recruitment and activity of γ-TuRC at the centrosome.
ACKNOWLEDGMENTS
We thank Y. Nishizuka for encouragement during this work.
Abbreviations used:
- SPB
spindle pole body
- γ-TuRC
γ-tubulin ring complex
- GCP
γ-tubulin complex protein
- PCM
pericentriolar material
- PKA
protein kinase A
- HA
hemagglutinin
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–02–0112. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–02–0112.
This work was supported in part by research grants from the Ministry of Education, Culture, Science, Sports, and Technology, Japan, and the “Research for the Future” program of the Japan Society for the Promotion of Science.
REFERENCES
- Amano M, Mukai H, Ono Y, Chihara K, Matini T, Hamajima Y, Okawa K, Iwanatsu A, Kaibuchi K. Identification of aputative target at Rho as the serine-threonine kinase protein kinase N. Science. 1996;271:648–650. doi: 10.1126/science.271.5249.648. [DOI] [PubMed] [Google Scholar]
- Berchtold MW, Egli R, Rhyner JA, Hameister H, Strehler EE. Localization of the human bona fide calmodulin genes CALM1, CALM2, and CALM3 to chromosomes 14q24–q31, 2p21.1-p21.3, and 19q13.2-q13.3. Genomics. 1993;16:461–465. doi: 10.1006/geno.1993.1211. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diviani D, Langeberg LK, Doxsey SJ, Scott JD. Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr Biol. 2000;10:417–420. doi: 10.1016/s0960-9822(00)00422-x. [DOI] [PubMed] [Google Scholar]
- Flory MR, Moser MJ, Monnat RJ, Jr, Davis TN. Identification of a human centrosomal calmodulin-binding protein that shares homology with pericentrin. Proc Natl Acad Sci USA. 2000;97:5919–5923. doi: 10.1073/pnas.97.11.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, Kern JW, Huneycutt BJ, Vinh DB, Crawford DK, Steiner E, Scheiltz D, Yates J, III, Resing KA, Ahn NG, Winey M, Davis TN. Yeast Mps1p phosphorylates the spindle pole component Spc110p in the N-terminal domain. J Biol Chem. 2001;276:17958–17967. doi: 10.1074/jbc.M010461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould RR, Borisy GG. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellog DR, Moritz M, Alberts B. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Microtubule-nucleating activity of centrosomes in Chinese hamster ovary cells is independent of the centriole cycle but coupled to the mitotic cycle. J Cell Biol. 1981;91:822–826. doi: 10.1083/jcb.91.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, Heim R, Lu P, Pu Y, Tsien RY, Chang DC. Dynamic redistribution of calmodulin in HeLa cells during cell division as revealed by a GFP-calmodulin fusion protein technique. J Cell Sci. 1999;112:1567–1577. doi: 10.1242/jcs.112.10.1567. [DOI] [PubMed] [Google Scholar]
- Li Q, Hansen D, Killilea A, Joshi HC, Palazzo RE, Balczon R. Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J Cell Sci. 2001;114:797–809. doi: 10.1242/jcs.114.4.797. [DOI] [PubMed] [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a gamma tubulin-associated protein with an essential role in gamma tubulin ring complex (gammaTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Moudjou M, Bornens M. Isolation of centrosomes from cultured animal cells. In: Celis J E, editor. Cell Biology: A Laboratory Handbook. New York: Academic Press; 1994. pp. 595–604. [Google Scholar]
- Mukai H, Toshimori M, Shibata H, Kitagawa M, Shimakawa M, Miyahara M, Sunakawa H, Ono Y. PKN associates and phosphorylates the head-rod domain of neurofilament protein. J Biol Chem. 1996;271:9816–9822. doi: 10.1074/jbc.271.16.9816. [DOI] [PubMed] [Google Scholar]
- Murphy SM, Preble AM, Patel UK, O'Connell KL, Dias DP, Moritz M, Agard D, Stults JT, Stearns T. GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol Biol Cell. 2001;12:3340–3352. doi: 10.1091/mbc.12.11.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Fujii T, Ogita K, Kikkawa U, Igarashi K, Nishizuka Y. The structure, expression, and properties of additional members of the protein kinase C family. J Biol Chem. 1988;263:6927–6932. [PubMed] [Google Scholar]
- Pereira G, Knop M, Schiebel E. Spc98p directs the yeast gamma-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol Biol Cell. 1998;9:775–793. doi: 10.1091/mbc.9.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyner JA, Koller M, Durussel-Gerber I, Cox JA, Strehler EE. Characterization of the human calmodulin-like protein expressed in Escherichia coli. Biochemistry. 1992;31:12826–12832. doi: 10.1021/bi00166a017. [DOI] [PubMed] [Google Scholar]
- Schiebel E. γ-Tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr Opin Cell Biol. 2000;12:113–118. doi: 10.1016/s0955-0674(99)00064-2. [DOI] [PubMed] [Google Scholar]
- Schmidt PH, Dransfield DT, Claudio JO, Hawley RG, Trotter KW, Milgram SL, Goldenring JR. AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J Biol Chem. 1999;274:3055–3066. doi: 10.1074/jbc.274.5.3055. [DOI] [PubMed] [Google Scholar]
- Shibata H, Mukai H, Inagaki Y, Homma Y, Kimura K, Kaibuchi K, Narumiya S, Ono Y. Characterization of the interaction between RhoA and the amino-terminal region of PKN. FEBS Lett. 1996;385:221–224. doi: 10.1016/0014-5793(96)00385-7. [DOI] [PubMed] [Google Scholar]
- Sundberg HA, Goetsch L, Byers B, Davis TN. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Mukai H, Oishi K, Isagawa T, Ono Y. Association of immature hypophosphorylated protein kinase c epsilon with an anchoring protein CG-NAP. J Biol Chem. 2000;275:34592–34596. doi: 10.1074/jbc.M005285200. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Shibata H, Shimakawa M, Miyamoto M, Mukai H, Ono Y. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the Golgi apparatus. J Biol Chem. 1999;274:17267–17274. doi: 10.1074/jbc.274.24.17267. [DOI] [PubMed] [Google Scholar]
- Tassin AM, Celati C, Moudjou M, Bornens M. Characterization of the human homologue of the yeast spc98p and its association with gamma-tubulin. J Cell Biol. 1998;141:689–701. doi: 10.1083/jcb.141.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Celati C, Paintrand M, Bornens M. Identification of an Spc110p-related protein in vertebrates. J Cell Sci. 1997;110:2533–2545. doi: 10.1242/jcs.110.20.2533. [DOI] [PubMed] [Google Scholar]
- Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S. Protein kinase N (PKN) and PKN-related protein rhophilin as targets of small GTPase Rho. Science. 1996;271:645–648. doi: 10.1126/science.271.5249.645. [DOI] [PubMed] [Google Scholar]
- Witczak O, Skalhegg BS, Keryer G, Bornens M, Tasken K, Jahnsen T, Orstavik S. Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J. 1999;18:1858–1868. doi: 10.1093/emboj/18.7.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]