Summary
Inborn errors of immunity (IEI), as congenital chronic disorders, are often associated with neurobehavioral symptoms, traditionally considered secondary to patient burden. Their origin, however, has yet to be addressed. Here, we found that IEI-associated genes are expressed in neural lineages during human brain development, and in the absence of immunological challenges, IEI mutations directly impair neurodevelopmental trajectories, leading to psychomotor defects. Warts hypogammaglobulinemia immunodeficiency myelokathexis (WHIM) mice—bearing a mutation causing Cxcr4 hyperactivation—show developmental foliation defects of the cerebellum correlating with sensorimotor and affective dysfunctions, which recapitulate the alterations described in patients. WHIM cerebella single-cell profiling revealed major transcriptional deregulation in granule cell progenitors, whose aberrant proliferation and migration induce foliation and circuit defects. AMD3100 intracerebroventricular injection rescues both morphological and behavioral defects, demonstrating their brain-specific and Cxcr4-dependent origin. Collectively, our findings highlight the relevance of neurodevelopmental implications underlying psychomotor IEI manifestations, broadening our understanding of these conditions beyond immune dysfunctions.
Keywords: inborn errors of immunity, WHIM syndrome, neurodevelopment, cerebellum, neuroimmune interactions, plerixafor, psychomotor symptoms, CXCR4, granule cells, immunodeficiency
Highlights
-
•
IEI genes show region- and stage-specific expression in the developing human brain
-
•
WHIM mutation alters cerebellar progenitor dynamics, causing behavioral defects
-
•
Early brain-targeted CXCR4 inhibition rescues morphological and functional outcomes
Demenego et al. show that inborn errors of immunity (IEI) directly impact neurodevelopment independently of immune dysfunction. In WHIM syndrome, CXCR4 hyperactivation disrupts cerebellar assembly and behavior via cell-intrinsic mechanisms. Early brain-targeted CXCR4 antagonist rescues both structural and psychomotor defects. The study redefines the neurodevelopmental origins of IEI-associated symptoms.
Introduction
Interactions between the nervous and immune systems are reciprocal, tightly controlled, and vital for maintaining physiological homeostasis.1,2 A growing body of evidence highlights the immune system’s influence on brain development and function under normal conditions,3 while disruptions in neuroimmune communication contribute to a range of disorders, including neurodevelopmental,4,5,6,7,8,9 psychiatric,4,6,10,11 and neurodegenerative diseases.12,13,14,15 Furthermore, individuals with chronic inflammatory conditions such as inflammatory bowel disease,16,17 psoriasis,18,19 and rheumatoid arthritis,20,21,22,23 frequently exhibit comorbid anxiety and depression—symptoms often regarded as secondary to their underlying immune dysfunction.
Inborn errors of immunity (IEI) are genetic disorders characterized by partial or complete dysfunction of components of the immune system.24,25,26 These patients experience heightened susceptibility to infections and malignancies, often alongside complex comorbidities.27,28 Importantly, IEI patients also exhibit a higher prevalence of neuropsychiatric symptoms, particularly anxiety and depression.26,29,30,31,32 Whether these manifestations arise as indirect consequences of chronic immune dysregulation or result directly from the underlying genetic mutations affecting brain development remains unresolved, largely due to challenges in modeling and limited cross-disciplinary research.
Among IEI, warts hypogammaglobulinemia immunodeficiency myelokathexis (WHIM) syndrome is characterized by recurrent infections, osteoporosis, and cardiac defects.33,34,35,36,37,38,39 A recent neuroimaging and neuropsychiatric assessment of a small cohort of WHIM patients reported subtle yet consistent deficits in motor coordination, including fine and gross motor dysfunction and ataxia, along with anxiety, depression, and social withdrawal.40 Notably, magnetic resonance imaging (MRI) revealed atypical cerebellar foliation in 66% of cases, particularly affecting the inferior hemispheres, with abnormal laminar orientation.40 While these observations suggest cerebellar involvement in WHIM pathology, a mechanistic connection has yet to be established.
WHIM syndrome is caused by autosomal dominant mutations in the CXC chemokine receptor 4 (CXCR4), specifically within the gene’s C-terminal cytoplasmic domain (OMIM: #193670).35,41 CXCR4 is uniquely activated by its ligand CXC chemokine ligand 12 (CXCL12),42 and upon binding, undergoes conformational changes that initiate downstream signaling cascades regulating cell migration and proliferation. Under normal conditions, β-arrestin-mediated receptor internalization ensures controlled CXCR4 signaling.43 In WHIM syndrome this internalization is impaired, leading to prolonged receptor exposure on the cell surface, named CXCR4 hyperactivation,39,42 and causing retention defects in hematopoietic stem cells41 as well as mature neutrophils in the bone marrow.35
Current therapies for WHIM syndrome are broad and costly, highlighting a pressing medical need. Phase III trials of Plerixafor (AMD3100), a CXCR4 antagonist, have shown efficacy in increasing circulating leukocyte subsets.44,45,46 However, these trials have only included individuals over 10 years of age, despite evidence that WHIM symptoms are present from birth, and the potential benefits of early intervention, particularly for non-immune manifestations, remain unknown.
In this study, we reveal a neurodevelopmental origin for the psychomotor deficits associated with IEI, decoupling these phenotypes from systemic immune dysfunction. We show that IEI-linked genes are actively expressed in cerebellar neural lineages during early brain development. By leveraging a WHIM mouse model, we demonstrate that Cxcr4 hyperactivation directly impairs cerebellar cytoarchitecture, triggering long-lasting behavioral and functional defects that mirror clinical observations. Finally, we show that early intracerebroventricular administration of Plerixafor rescues both structural and behavioral deficits in WHIM mice, establishing a direct link between brain-intrinsic Cxcr4 dysfunction and neurobehavioral symptoms.
Results
IEI-associated genes display time- and region-specific expression patterns in human developing brain
IEI patients experience early-onset impairments extending beyond immune system dysfunction.47 While numerous neurological alterations have been associated with IEI,32,48,49,50 there remains a lack of comprehensive understanding regarding their extent. By leveraging a validated classification of IEI-associated genes,27,28 we compiled a list of 517 IEI conditions, their 446 causative genes (presenting distinctive pathological variants), and associated clinical features—including a detailed description of the reported neurological symptoms where available (Table S1). This approach enabled us to systematically map the frequency and range of neural deficits associated with IEI. We found that 41.6% of IEI was associated with, among others, intellectual disability,51,52,53 neurodevelopmental delay and microcephaly,54,55,56 anxiety57,58,59,60 and recurrent encephalopathies, even without evident manifestation of viral infections61,62,63,64 (Figure 1A), suggesting that IEI genes can impact neurodevelopment. We therefore sought to investigate the expression of IEI genes in the human brain by taking advantage of the BrainSpan dataset,65 a comprehensive resource that provides gene expression patterns across different brain regions and developmental stages. By performing hierarchical clustering of IEI genes (444/446 causative genes were present in the dataset, Figure S1A), we identified three gene subsets exhibiting a time-specific expression pattern of IEI genes (Figures 1B and S1A) in all the region analyzed. Interestingly, one subset is enriched during prenatal and early infancy stages (early); another one peaks during late infancy (mid); the last one is enriched from childhood through adulthood (late) (Figures 1B and S1A; Table S2). Among the different brain regions, the cerebellum (CB) and thalamus (TH) exhibited the highest expression of IEI genes. The number of IEI genes enriched in the CB was more than twice that observed in the TH (Figures 1C, S1A, and S1B), suggesting that the CB may be particularly vulnerable to the effects of IEI mutations.
Figure 1.
IEI-associated genes display time- and region-specific expression patterns in human developing brain
(A) Alluvial plot showing the percentage of IEI with neurological symptoms across the clinical major IEI categories.
(B) Schematics of human developmental time points and line plots of average scaled expression of genes showing time-specific expression patterns (early, 85 genes, p < 2e−16; mid, 106 genes, p = 9.1e−10; late, 38 genes, p = 6.56e−9).
(C) Schematic of human brain sections showing tissue localization and line plots of average scaled expression of genes showing tissue-specific expression patterns (CB, 98 genes, p = 5.28e−12; TH, 39 genes, p = 2.33e−11).
(D) UMAP colored by cell type of single nuclei derived from healthy donor cerebella.66
(E and F) UMAP showing single nuclei colored by developmental stage and lineage.
(G) Dot plot showing subtype-specific expression of selected IEI genes.
∗ indicates genes present in SFARI database.
Therefore, we took advantage of a recently published human cerebellar single-nuclei RNA sequencing dataset66 to investigate the expression pattern of IEI genes at single-cell resolution. We analyzed 143,390 nuclei derived from healthy donor cerebella across 9 developmental stages from 7 post-conceptional weeks (pcw) to toddler stages (2.8–3.5 years old) and examined the expression of IEI genes (Figures 1D–1F and S1C–S1E). The single-nuclei analysis confirmed cellular heterogeneity across time points and lineages (Figures 1E and 1F) during human cerebellarmaturation.67,68,69,70 86% of the IEI-associated genes were expressed in this dataset with specific expression dynamics (Table S2): 23% belong to the BrainSpan CB cluster, 22% to the early cluster, 22% to the mid cluster, 8% to the TH cluster, and 7% to the late cluster. This result further confirms the previously observed enrichment in early, mid, and CB clusters. IEI genes showed expression in immune cell types resident in the parenchyma and circulating in brain vessels (Figures 1G, 40 most variable genes and S1F). However, only a small fraction was exclusive of the immune lineage (e.g., PTPRC and IKZF1), and we detected IEI genes in neural cells (Figure 1G). We identified genes with known expression and role in brain development and maturation, such as nuclear factor kappa B subunit 1 (NFKB1),71,72 B cell lymphoma/leukemia transcription factor 11B (BCL11B)73,74,75 and CXC chemokine receptor 4 (CXCR4),76,77 associated with NfkB1-deficiency, Bcl11b deficiency and WHIM syndrome, respectively, which present neurological symptoms (Tables S1 and S2). Interestingly, several IEI genes (25 genes, e.g., BCL11B, PRKDC, and CARD11B) are also present in the Simons Foundation Autism Research Initiative (SFARI) database and present high confidence scores78 (Table S2), highlighting how they are implicated in neurodevelopmental disorders. Overall, this pattern suggests a broader role for IEI-associated genes in different cell types of the human developing brain.
Magnetic resonance imaging reveals selective alterations in cerebellar architecture of WHIM mice
To investigate how IEI genetic alterations could impact brain assembly, we leveraged the well-established mouse model harboring the WHIM-linked heterozygous CXCR4S338X mutation (Cxcr4+/1013 or WHIM), which has been extensively employed to characterize immune and skeletal defects.33,36,38,39,79,80,81,82
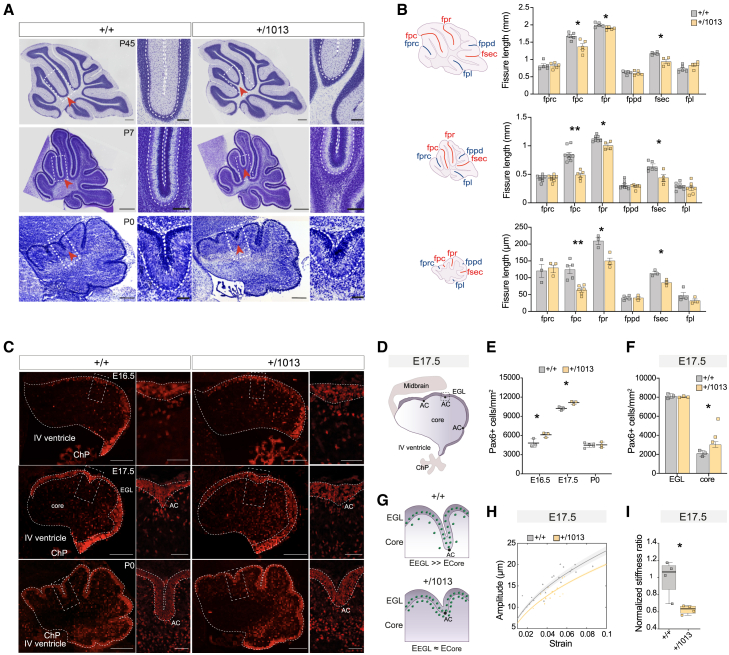
Given the structural alterations observed in WHIM patients,40 we performed MRI on post-natal day (P) 45 WHIM and control brains (Figures 2A and S2A). Ex vivo MRI voxel-wise analysis revealed structural alterations selectively in WHIM cerebellum (Figures 2A and S2A; Tables S3 and S4). The most pronounced defects were found in the vermis, where we observed a significant volume increase along the primary fissure (fpr), separating LIV–V and LVI, and a significant decrease along the preculminary fissure (fpc), separating LIV–V and LIII (Figures 2A and S2A). The white matter arbor vitae’s branch length, composed by cerebellar afferent and efferent projections, was reduced in LIII and LIX (Figure 2B), suggesting changes in pathways conveying sensorimotor information to/from the cerebellum in Cxcr4+/1013 animals.
Figure 2.
WHIM mutants display cerebellar morphological alterations and anxiety-like behavioral phenotypes
(A) Voxel-wise comparison of cerebellar brain volumes showing gross morphological abnormalities in P45 WHIM mice. Red: WHIM larger than controls; blue: WHIM smaller than controls; black contours: regions where FWE-corrected ∗p < 0.05; n > 9 animals/condition.
(B) 2D skeleton analysis of white matter arbor vitae. ∗∗p < 0.01; n > 9 animals/condition.
(C) Schematic of mouse dorsal cerebellum. Bar plots and sagittal cerebellar sections at different interaural levels in the vermis and hemisphere. Multiple t test, ∗p < 0.05, ∗∗p < 0.01; n > 4 animals/condition.
(D) Boxplots showing the time WHIM mice spend in arena and the number of vertical counts. Student’s t test, ∗∗p < 0.01; n = 16 animals/condition.
(E) Line plot indicating the latency to fall (s) from the accelerating rotarod. Two-way ANOVA, ∗p < 0.05; n = 11 animals/condition.
(F) Boxplots showing the number of transitions, the ratio between time in the dark chamber and number of transitions. Student’s t test, ∗p < 0.05, ∗∗p < 0.01; n = 16 animals/condition.
(G) Boxplot indicating the immobility time (s). Student’s t test, ∗∗p < 0.01; n = 16 animals/condition.
(H) Bar plot indicating the percentage of beads buried. Student’s t test, ∗p < 0.05; n > 6 animals/condition.
(I) Schematics of the experimental plan.
(J) Line plot showing body weight from P0 to P45. Multiple t test, ∗p < 0.05, ∗∗p < 0.01; n > 17 animals/condition.
(K) Line plot showing the time pups gripped the stick. Multiple t test, ∗p < 0.05; n > 14 animals/condition.
(L) Boxplot quantifying the time pups take to turn and face upward. Multiple t test, ∗p < 0.05; n > 30 animals/condition.
(M) Bar plot showing the percentage of pups that turn away from the cliff. Multiple t test, ∗∗p < 0.01; n > 30 animals/condition.
We then evaluated the cerebellar cytoarchitecture by Nissl staining in P45 animals. When measuring the length of the Purkinje cell layer (PCL), defined by Purkinje cells (PCs) arranged in a row along cerebellar lobules,67 we observed a reduction in WHIM cerebella in LIII, LVIII and LIX at the vermis level (Figures 2C and S2B). We also evaluated the PCL in the hemispheres at different interaural planes, and found a significant reduction in several lobules, including the paramedian lobule (PML, Figures 2C and S2B). Notably, the PML in rodents is thought to anatomically match the human gracile lobule,83 which is the most altered cerebellar region in WHIM patients.40
These data collectively support the presence of regional cerebellar abnormalities and selective lobulation defects in WHIM mice, closely resembling those reported in WHIM patients.
WHIM mutants display early-onset behavioral alterations with enduring effect on anxiety-related phenotype
We therefore investigated whether the aberrant morphological defects in WHIM mutants could lead to altered behaviors. First, we evaluated the activity of WHIM mice in the open-field arena (Figure 2D). They displayed increased thigmotaxis (tendency to remain close the walls, Figure 2D) and showed a significant increase of the ambulatory time in the periphery (Figure S2C), while reducing the resting time in the center (Figure S2C). They exhibited a lower number of vertical counts (Figure 2D) and traveled a reduced distance and at a lower speed compared with controls (Figure S2C).
To specifically assess whether Cxcr4+/1013 mice displayed an altered motor phenotype, we performed the accelerating rotarod test. We observed no significant difference in the latency to fall between WHIM and control mice (Figures 2E and S2D), indicating that the defective activity shown by Cxcr4+/1013 animals in the open field does not result from motor defects, and suggesting the presence of an anxiety-like phenotype. Indeed, Cxcr4+/1013 mice displayed reduced exploratory behavior in the dark-light chamber test, as indicated by the reduced number of transitions between the chambers and by the increased time spent in the dark chamber (Figure 2F). Furthermore, in the forced swim test, they showed decreased immobility time (Figure 2G), suggesting dysfunctional coping strategies associated to anxiety.84 No differences were found in the self-grooming test (Figure S2E); however, a significant increase in marble-burying activity was observed (Figure 2H). Overall, these data indicate the presence of an anxiety-like behavior in Cxcr4+/1013 mice.
To understand the impact of Cxcr4 hyperactivation on sensory and motor development, we leveraged a standardized battery of tests adapted from the Fox scale85 (Figures 2I–2M and S3). WHIM pups exhibited lower weight from P7 onward compared with control littermates (Figure 2J). They showed no locomotor deficits in the ambulation (Figure S3B) and air righting (Figure S3E) tests. However, they exhibited transient significance difference in the open-field traversal (Figure S3C) and surface righting tests (Figure S3D). Moreover, WHIM pups displayed impaired behavior in the forelimb grasp test (Figure 2K), suggesting a delay in neuromuscular development. Their slower motor response to a vestibular cue of gravity, measured in the negative geotaxis test,86 confirmed impairments in coordination and directionality (Figure 2L). Additionally, a delay in the response to sensory and auditory stimuli was found in the ear twitch (Figure S3F) and auditory startle tests (Figure S3G). Similarly, the cliff aversion test, evaluating the integration of motor outputs driven by sensory stimuli, revealed a defective response of WHIM pups (Figure 2M). In summary, our findings indicate that Cxcr4+/1013 pups follow atypical trajectories of physical, sensory, and motor development.
Overall, our analysis indicates that WHIM mice exhibit an anxiety-like phenotype, resembling those reported in WHIM patients.40 Our results also suggest that Cxcr4 hyperactivation disrupts the development of emerging circuits, leading to deficits in motor coordination and sensory integration during early post-natal development. These findings provide new insights into the role of Cxcr4 in neurodevelopmental disorders.
WHIM cerebellar architecture impairment originates at early prenatal stages
Cerebellar neuronal circuits, critical to the proper execution of complex functions,87,88,89 emerge from a prolonged post-natal neurogenesis and highly coordinated migration processes.90 We sought then to investigate whether the lobulation defects observed in WHIM mutants reflect early alterations in cerebellar assembly. We measured the length of cerebellar fissures, demarcating different lobules,70 at the vermis level in adult (P45), early post-natal (P7), and newborn (P0) cerebella (Figure 3A). At each stage, we observed consistent gross morphology alterations, with a significantly decreased length in the fpc, fpr, and secondary fissure (fsec, separating LVIII from LIX) of WHIM mice compared with littermates (Figures 3B and S4A), indicating that Cxcr4 mutation severely affects cerebellar architecture.
Figure 3.
Cerebellar architecture impairment of WHIM mutants originates in early prenatal stages
(A) Representative sagittal Nissl sections of Cxcr4+/1013 and control animals at P45, P7, and P0. Scale bar P45, 500 μm; scale bar crop, 120 μm; scale bar P7 and P0, 200 μm; scale bar crop, 50 μm.
(B) Schematic of mouse sagittal cerebellar sections at P45, P7, and P0 indicating fissure names and bar plots showing fissure length. Multiple t test, ∗p < 0.05, ∗∗p < 0.01; n > 3 animals/condition.
(C) Representative sections of Cxcr4+/1013 and control cerebella at E16.5, E17.5, and P0 stained with anti-Pax6 antibody. Scale bar, 200 μm; scale bar crop, 50 μm.
(D) Schematic of E17.5 cerebellum, highlighting the EGL, core, AC.
(E) Dot plot showing the number of Pax6+ cells/mm2 at E16.5, E17.5, and P0. Multiple t test, ∗p < 0.05; n > 3 animals/condition.
(F) Bar plot showing the number of Pax6+ cells/mm2 in the core and EGL at E17.5. Multiple t test, ∗p < 0.05; n > 3 animals/condition.
(G) Schematic of E17.5 cerebellar foliation process.
(H) Decreased folding amplitude at E17.5, modeled employing a low deformation wrinkling model (solid lines).
(I) Boxplot showing a decreased normalized stiffness ratio in WHIM cerebellum at E17.5. Student t test, ∗p < 0.05; n > 3 animals/condition.
In the developing cerebellum, the external granule layer (EGL) and the inner core region exhibit different growth rates, leading to different cellular densities. This process drives the formation of anchoring centers (ACs) and subsequent fissure elongation.91 To gain insights into the mechanisms underlying abnormal cerebellar foliation,92,93 we evaluated the proper differential expansion of EGL and core in WHIM mutants. We used the bona fide marker Pax6 to assess the density and number of granule cell progenitors (GCPs) and granule cells (GCs) in Cxcr4+/1013 cerebellum. We found that Pax6+ cell density significantly increased in WHIM cerebellum before foliation occurs (E16.5, E17.5; Figures 3C–3E), while the total number remained unaltered (Figure S4B). Remarkably, at E17.5, this variation can be attributed to an increased density of Pax6+ cells in the core cerebellar region (Figures 3F and S4C). Our results align with a previously proposed surface wrinkling model interpreting folding dynamics in the developing cerebellum, suggesting that a differential expansion occurs between the EGL and the core.91,94 We therefore exploited a mathematical model used for soft bilayer materials95 to interpret morphometric data from cerebellar sections at E17.5 and P0 and describe the evolution of wrinkling instability (Figure 3G). At E17.5, an increase in cell density in the core reduces the amplitude of the nascent folia (Figure 3H), which originates from a reduced stiffness ratio between the EGL and the core (Figures 3I and S4D). No differences in Pax6+ cell density (Figures 3C and 3E) nor in the wrinkling analysis (Figure S4E) were observed at P0, despite the evident morphological changes (Figures 3A and 3C). These mechanical alterations result in the affected folding pattern observed in WHIM mutants and underscore the critical prenatal dynamics of GCPs behavior.
Single-cell sequencing analysis reveals hyperactive Cxcr4 signaling in cerebellar precursors
To investigate the cellular and molecular underpinnings involved in the abnormal foliation observed in Cxcr4+/1013 mice, we performed single-cell RNA sequencing on micro dissected lobules LIII-VI-V from P0 Cxcr4+/1013 and controls (Figure 4A). By profiling 10,742 cells, proportionally distributed between genotypes (Figures S5A–S5C), we carried out unsupervised clustering, which revealed 26 clusters assigned to 19 distinct cell types (Figures 4B and S5A–S5C). In line with recently published datasets,66 granule progenitors (GPC, markers: Top2a, Mki67, and Cxcr4), bipotent progenitors (BPs, markers: Glis3, Sox2, and Pax3), gliogenic precursors (GPs, markers: Tnc, Slc4a4, Apoe, and Aldh1l1) and ependymal precursors (epy, markers: Spag17, Foxj1, and Tmem212) were identified. We depicted distinct excitatory cerebellar neuronal types, including unipolar brush cells (UBCs, enriched for Kirrel3, Eomes, Lmx1a) and GCs (expressing Reln, Neurod1, Eps8, St18). Molecularly distinct subpopulations of GC precursors (GCP-A Top2a+ and GCP-B Mfap4+) and GCs (GCI Neurod1+ and GCII Cadps2+) were found. As for inhibitory cell types, we identified PCs (marked by Pcp2, Calb1) and interneurons (INs, Gad1/2, Pax2, Slc6a5), including a somatostatin (Sst)-expressing IN subset. Neuronal populations of the isthmus, a boundary region between the cerebellum and midbrain (isthmic neurons, isth-N, enriched in Lhx9, Otx2 and Asic4) and GABAergic neurons of the deep cerebellar nuclei (γ-aminobutyric acid [GABA] DN, enriched in Sox14, Gad1/2) were also identified. Non-neuronal cell populations and mesoderm-derived cell types, including microglial cells (Cx3cr1, Trem2), vascular cells (Cldn5, Flt1, Cdh5, and Pecam1), meningeal endothelium (Cxcl12 and Vtn), and mesenchymal cells (Col1a1, Col3a1, and Dcn) were captured.
Figure 4.
Single-cell sequencing of developing cerebellar lobules revealed hyperactive Cxcr4 signaling of GC precursors inducing aberrant migration
(A) Experimental plan.
(B) Sequencing Uniform Manifold Approximation and Projection (UMAP) of control and Cxcr4+/1013 cerebella colored by cell type.
(C) Feature and dot plots showing Cxcr4 expression among cell types.
(D) Bar plot showing the number of deregulated genes in WHIM mutants colored by cell type. MAST, ∗p < 0.05, average log2 fold change > 0.1.
(E) Heatmap showing the scaled expression of representative top deregulated genes.
(F) Upset plot showing the degree of overlap between upregulated and downregulated genes in progenitors.
(G and H) (G) Dot plots of selected deregulated canonical pathways and biological functions in cerebellar precursors and (H) in gliogenic progenitors.
(I) Representative immunofluorescence staining of P7 cerebellum showing the orientation of BLBP+ fibers (red), and boxplots indicating the number of processes/100 μm and the angle of processes at the AC. Student t test, ∗p < 0.05; n > 3 animals/condition. Scale bar, 50 μm; scale bar crop, 25 μm.
We outlined Cxcr4 expression and confirmed it in GCPs, GPs, BPs, microglial cells, and epy (Figure 4C). By performing differential gene expression analysis (Figures 4D and 4E), we discovered a significant difference in the transcriptional footprints of both neuronal and glia progenitors (Table S5), coupled with alterations of neuronal molecular signatures. GCPs exhibited the highest number of differentially expressed genes (DEGs); in contrast, isth-N, GABA-DN and non-neural cell types, including microglia, albeit less abundant, displayed negligible or no transcriptional changes in WHIM cerebellum (Figures 4D and 4E). A distinct pattern of transcriptional deregulation was observed in BP, GP and GCPs, while GCPs A and B shared several deregulated genes (Figure 4F), suggesting that lineage-specific responses are triggered in WHIM cerebellar progenitors.
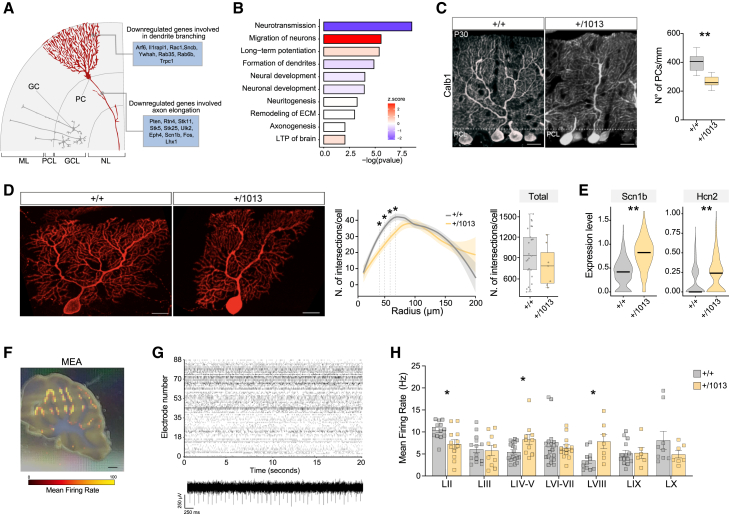
Enrichment analysis across progenitor clusters unveiled a pervasive and coordinated activation of Cxcr4 signaling pathway (Figures 4G and S5D; Table S6), highlighting potential implications for enhanced cell migration and survival. To confirm this hypothesis, we performed a comprehensive analysis of biological functions (using ingenuity pathway analysis), which revealed a significant enrichment of terms associated with enhanced migration, invasion, and cytoskeletal dynamics (Figure 4G; Table S7), providing valuable insights into the multifaceted impact of Cxcr4 hyperactivation in cerebellar progenitors.
Interestingly, GPs, the precursors of Bergmann glia cells (BG)—a glial subtype that aligns their fibers around the base of each fissure (AC) during cerebellar surface foliation, acting as tracks for inwardly migrating GCs94—showed upregulation of Cxcr4 pathway alongside deregulation of terms related to the formation of cellular protrusions and cytoskeletal organization (Figures 4G and 4H). To validate the molecular changes observed in GP, we performed immunostaining with brain-lipid-binding protein (BLBP) marker to selectively label BG.96 Tracing analysis of BLBP+ fibers revealed that at P7 the arrangement of BG fiber is altered in Cxcr4+/1013 mutants: we found a significant reduction in the number of BLBP+ processes in the molecular layer of WHIM animals, along with an increased angle formed by these fibers at the AC between LIII and LIV compared with controls (Figures 4H and 4I). This atypical fanning pattern of BG fibers likely contributes to the altered development of cerebellar lobules.
Overall, our single-cell analyses indicate that Cxcr4 mutation impinges on the landscape of multiple cerebellar progenitors during development, supporting a direct effect on the differentiation of distinct lineages.
Hyperactive Cxcr4 signaling affects GC differentiation and spontaneous activity
Given the transcriptional changes observed in GCPs, we investigated whether the differentiation and maturation of GCP derivatives, the mature GCs (Figure 5A), was affected in WHIM mice. We examined the developmental route of the granule lineage by inferring trajectory topology and performing pseudotime analysis (Figure 5B). We identified a common starting point (root) at GCP-B —the cluster with the highest entropy score as mean of the lowest differentiation level97 (Figure S5E)—from which the trajectory progresses through GCP-A and GCI, terminating in GCII (Figures 5B, 5C, and S5F). No changes in the lineage topography were observed between Cxcr4+/1013 and controls (Figure 5B).
Figure 5.
GC defects arise independently from the immunological phenotype
(A) Schematic of GC lineage development.
(B) UMAPs of the GC lineage colored by cell type and pseudotime score highlighting inferred trajectory (red).
(C) Line plot showing the expression of selected positional genes (Atoh1 and Plxna2) in control cerebellum with relative P4 ISH images form Allen Brain Atlas and density plot showing the distribution of the cells along the pseudotime, colored by cell type.
(D) Cell distribution plots showing cell density along the pseudotime score colored by genotypes (up, Density plot, Kolmogorov-Smirnov test, p = 0.028) and cell types (bottom, dot plot).
(E) Line plots showing the expression of selected upregulated genes across pseudotime of Cxcr4+/1013 cerebellum.
(F) Schematic of the experimental plan.
(G) Representative image of Cxcr4+/1013 and control mCherry positive GCs at DIV7, boxplots showing quantification of neurites’ number and length. Student’s t test, ∗p < 0.05, ∗∗p < 0.01; n > 10 cells/condition. Scale bar, 10 μm.
(H) Maximal intensity projection image of GCamP6s fluorescence at DIV7 and representative normalized traces (ΔF/F0). Boxplots showing the quantification of single-neuron spike frequency and synchronicity rate across GCs, density plot showing the distribution of amplitude values between genotypes. Student’s t test, ∗p < 0.05, ∗∗p < 0.01; n > 10 cells/condition. Data are presented as mean ± SEM. Scale bar, 100 μm.
Notably, GCPs originate in the rhombic lip, migrate to the EGL, and subsequently move inward to the inner granular layer (IGL) as they differentiate into mature GCs92 (schematic in Figure 5A). This developmental trajectory is known to be associated with dynamic changes in gene expression that identify the differentiation stages of GCs migrating along the cerebellar cortical layers.98 Our pseudotime analyses confirmed that the expression pattern of position-specific markers (such as Atoh1 and Plxna2) follows a differentiation sequence that corresponds to the distribution of GCs along their developmental trajectory (Figures 5C and S5F). This allowed us to compute the pseudotime scoring as a proxy for both differentiation and GC migration and simultaneously assess the two processes across genotypes. We observed significant differences in the cell distribution, with a divergent frequency associated with GCI state (pseudotime score 20–30, Figure 5D). Despite the exclusive expression of Cxcr4 in progenitors, we found transcriptional changes in across the entire pseudotime trajectory (Figures 5D and 5E). These findings confirm our previous observation of impaired GCs migration in WHIM mice (Figure 3), while also revealing a broader differentiation defect affecting this lineage.
However, it remains to be elucidated whether the disrupted migration interferes with differentiation progression, or whether both impairments arise independently as consequences of hyperactive Cxcr4 signaling.
To begin disentangling the intrinsic effects of Cxcr4 dysregulation on GCs, we isolated them from P5 Cxcr4+/1013 and control cerebella and culture them ex vivo, thereby excluding the contribution of local migratory cues. At 1 day in vitro (DIV), GCs were infected with pAAV-hSyn-mCherry to sparsely label neurons for morphological analysis (Figures 5F and 5G), and pAAV-hSyn-GCaMP6s, to visualize their spontaneous electrical activity (Figures 5F–5H). At DIV7, we observed that Cxcr4+/1013 neurons show a significant increase in the number of primary and secondary branches compared with controls, while neurite length remained unchanged (Figure 5G), suggesting a direct impact on cerebellar morphological complexity. At the same stage, we found that WHIM GCs had a significantly increased spike frequency and spike synchronicity rate across multiple neurons compared with controls (Figure 5H). These findings are consistent with previous studies showing that alterations of Cxcr4/Cxcl12 signaling—such as in Cxcr4 knockout models—impinges not only on neuronal positioning but also intrinsic properties like excitability, dendritic growth, and synaptic plasticity.99
Collectively, our results demonstrate that Cxcr4 hyperactivation directly affects GC development and maturation. While neuronal differentiation trajectories and migratory behaviors are deeply interwoven processes, our ex vivo data indicate that WHIM altered morphological and functional phenotypes arise from a cell-autonomous defect of the GC lineage, independent from the three-dimensional cytoarchitecture of the developing cerebellum and from any peripheral alteration.
Defective granule lineage development in WHIM mice impacts PC differentiation and functional properties
Proper development of GCs is required for PCs maturation.90 PCs have a characteristic dendritic arbor (Figure 6A) and represent the sole output of the cerebellar cortex.100 In our transcriptional analysis, PCs, as well as other Cxcr4-negative cells, exhibited significant deregulation in gene expression (Figure 4D). Enrichment analysis revealed downregulation of genes associated with dendritic arborization and axonogenesis, as well as synaptic neurotransmission and LTP (Figure 6B), highlighting an impairment of PC development and synaptic maturation. Consistently, we observed a decrease in the overall PC count within the impacted lobules at P30 (Figure 6C). Furthermore, we found a significant reduction in PC dendritic complexity at specific distances from the soma (40–70 μm) (Figures 6D and S6A), despite the total number of intersections is not significantly changed (data not shown). This suggests that Cxcr4 hyperactivation selectively and non-cell autonomously controls the number of PCs and their proximal dendritic branching, potentially disrupting circuit assembly.
Figure 6.
Non-cell-autonomous effects of Cxcr4 mutation
(A) Schematic of cerebellar layers showing PC localization and selected downregulated genes.
(B) Bar plot showing top deregulated biological functions in PCs colored by activation Z score.
(C) Immunofluorescence staining of P30 cerebellum showing Calb1+ PCs (red) in Cxcr4+/1013 and boxplots indicating PC density. Student’s t test, ∗∗p < 0.01; n > 7 animals/condition. Scale bar, 20 μm.
(D) Single PC at P21 of control and WHIM mice. Line plot showing the radius (μm) and number of intersections of PC dendrites and boxplot with total number of intersections. Student’s t test, ∗p < 0.05; n > 7 cells/condition. Scale bar, 20 μm.
(E) Violin plots showing the expression of Scn1b and Hcn2 in PCs, ∗∗p < 0.01.
(F) Optical image of the cerebellar slice superimposed with the activity map. Scale bar, 2 mm.
(G) Representative raster plot (upper) recorded in control cerebellar activity with action potentials recorded in single active electrodes over time (lower).
(H) Bar plot showing quantitative analysis of mean firing rate (MFR) recorded in each lobe. Dots show the number of analyzed slices. Mann-Whitney test, ∗p < 0.05; n > 6 animals/condition.
Transcriptomic analysis revealed increased expression of several ion channel genes, including sodium voltage-gated channel β1 subunit (Scn1b) and hyperpolarization-activated cyclic-nucleotide-gated channel 2 (Hcn2), in WHIM PCs (Figure 6E). To investigate potential functional consequences of the observed aberrant features of WHIM cerebellum, we monitored the spontaneous activity of Cxcr4+/1013 and control cerebellar slices using a high-density multielectrode array (HD-MEA; Figures 6F–6H and S6B–S6E), which in basal conditions records the activity of PCs.101 The overall mean firing rate (MFR) and the number of active units in WHIM slices were not significantly different from controls (Figure S6C), while notable differences in MFR were evident across specific lobules (Figures 6H and S6E). In WHIM cerebella, lobules LIV–V and LVIII, adjacent to the altered fissures, displayed a significant increase of the MFR and, consistently, the amplitude of spike signals was increased in LVIII (Figure S6D).
Overall, these data provide evidence of the close interplay between excitatory and inhibitory neurons during cerebellar development, resulting in an abnormal lobule-specific network activity and likely impairing the overall output of WHIM cerebellar cortex.
WHIM mutants show congenital leukopenia but no altered immune state in the cerebellum
While extensively characterized in adulthood,36,38 the immune phenotype of WHIM mice during development has never been explored. This represents a critical gap, especially given the early onset of immunodeficiency observed in WHIM patients102,103 and our neurodevelopmental findings in Cxcr4+/1013 pups. To address this, we performed a longitudinal immune profiling of WHIM mice from embryonic day 16.5 (E16.5) to P45, using multi-parameter flow cytometry on liver, spleen, blood, and cerebellar tissues (Figure 7A).
Figure 7.
WHIM mutants show congenital leukopenia, but no altered immune state in the cerebellum
(A) Experimental plan.
(B) Bar plots showing immune cell subsets in E16.5 WHIM liver analyzed by fluorescence-activated cell sorting (FACS). Student’s t test, ∗p < 0.05; n > 4 animals/condition. Data are presented as mean ± SEM.
(C) Boxplots showing circulating immune cell subsets in P0 WHIM blood analyzed by FACS. Student’s t test, ∗p < 0.05; n > 4 animals/condition. Data are presented as mean ± SEM.
(D) Line plots showing circulating CD45+, CD19+, and CD3+ cells in Cxcr4+/1013 blood from P0 to P45 analyzed by FACS. Student’s t test, ∗p < 0.05, ∗∗p < 0.01; n > 4 animals/condition.
(E) Plots showing the percentage of microglia cells on CD45+ cells, MFI of CD86 and relative quantification in E16.5 WHIM cerebellum analyzed by FACS. Student’s t test, ∗p < 0.05; n > 4 animals/condition.
(F) Plots showing the immunological profile, MFI of CD86 and relative quantification in P0 WHIM cerebellum analyzed by FACS. Student’s t test, ∗p < 0.05; n > 3 animals/condition. Data are presented as mean ± SEM.
(G) Representative cerebellar sections of Cxcr4+/1013 and control at P0 stained for microglia marker Iba1 and density bar plot. Scale bar, 200 μm. Multiple t test, ∗p < 0.05; n > 3 animals/condition.
(H) 3D reconstruction and relative quantification of Iba1+ microglia cells in WHIM and control cerebella at P21. Scale bar, 10 μm. Multiple t test, ∗p < 0.05; n > 3 animals/condition.
As expected, no differences in the WHIM fetal liver at E16.5 were found in any of the immune populations studied (Figure 7B), fetal liver hematopoiesis takes place independently of the Cxcr4-Cxcl12 axis.104,105,106 Accordingly, no alterations were observed in the number of splenic immune populations at the same stage (Figures S7E and S7F).
Interestingly, as early as P0, animals bearing the WHIM mutation showed a pronounced neutropenia and lymphopenia (Figures 7C, 7D, S7A, and S7B). A widespread decrease in CD45+ cell count was observed in WHIM blood (Figures 7C, 7D, S7A, and S7B), while a significant decrease of CD11b+ immune cells, neutrophils and B cells was found in the spleen of WHIM newborns, along with a general trend toward decrease of the other analyzed cell types (Figures S7E and S7G).
No differences were detected in WHIM cerebellum in any of the immune populations studied (Figures 7E, 7F, and S7C), including resident microglia. Also, both microglial abundance and CD86 mean fluorescence intensity (MFI), a marker of activation, remained unchanged (Figures 7E and 7F). Consistently, Iba1 immunostaining revealed no difference in microglial density between Cxcr4+/1013 and control newborns (Figure 7G), and at P21, microglial morphology—measured as branch and junction counts/cell—was unaffected (Figure 7H). Notably, WHIM mice were maintained under specific pathogen-free (SPF) conditions, never exposed to any immune challenge. Furthermore, expression levels of Il17 and its receptor Il17ra, components of the γδ T cell-mediated interleukin (IL)-17 signaling pathway linked to anxiety-like behavior107 were unaltered in WHIM cerebellum (Figure S7D).
Together, these findings indicate that, under basal conditions, WHIM cerebellum does not exhibit signs of immune dysregulation, supporting a cell-autonomous origin of the observed morphological and behavioral phenotypes.
Early developmental intervention ameliorates WHIM behavioral and structural deficits
AMD3100, a CXCR4 antagonist, is a promising therapy currently under evaluation for the treatment of WHIM syndrome’s immune-related symptoms. However, clinical trials to date have exclusively enrolled patients older than 10 years,44,45,46 despite clinical manifestations of the disorder emerge at birth.102,103
To assess whether early, brain-specific CXCR4 inhibition could rescue WHIM phenotypes, we administered AMD3100 intracerebroventricularly at E12.5 and analyzed behavioral and cerebellar outcomes at P7 and P45 (Figure 8A). Treated Cxcr4+/1013 pups displayed a significant increase in body weight, compared with wild-type controls (Figure 8B), suggesting that AMD3100 impacts key developmental trajectories. In the cliff aversion test, nearly all AMD3100-treated pups exhibited an appropriate avoidance response, contrasting with the poor performance of vehicle-treated mutant littermates (Figure 8C). Morphological assessment revealed significant restoration of cerebellar fissure length at P7 (Figures 8D and S8A), with a partial improvement in fsec length also observed at P45 (Figure S8B). To consider an early post-natal therapeutic intervention with potential clinical relevance, we repeated intraventricular AMD3100 injections at P1 (Figure 8E). Similar to the in utero administration, AMD3100-treated Cxcr4+/1013 pups exhibited a significant increase in body weight by P7, comparable to wild-type levels (Figure 8F), and performed markedly better in the cliff aversion test compared with vehicle-treated mutant littermates (Figure 8G). At P45, treated mice showed increased time spent in the center of the open field and a higher number of vertical counts (Figure 8H), indicating improved exploratory behavior. Morphological analysis at this stage also revealed increased fsec length (Figure S8C), consistent with results from embryonic AMD3100 delivery.
Figure 8.
WHIM behavioral and structural alterations can be rescued at early developmental stages
(A) Experimental plan showing in utero injection of AMD3100 or vehicle.
(B) Boxplot showing the weight of P7 pups upon AMD3100 or vehicle injection at E12.5. Multiple t test, ∗∗p < 0.01; n > 4 animals/condition.
(C) Bar plot showing the percentage of P7 pups treated with AMD3100 or vehicle at E12.5 that turn in the cliff aversion test. Multiple t test, ∗∗p < 0.01; n > 4 animals/condition.
(D) Representative sagittal Nissl sections of WHIM and control cerebella at P7 upon AMD3100 or vehicle injection at E12.5 and bar plots showing fissure length. Scale bar, 200 μm. Multiple t test, ∗∗p < 0.01; n > 3 animals/condition.
(E) Experimental plan of post-natal injection of AMD3100 or vehicle.
(F) Boxplot showing the weight of P7 pups upon AMD3100 or vehicle P1 injection. Multiple t test, ∗p < 0.05; n > 4 animals/condition.
(G) Bar plot showing the percentage of P7 pups treated with AMD3100 or vehicle at P1 that turn in the cliff aversion test. Multiple t test, ∗p < 0.05; n > 4 animals/condition.
(H) Boxplot showing the time P45 mice upon AMD3100 or vehicle injection at P1 in the open field. One-way ANOVA. Boxplot showing the number of vertical counts in the open-field test. Student’s t test, ∗p < 0.05; ∗∗p < 0.01; n > 5 animals/condition.
Together, these findings provide compelling evidence that the structural and behavioral abnormalities observed in WHIM mice arise from intrinsic Cxcr4 dysfunction in the developing brain and can be rescued by early, targeted intervention.
Discussion
Our findings provide new insight into the complex neuroimmune interactions underlying neurobehavioral symptoms in chronic immune conditions. By identifying a developmental enrichment of IEI genes in cerebellar lineages, we propose that intrinsic, brain-specific contributions may underlie neurobehavioral features in IEI patients. Grounded in a comprehensive characterization of the WHIM mouse model—faithfully recapitulating the human genetic and immunological phenotype33,36,38,39—we show that GCPs are key mediators of the cerebellar foliation defects and circuit miswiring observed, even in the absence of immune challenge. Their molecular, morphological, and functional disruption impair PC development and cerebellar output, leading to enduring behavioral phenotypes consistent with those described in patients.
Charting IEI-associated genes in the developing human brain
While behavioral alterations are frequently reported in IEI and other chronic inflammatory conditions, especially anxiety and depression, these symptoms are often dismissed as secondary to systemic disease—particularly in pediatric patients. Yet, the long-term impact of undiagnosed or untreated neuropsychiatric symptoms can be profound. Although prior studies have linked immune signaling to brain function through non-cell-autonomous pathways,2,17,108 direct contributions of IEI genes to neurodevelopmental processes have not been systematically examined.
Here, we provide the first comprehensive analysis of IEI, their genetic architecture and associated clinical features. Our finding that over 40% of IEI genes are associated with neurobehavioral phenotypes may still be underestimated due to limited neurological characterization of patients. By identifying specific developmental windows and cerebellar vulnerability, we lay the groundwork for a deeper understanding of how immune-related genetic variants shape brain structure and function.
WHIM-linked mutation directly controls GC spatiotemporal dynamics
MRI studies in mouse models of immune dysfunction (e.g., IL-6, IL-10, and IL-18 deficient mice) have linked immune dysregulation to structural brain alterations, particularly in the cerebellum and lateral septum.109 Our results extend this work by demonstrating a direct, gene-specific mechanism: hyperactivation of Cxcr4 disrupts GCP behavior, impairing cerebellar foliation (assessed by MRI and histology) and GC circuit assembly. It is reasonable to suggest that the functional GC behavior in embryonic WHIM cerebellum is strongly influenced by their precise spatial positioning, given the dense, yet highly specific input received by their dendrites.110
Our results on the Cxcr4-dependent alterations in fanning pattern of BG processes impinge on the ultimate arrangement of mature GCs in the lobules of WHIM mice, likely affecting cell-cell contact cues and scaffold for their migration along the lobules. Altogether, this supports the existence of a defined prenatal developmental window that governs the foliation onset and progression, then mechanically sustained and affected by the hyperactive Cxcr4 pathway.
The coordinated migration of GCs across the layers is closely linked to their differentiation, establishing a clear relationship between maturation state and spatial positioning—an association that can be effectively captured through single-cell pseudotime/pseudospace analyses. This coupling is not unique to the GC lineage, as other neuronal populations—inhibitory INs of the cerebellum111 and of the cerebral cortex112—also progressively define their identity while migrating their final positions.
Our ex vivo culture experiments revealed that, even in the absence of migratory cues, WHIM GCs display distinct morphological and electrophysiological profiles. Previous studies on isolated GCs have shown that their migration follows an intrinsic program of sequential morphological changes, independent of external cues, ending in a stationary phase characterized by increased dendritic branching.99,113 It is therefore plausible that the observed increased branching in WHIM GCs in vitro stems from a premature migration arrest and anticipated neuritogenesis.
Considering the immunological longitudinal data, it emerges that the observed structural defects in WHIM mice originate from intrinsic alterations in GC migration and differentiation.
WHIM mutation impinges on cerebellar circuits assembly and behavior
The cerebellum is increasingly implicated in affective regulation, with altered cerebellar connectivity linked to anxiety and mood disorders.114,115 At the cellular scale, GCs, the largest population of neurons of the entire brain, encode a wide variety of sensory and motor information, with their activity patterns reflecting sensorimotor context.110 The pervasive alterations in the behavior of WHIM pups support the role of cerebellar GCs in integrating sensory and motor signals. GCs indeed receive inputs from mossy fibers, relaying information to PCs which in turn project to the cerebellar nuclei. This complex architecture eventually connects the cerebellum with the rest of the CNS.110 Here, we show that Cxcr4-linked alterations affect not only GCs, but also PCs in a non-cell-autonomous manner, altering their transcriptional profile and reducing their dendritic extension and arborization, eventually leading to changes in microcircuit activity. The dynamic interplay between excitatory and inhibitory neuronal subtypes has been shown to be crucial in brain development,116,117 but it remains to be elucidated whether a precise molecular code regulates the cell-type-specific and spatial assembly of GC-PC connections in different lobules.
Although early developmental behaviors in WHIM pups are disrupted, some improve by the second post-natal week, suggesting potential for compensatory mechanisms. Still, anxiety-like phenotypes persist into adulthood and can be reversed by CXCR4 antagonism, underscoring the limited plasticity of cerebellar affective circuits and reinforcing the existence of a developmental sensitive window.118 Future studies should explore whether these effects are cerebellum-specific or reflect broader disruptions in long-range connectivity.
Early AMD3100 treatment restores cerebellar foliation with enduring effects on behaviors in WHIM mice
The CXCR4 antagonist AMD3100 has been approved by the international drug agencies for pediatric hematopoietic stem cell mobilization,119,120 but it has only been tested in WHIM patients older than 10 years.44,45 Here, we demonstrate that intraventricular AMD3100 administration at both embryonic and early post-natal stages in mice rescues cerebellar morphology and behavioral deficits. These findings suggest a previously unrecognized potential for CNS-targeted early interventions in CXCR4-related pathologies. While previous work showed that in utero intracerebroventricular injection of AMD3100 can modulate the entrance into the cortical plate of Cxcr4-expressing GABAergic INs in mice,121 to our knowledge, his is the first evidence of AMD3100 acting directly on cerebellar developmental processes.
While our results offer intriguing insight into the developmental role of the Cxcr4-Cxcl12 axis in the cerebellum and support the feasibility of early-stage intervention, further work is needed to assess how such findings translate to human pathology. Thus, large patient cohorts, age-matched controls, and integrated morphometric-behavioral analyses are essential.
Our results support the possibility that brain structures, whose development has been primed by the direct effects of mutations, may become more vulnerable to functional changes when faced with an immunological insult. Further studies will elucidate whether in WHIM syndrome, and even in other IEI that involve neurobehavioral deficits, recurrent immunological challenges trigger an altered neuroimmune crosstalk, eventually exacerbating the existing phenotypes.
Our research overall uncovers a significant role of the CNS development, in particular of the cerebellum, in the neuroimmune interplay and its impact on behavior in IEI. Accurate diagnosis of neurobehavioral conditions in IEI is crucial for comprehensive patient care and would represent a significant shift in the clinical evaluation of these complex disorders.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Simona Lodato (simona.lodato@hunimed.eu).
Materials availability
This study did not generate any new unique reagents.
Data and code availability
-
•
Single-cell gene expression datasets of P1 WHIM and control cerebella have been deposited at GEO and are publicly available as of the date of publication. GEO: GSE266671.
-
•
MRI data of P45 WHIM and control cerebella have been deposited on OpenNeuro and are publicly available as of the date of publication. OpenNeuro: ds005137.
-
•
The BrainSpan Atlas is available at https://www.brainspan.org/.
-
•
The human single-nuclei dataset is available at https://cellxgene.cziscience.com/collections/72d37bc9-76cc-442d-9131-da0e273862db/.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Acknowledgments
We thank A. Fontanesi and D. Uralov for their technical assistance and C. Saulle for help in setting up FACS analysis. We wish to thank Prof. Arenzana-Seisdedos from Pasteur Institute for kindly sharing the Cxcr4+/1013 mouse line with our institution. We thank Dr. D. Kim and GENIE Project for the pAAV-hSyn-GCaMP6s-WPRE plasmid and Prof. K. Deisseroth for the pAAV-hSyn-mCherry plasmid. We are grateful for the technical support provided by the Genomics, Imaging, and Flow Cytometry Humanitas Facilities. We thank all the members of the Lodato laboratory for the fruitful discussion and suggestions. This work was supported by ERC Starting grant IMPACT 101043003, by Cariplo Giovani 2019-1785 and Ricerca Finalizzata 2019 by the Italian Ministry of Health to S.L., by ERC Advanced Grant MATILDA 101055323 and EraNET Neuron JTC2021 InflASD to M. Matteoli, by Cariplo Giovani 2019-1973 to I.C., by Cariplo-Telethon GJC21044A and PRIN-2022 2022JLA3EA to D.P., and by FRAXA 2021 to A.B.
Author contributions
S.L. and G.D. conceived the study, designed the experiments, and analyzed the data. G.D. performed all the experiments and colony maintenance; G.D. and L.Z. performed genotyping, sample collection, and histological staining; S.M. performed single-cell sequencing experiments and analysis of bulk transcriptional data; S.M. and M. Miotto analyzed and interpreted the single-cell/nuclei sequencing data; R.O.P. performed Nissl analyses and set milestone tests; A.B, I.C., and M. Matteoli performed and analyzed behavioral experiments; V.A., M.C., and M.K. performed and analyzed FACS experiments; E.K., K.I., and D.C. performed and analyzed MRI experiments; E.F. and D.P. performed and analyzed MEA recordings; L.P. and R.R. designed and analyzed the foliation model; S.L., G.D., and S.M. wrote the original draft; and all authors revised the manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Calbindin (mouse monoclonal) | Swant | Cat#300; RRID: AB_10000347 |
| Anti-Pax6 (rabbit polyclonal) | Sigma-Aldrich | Cat#AB2237; RRID: AB_1587367 |
| Anti-Iba1 (rabbit polyclonal) | FujiFilm | Cat#019-19741; RRID: AB_839504 |
| Anti-BLBP (rabbit polyclonal) | Merck Millipore | Cat#ABN14; RRID: AB_10000325 |
| Anti-RFP (rabbit polyclonal) | Rockland | Cat#600-401-379; RRID: AB_2209751 |
| Anti-CD45 | Biolegend / BD | Cat#103139/564225; RRID: AB_2716861 |
| Anti-CD19 | BD | Cat#560245/612971; RRID: AB_2870243 |
| Anti-CD3e | Invitrogen | Cat#15-0031-81; RRID: AB_3697163 |
| Anti-Ly-6C | BD | Cat#560596; RRID: AB_1727555 |
| Anti-CCR2 | Biolegend | Cat#150605; RRID: AB_2571913 |
| Anti-CX3CR1 | Biolegend | Cat#149027 RRID: AB_2565967 |
| Anti-CD49b | BD | Cat#740704; RRID: AB_2740388 |
| Anti-CD11b | BD | Cat#563553; RRID: AB_2738276 |
| Anti-CD4 | BD | Cat#612952; RRID: AB_2813886 |
| Anti-CD8 | BD | Cat#612898; RRID: AB_2870186 |
| Anti-Ly-6G | BD | Cat#560601; RRID: AB_1727562 |
| Anti-F4/80 | BD | Cat#567201/565613; RRID: AB_2734770 |
| Anti-CD86 | Biolegend | Cat#105016; RRID: AB_493602 |
| Bacterial and virus strains | ||
| pAAV-hSyn-GCaMP6s-WPRE | Plasmid was a gift from Douglas Kim & GENIE Project; Chen et al.122 | Addgene, Cat#100843 |
| pAAV-hSyn-mCherry | Plasmid was a gift from Karl Deisseroth Lab | Addgene, Cat#114472 |
| Biological samples | ||
| Cerebellar tissue from Cxcr4+/1013 and WT mice | This paper | N/A |
| Liver, spleen, blood from Cxcr4+/1013 and WT mice | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Poly-L-lysine | Sigma-Aldrich | Cat#P2636 |
| Neurobasal medium | Life Technologies | Cat#21103049 |
| Fetal Bovine Serum (FBS) | Life Technologies | Cat#1600004 |
| B-27 Supplement | Life Technologies | Cat#17504001 |
| Penicillin-Streptomycin | Euroclone | Cat#ECB3001 |
| Cytosine arabinofuranoside (AraC) | Sigma-Aldrich | Cat#C1768 |
| Papain Dissociation System | Worthington Biochemical | Cat#LK003150 |
| AMD3100 (Plerixafor) | Sigma-Aldrich | Cat#239825 |
| Eukitt Mounting Medium | Sigma-Aldrich | Cat#3989 |
| Violet stain | Sigma-Aldrich | Cat#C1791 |
| Glacial Acetic Acid | Sigma-Aldrich | Cat#A6283 |
| Fluoromount | Thermo Fisher Scientific | Cat#F4680 |
| LIVE/DEAD Aqua Dead Cell Stain Kit | Thermo Fisher Scientific | Cat#L34957 |
| DAPI | Thermo Fisher Scientific | Cat#D1306 |
| Critical commercial assays | ||
| RNeasy Mini Kit | Qiagen | Cat#74106 |
| QuantiTec Reverse Transcription Kit | Qiagen | Cat#205313 |
| PowerTrack SYBR Green Master Mix | Thermo Fisher Scientific | Cat#A4611 |
| Deposited data | ||
| Single cell gene expression datasets | GEO | GSE266671 |
| MRI data | OpenNeuro | ds005137 |
| Experimental models: Organisms/strains | ||
| Cxcr4+/1013 knock-in mice | Institut Pasteur; Balabanian et al.38 | N/A |
| Wild type C57BL/6J mice | Jackson Laboratory | Cat#000664 |
| Oligonucleotides | ||
| IL17ra primer fw: CCTCATCACACTCATCGCCA | This paper | N/A |
| IL17ra primer rev: GCCGAGTAGACGATCCAGAC | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism (v10.0.0) | GraphPad | www.graphpad.com |
| R (v4.3.1) | R Project | www.r-project.org |
| Seurat (v4.4.0) | Hao et al.123 | https://satijalab.org/seurat/ |
| FIJI (ImageJ) | NIH | https://imagej.net/Fiji |
| FlowJo (v10.10.0) | BD | https://www.flowjo.com/ |
| MATLAB | MathWorks | www.mathworks.com |
Experimental model and study participant details
Animals
All experiments were performed in accordance with the guidelines established by the European Community Council (Directive 2010/63/EU of September 22nd,2010) and the Italian D.Lg. 26/2014. The study was approved by the Institutional Animal Care and Use Committee (IACUC) of the Humanitas Research Hospital and by the Italian Ministry of Health. Mice were housed in a specific pathogen-free (SPF) facility under constant temperature (22 ± 1C) and humidity (50%) conditions with a 12-hour light/dark cycle with ad libitum access to food and water. SPF conditions ensured that animals were never exposed to immunological challenges. WHIM-associated mutant Cxcr4+/1013 knock-in mice were generously provided by Prof. Arenzana-Seisdedos and originally generated at the Institut Pasteur38 and were kept on a C57BL/6J background. Experimental cohorts included littermates from embryonic day 16.5 (E16.5) through postnatal day 45 (P45). No sex-based differences were observed in any of the measured parameters. Results were pooled across sexes in all the experiments with the exception of adult behavioral testing, where only age matched littermate male mice were used.
For timed pregnancies, Cxcr4+/1013 knock-in male was housed overnight with up to three C57BL/6J females (acquired by Charles River laboratories), and successful mating was confirmed by the presence of a vaginal plug the following morning. All animals used in this study were experimentally naive and had not been previously exposed to AMD3100 or other interventions.
Primary granule cell cultures
Granule cell cultures were obtained as previously reported.124,125,126 Each experiment utilized a minimum of 4 postnatal day 5 (P5) pups per genotype from at least two different litters. GCs were plated at approximately at 600,000 cells/mL on Poly-L-lysine-coated (#P2636, Sigma-Aldrich) cover glasses in Neurobasal medium (#21103049, Life Technologies) supplemented with 10% fetal bovine serum (FBS, #1600004, Life Technologies), 0.5% serum-free supplement B-27 (#17504001, Thermo Fisher Scientific), and 100 I.U./mL of Pen-Strep (#ECB3001, Euroclone). Cultures were incubated at 37°C/5% CO2. After 24 hours, the medium was replaced with fresh Neurobasal medium supplemented with 10% FBS, 0.5% serum-free supplement B-27, 100 I.U./mL of Pen-Strep, 10μM cytosine arabinofuranoside to eliminate proliferating non-neuronal cells (e. g., endothelial and microglial cells), and 25mM KCl, which promote GC survival through mild depolarization.
On day in vitro 4 (DIV4), the medium was replaced with fresh Neurobasal medium supplemented with 10% FBS, 0.5% serum-free supplement B-27, 100 I.U./mL of Pen-Strep, 10μM cytosine arabinofuranoside, and 5mM KCl to facilitate functional synapse formation.
Method details
Datasets and analysis of IEI-associated genes in human brain
To explore IEI-associated gene expression in the human brain, we initially assembled a roster of 526 genetic IEI and their corresponding 444 causative genes, utilizing a validated classification system (https://www.niaid.nih.gov/diseases-conditions/types-IEI, source: International Union of Immunological Societies, IUIS).27,28 Subsequently, we leveraged the BrainSpan Developmental Transcriptome65 and single nuclei RNA-seq66 datasets to analyze gene expression patterns.
We leveraged the BrainSpan dataset, comprising normalized expression data (RPKM) summarized to genes across 25 brain structures and 31 ages, which were systematically grouped into 7 distinct region types (PFC, prefrontal cortex; CTX, cerebral cortex; HP, hippocampus; CB, cerebellum; STR, striatum; AMY, amygdala; and TH, thalamus) and 8 developmental periods (early prenatal, late prenatal, late infancy, early childhood, late childhood, adolescence, and adulthood). We subset the expression of our 444 gene of interest (GOI, Table S2) and performed gene clustering using Hierarchical Cluster Analysis, incorporating both region types and developmental periods as key grouping variables. To determine the optimal number of clusters (k), we employed the silhouette score, selecting the value that maximized this metric. To allow for the identification of distinct gene expression patterns, we plotted average scaled gene expression across different regions and developmental stages. To capture dynamic changes in gene expression throughout critical developmental periods in the single-nuclei analysis, we focused on a subset comprising nine out of ten timepoints, excluding the adult stage, of only healthy subjects. The data were normalized by individual timepoint using “SCTransform()”, then dimensionality reduction was performed on the scaled data with “RunHarmony()” to integrate the data for visualization purposes without altering the original annotations. To specifically analyze the expression patterns of our GOIs, we computed the average gene expression across cell types. The results are presented in dot plot in Figure 1G.
Ex-vivo magnetic resonance imaging
Animals were transcardially perfused first with heparinized saline and then with 4% paraformaldehyde (PFA) dissolved in PBS and decapitated. The skin, ears, and lower jaw were removed, and the brain was left within the cranium to minimize tissue damage and deformations due to dissection. Each brain was then incubated for 24 h in 4% PFA solution and then placed in 0.1 M PBS containing 0.05% (weight/vol) sodium azide. Samples were then shipped to King’s College London and stored at 4°C in this solution for 3-4 months to allow tissue rehydration. Sixty days prior to ex vivo MRI acquisition, the samples were transferred to an 8 mM solution of Gadobutrol (Gadovist, Bayer AG) in 0.1 M PBS and 0.05% sodium azide. MRI examiners were blinded to the experimental groups.
The samples were scanned four at a time, secured in a custom 3D-printed holder inside a 50-ml Falcon tube filled with perfluoropolyether (Galden SV80, Apollo Scientific), using a 39-mm quadrature transmit/receive volume coil (RAPID Biomedical) in a 9.4T BioSpec 94/20 (Bruker BioSpin) equipped with a B-GA12SHP gradient set (1000 mT/m) and an Avance™ III HD console controlled by ParaVision 7.0.0. Images with 40-micron isotropic resolution were acquired using a 3D FLASH sequence with the following parameters: echo time = 6 ms, repetition time = 20 ms, flip angle = 33°, readout bandwidth = 100 kHz, averages = 7, matrix size = 625×625×500, field of view = 25×25×20 mm, scan time = 14 h.
Magnitude and phase images were reconstructed in ParaVision 7.0.0 and converted to NIfTI format using BrkRaw. The phase images were unwrapped using the unwrap_path command in Quantitative Imaging Tools (QUIT).127 The magnitude and phase images of all 22 subjects were used to create a multivariate study-specific template using the antsMultivariateTemplateConstruction2.sh script in Advanced Normalization Tools (ANTs).128 All subjects were then registered to the template via sequential rigid-body, affine, and diffeomorphic (SyN algorithm) transforms using antsRegistration.129 Log-transformed Jacobian determinant maps of these composite transforms were generated using the ANTs command CreateJacobianDeterminantImage. For voxel-wise comparison of local volumes of WHIM and WT brains, FSL randomize130 was used to perform permutation tests on the log-Jacobian images (5000 permutations, threshold-free cluster enhancement, and family-wise error rate controlled).
Each individual magnitude image was rigidly aligned to the template using antsRegistration and cropped around the cerebellum. In Fiji, a 3D median filter with a two-voxel (80 μm) radius was applied, then a minimum intensity projection of the central 10 voxels in the sagittal plane was created, and finally unsharp masking was performed with a radius of 3 and mask weight of 0.7. The arbor vitae was semi-automatically segmented from this filtered image using the wand tool. The same procedure was done on the template image, and each subject’s segmented central sagittal arbor vitae was rigidly registered to the template’s using antsRegistration. In python, the skeleton of the arbor vitae was created using scikit-image 0.21.0 (skimage.morphology.skeletonize),131 and the branch lengths of the skeleton were measured using Skan 0.11.1 (skan.summarize).132 Branches were automatically assigned to a cerebellar lobule based on junction/endpoint coordinates, which are also returned by skan.summarize.
Behavioral testing in adult mice
Behavioral testing was performed during the light phase, P45 mice were habituated to the testing room for 30 min before starting. Tests were conducted on the same day for the two genotypes in randomized order and data were recorded and analyzed blind to genotype.
For the open field test
Mice activity was recorded by video tracking in a 30 min-open field session. The area was virtually divided into periphery and center. For each of these regions, the time spent within (s), the distance covered (cm), the number of vertical counts (free-standing animal or touching the side of the field), the vertical, ambulatory and resting times (s) and the ambulatory rate (cm/s) were analyzed.
For the accelerating rotarod test
We used an accelerating rotarod (Panlab, #76-0770). Mice were tested starting from P45. They were positioned on the rotating bar and allowed to become acquainted with the environment. The rod motor was started initially at 4 rpm and accelerated by one rpm every 8s to a maximum of 40 rpm for 300 s. Animals were given 5 trials per day for 4 days. The performance was scored as latency to fall in seconds, and the average was used for statistical analysis.
For the dark-light chamber test
Mice were placed in the center of the light chamber and recorded while allowed to explore the two chambers for 10 min. The following behavioral performances were analyzed: time spent in the light chamber (s), time spent in the dark chamber (s), latency (s), and number of transitions between chambers.
For the forced swim test
Animals were placed in an open water container, with water temperature maintained at 23° ± 2°C; after two minutes of adaptation period, the time of activity and immobility (s) were measured in a 4 minutes session.
For the marble burying test
Mice were tested in a regular clean cage with twelve glass marbles positioned under the bedding and equidistant in a 3 x 4 arrangement, covering the whole cage area. The subjects were placed in the test cage and allowed to explore and bury the marbles for 10 min. At the end of the session, the number of marbles buried (>50% marble covered by bedding material) was considered.
For the self-grooming test
Animals were placed in an empty, clean arena with a thin layer of bedding and allowed to freely move for 10 min. The time each mouse spent self-grooming during the session was measured.
Testing of developmental milestones
Developmental milestones were assessed following established protocols,133,134 and adapted from the Fox scale.85 Pups of independent litters were handled daily at a consistent time to evaluate their physical development. All assessments were conducted and scored blindly to genotype. Parameters were categorized into weight, motor skills and sensory reflexes, and were evaluated as described below. Body weight (g) was measured at P0, P7, P15, P20, P30 and P45. Other developmental milestones were tested between P7 and P15.
Motor skills
Motor activity was assessed using ambulation, negative geotaxis, surface righting, air righting and traversal open field tests. In the ambulation test, pups were observed in a clear enclosure for 3 min, and movement was scored on a scale from 0 to 3. The negative geotaxis test measured the time spent to turn 180° on an inclined plane in 30-second trials. Surface righting assessed the time for pups placed on their back to turn to a prone position within 30 seconds. Air righting measured the ability of pups dropped form a height of 15 cm to land on all paws. In the open field traversal test, we measured the time taken by a mouse to move out of a 13 cm diameter circle on a paper sheet.
Sensory reflexes
Developing reflexes were evaluated through cliff aversion, forelimb grasping, ear twitch and auditory startle tests. In the cliff aversion test, pups were placed at the edge of a box, and the latency time to turn and crawl away was measured in 3 trials of 30 seconds each. The test was considered failed if the pup did not turn away from the cliff in the given time.
Ear twitch reflex development in response to ears stroking with a cotton swab was recorded. For the forelimb grasping pups were released with forepaws resting on a suspended stick, and the duration of grasping was measured. The auditory startle test recorded the involuntary jump response to a recorder hand clap sound.
Nissl staining
Tissues were fixed with 4% paraformaldehyde (PFA) in 1x PBS and 50μm sections were obtained with a Leica VT1000S vibratome, 15μm sections with Histo-Line MC4000 Cryostat.
Nissl staining was prepared one day prior to use with 1% Violet (Sigma-Aldrich, #C1791), 1% Glacial Acetic Acid (#A6283, Sigma-Aldrich) in bidistilled water, and filtered. Sagittal cerebellar sections at different interaural levels (vermis: from 0.00 to 0.24; hemisphere: from 1.56 to 2.04, from 2.16 to 2.52) were selected and dried, hydrated in bidistilled water, stained with Nissl staining for 10’, washed in bidistilled water, dehydrated in increasing ethanol gradient, cleared in xylene and mounted with Eukitt Quick-hardening mounting medium (#03989, Sigma-Aldrich).
Real-time quantitative PCR
Cerebella from P45 mice were lysed in TRIzol (# 15596018, Thermo Fisher Scientific), then total RNA was extracted using manufacturing instructions. Purified RNA was quantified spectrophotometrically using Nanodrop (Thermo Fisher Scientific) and 500 ng was reverse-transcribed into cDNA using the QuantiTec Reverse transcription Kit (#205313, Qiagen) following manufacturer’s specifications. RT-qPCRs were performed using PowerTrack™ SYBR Green Master Mix (#A4611, ThermoFisher Scientific) with GAPDH as an internal control. Cycling was executed using the Viia7 Real-Time PCR System (ThermoFisher Scientific) and analysis of relative gene expression was performed using the 2−yste method (Livak and Schmittgen, 2001135). Three technical replicates were averaged for their cycling thresholds for all the independent biological replicates and further calculations were performed with those means. Each RNA sample was analyzed for IL17ra using the following primers: I17ra: fw = CCTCATCACACTCATCGCCA, rev = GCCGAGTAGACGATCCAGAC.
Foliation analysis
A morphometric analysis on immunofluorescence images of sagittal sections of the cerebellum at E17.5 and P0 was performed. This analysis entailed measuring the precise contour of the sections to obtain the EGL length, along with the convex length representing the EGL’s smooth length or EGL0. Additionally, we determined the total area defined by the EGL boundary, as well as the areas enclosed by the external and internal smooth boundaries of the cerebellar sections. These parameters facilitated the assessment of mechanical strain exerted on the cerebellum’s bilayer structure
and the amplitude of resulting wrinkles. Specifically, we examined the relationship between strain () and wrinkle amplitude () arising upon induction of a sinusoidal wrinkling instability in a soft bilayer material. The relationship between amplitude and strain of folding patterns emerging under low mechanical deformation (i.e., at E17.5) were modelled using the following equation (https://doi.org/10.1039/C9SM02124H):
where is the thickness of the EGL layer and the critical strain that must be exceeded to trigger the instability. The critical strain depends on the ratio of the stiffness of the two layers through the relation
where and are the in-plane elastic modulus of the external layer and the in-plane elastic modulus of the core, respectively.
Patterns emerging beyond the low deformation limit (), and therefore in the so-called ‘high deformation (HD)’ regime (i.e., at P0), were modelled using the following equation (https://doi.org/10.1039/C9SM02124H):
where accounts for the non-linearity in the stress-strain response (neo-Hookean behavior). The high-deformation model also better represents the evolution of the wrinkling pattern into a fold-like instability, typical of developed cerebellar folds, diverging from the purely sinusoidal motif described by the low deformation model better representing the onset and initial foliation stages.
Single-cell RNA sequencing and analysis
Sample preparation
P0 cerebellar region around fpc (LIII and LIV-V) from control and Cxcr4+/1013 pups (n = 8 embryos per sample per litter) was rapidly dissected in ice cold HBSS solution under microscopic control in sterile conditions. To minimize the sex effect, a balanced pool of males and females in both control and Cxcr4+/1013 samples was employed. Dissected tissue was dissociated using Papain Dissociation System kit (#LK003150, Worthington) following the manufactured instructions. Around 10,000 cells per sample were loaded in the Single Cell Chip G using the Single Cell 30 v2.1 single cell reagent kit (10X Genomics) for Gel Bead Emulsion generation into the Chromium system. Following capture and lysis, cDNA was synthesized and amplified for 14 cycles following the manufacturer’s protocol (10X Genomics). 50 ng/sample of the amplified cDNA were then used to construct Illumina sequencing libraries. Sequencing was performed on the NovaSeq 6000 Illumina sequencing platform following 10X Genomics instruction for reads generation.
Single-cell RNA sequencing analysis
Raw sequencing data were converted to fastq files using Illumina bcl-convert tool. CellRanger (10X Genomics, version 6.1.1.113) count module was used to map and count reads with default settings and with sequence length set to r1-length = 28 and r2-length = 90, using pre-build mouse genome (version mm10-1.2.0) as reference. A mean of 45,000 reads/cell were produced for each sample. The raw digital gene expression matrix (UMI counts per gene/cell) was imported in R (version 4.3.1) using Seurat R123 package (version 4.4.0). Briefly, sample quality control was assessed by filtering out, for each sample, cells meeting any of the following criteria: log10(MAD) (Median Absolute Deviation) was calculated for both expressed genes and UMI counts and cells outside 2.5 MAD (upper and lower limit) from median were filtered out, as well as cells with more than 10.0% of reads mapping to mitochondrial genes or less than 5.0% or reads mapping to ribosomal genes. After computing MAD, gene filtering was applied by removing genes expressed in less than median - 2.5 MADs cells, as well as genes expressed by Y chromosome, and XIST and TSIX, to remove sex-related gene expression, and ribosomal genes.
For each cell, feature counts are divided by the total counts for that cell and multiplied by 10,000, and this is then natural-log transformed using log1p. This was followed by assignment of cell cycle scores to individual cells based on the expression of G2/M and S phase markers.
To integrate the data, SCT v2 pipeline from Seurat was used: samples were normalized individually using “SCTransform (vst.flavor = "v2", assay = "RNA")”, then integrating features were selected through “SelectIntegrationFeatures” function, followed by “PrepSCTIntegration”, “FindIntegrationAnchors” and “IntegrateData(normalization.method = "SCT")”.
We then performed principal component analysis (PCA) on the scaled data, and used a graph-based method (elbow plot) to choose the number of principal components to use. We then clustered the cells using the Louvain algorithm (within Seurat) to iteratively group cells together. Optimal resolution was chosen by analyzing the cluster tree plot (clustree). To annotate clusters, we determined differentially expressed genes using FindAllMarkers function embedded in Seurat (Wilcoxon Rank Sum test with Bonferroni correction for multiple testing; adjusted P< 0.05). We tested only genes that were detected in a minimum of 40% of cells within the cluster and that showed, on average, at least a 0.25-fold difference (log-scale) between the cells in the cluster and all remaining ones. By reviewing the resulting markers, as well as the expression of canonical marker genes and additional markers from the literature,66,136 we assigned a cell-type identity.
Cell-type-specific differential gene expression analysis
To determine differentially expressed genes, we used FindMarkers from Seurat function implemented with default two-part hurdle model (MAST) testing only genes found in at least 20% of cells within the cluster and that showed, on average, at least a 0.10-fold difference (log-scale) across genotypes. The list of differentially expressed genes was analyzed using the Ingenuity Pathway Analysis (IPA - Qiagen Ingenuity Systems) canonical pathways and biological functions enrichment.
Trajectory inference and pseudotime scoring
For trajectory inference and pseudotime scoring, we first subsetted the Seurat object to retain only GCPs and GCs. Dimensionality reduction was performed using UMAP on this subsetted object to visualize the high-dimensional data in a reduced space. To determine the root of the trajectory, we used an entropy-based method, which calculates entropy for each cell using the “perCellEntropy()” function from TSCAN package 1.40.0. To infer the trajectory and assign pseudotime scores, we sed the Slingshot 2.10.0 algorithm. The GCP-B cluster was selected as the starting point ("root"), and pseudotime scoring was performed based on UMAP-reduced dimensions. The slingshot() function was used to fit the trajectory, and “slingCurves()” was applied to obtain the lineage curve data. To assess the significance of differences in pseudotime distributions between genotypes, we performed a Kolmogorov-Smirnov (KS) test using the “ks.test()” function. This test compared the density distributions of pseudotime scores between the wildtype and Cxcr4+/1013 mutant genotypes.
Differential expression analysis along pseudotime
We applied the “fitGAM()” function from Tradeseq packages 1.16.0 to model gene expression changes along pseudotime and across different genotypes, fitting the model using 6 knots for each condition. Differential expression analysis across pseudotime was performed using the “associationTest()” function to evaluate genes with significant changes in expression along the trajectory. To assess the genes that passed the false discovery rate (FDR) threshold (FDR ≤ 0.05) for each condition, we applied the “p.adjust()” function to correct p-values. The final gene lists for both genotypes were generated by filtering for those that met the FDR threshold.
Multielectrode array analysis
Ex vivo acute cerebellar slices preparation
Animals at P21 were deeply anesthetized with isoflurane and decapitated. Brains were dissected immediately and placed in ice-cold solution with the following composition (mM): 87 NaCl, 21 NaHCO3, 1.25 NaH2PO4, 7 MgCl2, 0.5 CaCl2, 2.5 KCl, 25 D-glucose, and 7 sucrose, equilibrated with 95% O2 and 5% CO2 (pH 7.4). Parasagittal slices of the cerebellar vermis (300 μm) were cut using vibratome (Leica VT1000S) and then incubated at RT for at least 1 h at 33°C in artificial cerebrospinal fluid (ACSF) with the following composition (mM): 129 NaCl, 21 NaHCO3, 1.25 NaH2PO4, 1.8 MgSO4, 1.6 CaCl2, 3 KCl, 10 D-glucose, equilibrated with 95% O2 and 5% CO2 (pH 7.4). During the recordings, slices were perfused with ACSF solution at 3.0 mL/min using a peristaltic pump (Shenchen).
MEA electrophysiological recordings
Electrophysiological recordings were performed using 2D high-density multi-electrode arrays (HD-MEA, 3Brain) made of 4096 electrodes distributed with 81 μm pitch in 5.1 x 5.1 mm2 total recording area (HD-Stimulo). Slices were positioned on the chip using a platinum anchor with wires of 100 μm width. After 10 minutes of equilibration, spontaneous activity cerebellar slices were recorded for 5 minutes at a sampling rate of 20kHz in standard ACSF solution, using an 50Hz high-pass filter. At the end of each recording session, a high-definition (HD) image of the slice was taken using HD digital microscope (Bysameyee) and subsequently superimposed to the activity map ensuring a precise electrical-anatomical correlation for each lobe.
MEA data analysis
Recordings, spike detection and spike sorting were performed using BrainWave 4 software from 3Brain. To evaluate spike detection, we used the Hard Treshold algorithm set at -70μV, (refractory period = 1ms, AI-validation) while spike sorting on individual electrodes was computed using a Principal Component Analysis (PCA) with 3 features and a standard k-means clustering algorithm. Each recording session included both control and Cxcr4+/1013 genotypes. Only recordings with a spike rate higher than 0.8 spike/s and more than 10 active units per lobes were considered for the analysis. If none of the slices recorded from one genotype met the two criteria, the entire experiment was not considered. Median values of mean firing rates (MFR) and amplitude have been measured for each lobe. Additionally, Kernel Density Estimation (KDE) has been used to visualize and analyze the data distribution. Data analysis was conducted using a non-parametric variant of the t test (Mann-Whitney test) to evaluate both frequency and amplitude metrics within the same cerebellar lobe for control and Cxcr4+/1013 slices.
Liver-, spleen-, blood-, and cerebellum-derived collection of immune cells
Embryos at E16.5 were dissected and livers, spleens and cerebella were collected and smashed on a 40 μm cell strainer. Smashed tissues were left in lysis buffer 1x (8.29g/L ammonium chloride #A9434, 1g/L potassium bicarbonate #60339, 0,0372g/L ethylenediaminetetraacetic acid disodium salt dihydrate #ED2SS) for 10 minutes and washed, then cells were stained with vitality markers and directly conjugated primary antibodies. Cells were fixed with 4% PFA and samples were acquired at the BD LSR Fortessa or FACSymphony Cell Analyzer.
Pups at P0 were dissected and spleens and cerebella were collected and smashed on a 40 μm cell strainer. Smashed tissues were left in lysis buffer 1x for 10 minutes and washed, then cells were stained with vitality markers and directly conjugated primary antibodies. Cells were fixed with 4% PFA and samples were acquired at the BD LSR Fortessa or FACSymphony Cell Analyzer.
Blood was collected at P0 and P7 by decapitation. At P45, blood was collected from the lateral tail vein: following collection site cleaning with 70% alcohol, the mouse was restrained, the lateral tail vein was gently nicked and sample was obtained. Upon collecting 80-100 μL, bleeding was stopped by applying pressure with sterile gauze to achieve hemostasis. Red blood cells present in the sample were removed with lysis buffer 1x and stained with vitality markers and directly conjugated primary antibodies. Cells were fixed with 4% paraformaldehyde, counting beads were added to the samples, and the samples were acquired at the BD LSR Fortessa or FACSymphony Cell Analyzer. The vitality marker used was LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (#L34957, ThermoFisher Scientific) and the primary antibodies used were: anti-CD45 (#103139, Biolegend), anti-CD45 (#564225, BD), anti-CD19 (#560245, BD), anti-CD19 (#612971, BD), anti-CD3e (#15-0031-81, eBioscience), anti-Ly-6C (#560596, BD), anti-CCR2 (#150605, Biolegend), anti-CX3CR1 (#149027, Biolegend), anti-CD49b (#740704, BD), anti-CD11b (#563553, BD), anti-CD4 (#612952, BD), anti-CD8 (#612898, BD), anti-Ly-6G (#560601, BD), anti-F4/80 (#567201, BD), anti-F4/80 (#565613, BD), CD86 (#105016, Biolegend). The data were analyzed via FlowJo (version 10.10.0).
Cerebellar granule cells culture
Granule cell cultures were obtained as previously reported.124,125,126 Briefly, postnatal day 5 (P5) pups were anesthetized on ice and subsequently sacrificed. Each experiment utilized a minimum of 4 pups per genotype from at least two different litters. The brains were rapidly removed from the skull, and the meninges were carefully peeled off the cerebellum. The cerebellar cortex was then dissected in Hank’s Balance Salt Solution (HBSS) under sterile conditions with stereomicroscopic guidance. Tissue dissociation was performed using the Papain Dissociation System kit according to the manufacturer’s instructions (#LK003150, Worthington Biochemical). The resulted single cell suspension was subsequently filtered through a 70 μm cell strainer to remove large non-neuronal cells, ensuring a uniform neuronal preparation. Cerebellar cells were plated at approximately at 600,000 cells/mL on Poly-L-lysine-coated (#P2636, Sigma-Aldrich) cover glasses in Neurobasal medium (#21103049, Life Technologies) supplemented with 10% fetal bovine serum (FBS, #1600004, Life Technologies), 0.5% serum-free supplement B-27 (#17504001, Thermo Fisher Scientific), and 100 I.U./mL of Pen-Strep (#ECB3001, Euroclone). Cultures were incubated at 37°C/5% CO2. After 24 hours, the medium was replaced with fresh Neurobasal medium supplemented with 10% FBS, 0.5% serum-free supplement B-27, 100 I.U./mL of Pen-Strep, 10μM cytosine arabinofuranoside to eliminate proliferating non-neuronal cells (e. g., endothelial and microglial cells), and 25mM KCl, which promote GC survival through mild depolarization.
To visualize spontaneous GCs activity, pAAV-hSyn-GCaMP6s-WPRE (1μL/mL, titer 2.2 x 1013 vg/mL) was added to the medium. pAAV-hSyn-GCaMP6s-WPRE was a gift from Douglas Kim & GENIE Project (#100843, Addgene viral prep).122 Additionally, pAAV-hSyn-mCherry (1μL/ml, titer 1012 vg/mL) was included to enable sparse neuronal labelling for morphological analysis. pAAV-hSyn-mCherry was a gift from Karl Deisseroth (#114472, Addgene viral prep).
On day in vitro 4 (DIV4), the medium was replaced with fresh Neurobasal medium supplemented with 10% FBS, 0.5% serum-free supplement B-27, 100 I.U./mL of Pen-Strep, 10μM cytosine arabinofuranoside, and 5mM KCl to facilitate functional synapse formation. Calcium imaging and morphological analysis were performed on DIV7 neurons.
Granule cell culture calcium imaging recordings and analysis
Calcium (Ca2+) imaging of DIV7 GCs expressing GCaMP6s was performed at the Thunder Imaging System (Leica) at 20x magnification. Live-cell imaging culture was performed in a recording chamber maintained at 37°C/5% CO2 in artificial cerebrospinal fluid (aCSF containing in mM: 125 NaCl; 5 KCl; 1,2 KH2PO4; 25 HEPES; 6 Glucose; 2 CaCl2; pH 7,4). Recordings were acquired over 5 minutes, with imaging sampling rate of 300 milliseconds per frame. For each slide, 3-4 view field of view were randomly selected for imaging and analysis. Time-lapse image files were analyzed using a MATLAB (MathWorks) script adapted from Sun and Südhof.137 The analysis was conducted using a custom function (GCAMP_multiROI.m) designed to extract fluorescence intensity from selected regions of interest (ROIs) and quantify calcium activity. To determine ROIs, a median intensity projection of frames 10 to 40 from the first 50 frames was generated for data preview. ROIs were manually selected based on fluorescence intensity using the impixel function. Circular ROIs were created, and the mean intensity within each ROI was extracted across all frames. The raw intensity traces were then plotted over time to visualize fluorescence fluctuations. Calcium spikes were detected using a threshold-based method, where the threshold was set as the mean plus 1.5 times the standard deviation of the background signal. The spike_detection function identified spikes by detecting local maxima above this threshold. Synchronous calcium peaks were defined as peaks occurring across multiple ROIs within a specified time window. For the analysis of synchronous activity, the synchronous firing rate was calculated as the number of synchronous peaks per minute. The amplitude of synchronous peaks was determined by computing the mean ΔF/F₀ across neurons. Baseline fluorescence (F₀) was estimated as the mean of the three lowest intensity values over various time windows. The relative fluorescence change (ΔF/F₀) was computed using the formula (F - F₀) / F₀ for each detected peak, and the coefficient of variation (CV) of ΔF/F₀ was determined.
For visualization purposes, synchronous peaks were plotted over time using sparse matrices, providing a clear representation of synchronized activity. The distribution of spike amplitudes and frequencies was analyzed across ROIs, and ΔF/F₀ traces were overlaid to compare individual cell responses with the population mean.
Immunostaining on cell culture
DIV7 GCs were fixed with 4% PFA/10% sucrose in 1x PBS. Cells were blocked and permeabilized in 10% goat serum, 0.3% Triton X-100 in 1x PBS for 1h at room temperature (RT). Next, cells were incubated with primary antibodies overnight at 4°C. After washing in 1x PBS, cells were incubated with secondary antibodies (1:750, Molecular Probes Alexa Fluor, Life Technologies) at RT for 2h, washed with1x PBS, counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, 1:5000 in 1x PBS) for 10’ and mounted with Fluoromount aqueous mounting medium (# F4680, ThermoFisher Scientific). The primary antibodies used were mouse anti-RFP (1:500, #600-401-379, Rockland).
Immunostaining on sections
Animals were transcardially perfused with 4% PFA in PBS and decapitated. Brains were fixed with 4% PFA in 1x PBS and fluorescence immunostaining was performed on 50 μm sections obtained with a Leica VT1000S vibratome or on 15 μm sections obtained with Histo-Line MC4000 Cryostat.
Sagittal cerebellar sections were blocked and permeabilized in 10% goat serum, 0.3% Triton X-100 in 1x PBS for 1h at room temperature (RT), then incubated with primary antibodies overnight at 4°C. After washing in 1x PBS, sections were incubated with secondary antibodies (1:750, Molecular Probes Alexa Fluor, Life Technologies) at RT for 2h, washed and counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, 1:5000 in 1x PBS) for 10’ and mounted with Fluoromount aqueous mounting medium (# F4680, ThermoFisher Scientific).
The primary antibodies used were mouse anti-Calbindin (1:1000, #300, Swant), rabbit anti-Pax6 (1:500, #AB2237, Sigma-Aldrich), rabbit anti-Iba1 (1:2000, #019-19741, Fisher Scientific), rabbit anti-BLBP (1:200, #ABN14, Merck Millipore), anti-RFP (1:500, #600-401-379, Rockland).
AAV injection in neonatal mouse cerebellum
Injection of AAV into the cerebellum of neonatal WHIM and control mice was performed following established protocols.138,139,140,141 P0 pups were anesthetized by hypothermia and positioned on a stage. The gauge of a microsyringe (Hamilton Company, #87930) was inserted 0.5 mm into the molecular layer of the cerebellar vermis and 1 μl of AAV-hSyn-mCherry (Addgene, #114472) at titer of 1012 vg/ml was injected over 60 s. Pups were revived at 37°C and returned to the litter. Subsequently, the brain of injected pups was collected by intracardial perfusion at P21. Fluorescence immunostaining was performed on 100 μm cerebellar sections.
Intraventricular injection of AMD3100
AMD3100 (currently used in clinical trial for WHIM patients: ClinicalTrials.gov: NCT02231879; NCT00967785; NCT01058993) or saline solution was injected into the fourth ventricle of WHIM and control embryos (E12.5) or pups (P1).
Specifically, 1 μL of AMD3100 (1mg/ml, Sigma-Aldrich, #239825) was mixed with 0.005% fast green in sterile saline solution and administered to WHIM and control embryos at E12.5.
P1 pups were anesthetized by hypothermia. 2 μL of AMD3100 (5mg/ml) mixed with 0.005% fast green in sterile saline solution was administered to WHIM and control pups. Pups were revived at 37°C and returned to the litter.
The injected embryos underwent behavioral analysis at P7, followed by collection for perfusion and histological characterization at P7 or P45.
Imaging
Fluorescent images were acquired at the Axioscan 7 Slide Scanner (Zeiss) at 10x and 20x magnifications or at the Thunder Imaging System (Leica) at 40x magnification. Confocal images were acquired at the SP8 confocal microscope (Leica) at 40x or 63x magnifications. Brightfield images were acquired at the Axioscan 7 Slide Scanner (Zeiss) at 20x magnification or at the DotSlide microscope (Olympus) at 20x magnification.
Image analysis
PCL length measurement
PCL length was assessed in sagittal cerebellar sections at different interaural levels comprising the vermis and the hemispheres stained with Nissl staining on P45 brain sagittal sections using QuPath (version 0.4.4). Lobules were delineated at the initiation of the fissure, and a line was drawn along the PCL of each lobule using QuPath. All experiments were conducted on a minimum of 3 biological replicates, considering at least 5 sections per animal at different interaural levels.
Fissure length measurement
Fissure length was measured on P0, P7 and P45 Nissl-stained sagittal cerebellar sections at different interaural levels comprising the vermis and the hemispheres using QuPath. Starting from the conjunction of adjacent lobules on the surface on the sections, a line was traced along each fissure. All experiments were conducted on a minimum of 3 biological replicates, considering at least 5 sections per animal at different interaural levels.
Cell counting
Cell counts were semi-automatically performed using Positive Cell Detection function in QuPath. In brief, regions of interest (ROIs) were manually defined, and intensity thresholds were manually adjusted to segment each marker accurately. Only marker-positive cells co-stained with DAPI were considered to ensure their identity as cells. The resulting counts were then normalized to the ROI area to obtain cell density. All experiments were conducted on a minimum of three biological replicates, considering at least 6 sections per animal. Regions of interest were manually defined and a threshold was set in order to delimitate positive from negative cells. The same threshold was maintained throughout the whole experiment analysis.
Microglia morphological analysis
Sagittal cerebellar sections of P21 animals at the vermis level were stained with anti-Iba1 antibody. Individual cells positive for the microglial marker Iba1 were binarized and segmented using Fiji (v2.14). The resulting images were skeletonized using the Skeletonize Plugin for Fiji142 and processed with the Analyze Skeleton function. The number of branches and junctions/cell were obtained from the Results tables.
Bergmann glia fibers analysis
Sagittal cerebellar sections of P7 pups at the vermis level were stained with anti-BLBP antibody. The number of processes present in a length of 100 μm of the molecular layer was counted with the Multi-point tool in Fiji. The angle between the line of the fissure and the closest BLBP+ fiber was calculated with the Angle tool in Fiji. All experiments were conducted on a minimum of three biological replicates, considering at least 6 sections per animal.
Purkinje cell morphology analysis
Sagittal cerebellar sections of P21 animals at the vermis level were stained with anti-RFP antibody, in order to enhance the signal of AAV-hSyn-mCherry. Individual cells positive for the marker Calb1 and RFP were selected and a mask was created using the Threshold function in Fiji. The resulting images were analyzed using the Neuroanatomy Plugin for Fiji, using the Sholl function. The number of branches/cell and the dendritic extension were obtained from the Results tables.
Granule cell morphological analysis
At DIV7, individual GCs expressing mCherry were selected for analysis using the Fiji plugin NeuronJ (8-bit images). Each selected neuron was semi-automatically traced to quantify neurite morphology, including total neurite length and the number of neurites/cell.
Quantification and statistical analysis
Data collection and data analysis were performed in a blinded fashion. All statistical tests are indicated where used. Statistical tests through the manuscript were performed using Prism 8 (GraphPad Prism version 10.0.0 for Windows, GraphPad Software, Boston, Massachusetts USA, www.graphpad.com) and R (version 4.3.1). After removing outliers and testing for normality (Shapiro–Wilk test), intergroup differences were analyzed by unpaired, two-tailed Student’s t-test or Mann–Whitney U-test for single comparison, or by one-way or two-way analysis of variance (ANOVA) with Bonferroni’s multiple-comparison test, as indicated in figure legends. Statistical significance is indicated as ∗p < 0.05, ∗∗p < 0.01, and n.s. > 0.05, as indicated in figure legends. All data are shown as mean ± standard deviation (SD) unless stated otherwise. The exact value of n, as well as what n represents (e.g., number of animals, number of cells, etc.), is specified in figure legends.
Published: June 6, 2025
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.neuron.2025.05.016.
Supplemental information
References
- 1.Ben-Shaanan T.L., Azulay-Debby H., Dubovik T., Starosvetsky E., Korin B., Schiller M., Green N.L., Admon Y., Hakim F., Shen-Orr S.S., et al. Activation of the reward system boosts innate and adaptive immunity. Nat. Med. 2016;22:940–944. doi: 10.1038/nm.4133. [DOI] [PubMed] [Google Scholar]
- 2.Haykin H., Rolls A. The neuroimmune response during stress: A physiological perspective. Immunity. 2021;54:1933–1947. doi: 10.1016/j.immuni.2021.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlov V.A., Chavan S.S., Tracey K.J. Molecular and Functional Neuroscience in Immunity. Annu. Rev. Immunol. 2018;36:783–812. doi: 10.1146/annurev-immunol-042617-053158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Haddad B.J.S., Jacobsson B., Chabra S., Modzelewska D., Olson E.M., Bernier R., Enquobahrie D.A., Hagberg H., Östling S., Rajagopal L., et al. Long-term Risk of Neuropsychiatric Disease After Exposure to Infection In Utero. JAMA Psychiatry. 2019;76:594–602. doi: 10.1001/jamapsychiatry.2019.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi G.B., Yim Y.S., Wong H., Kim S., Kim H., Kim S.V., Hoeffer C.A., Littman D.R., Huh J.R. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khandaker G.M., Zimbron J., Lewis G., Jones P.B. Prenatal maternal infection, neurodevelopment and adult schizophrenia: a systematic review of population-based studies. Psychol. Med. 2013;43:239–257. doi: 10.1017/S0033291712000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S., Kim H., Yim Y.S., Ha S., Atarashi K., Tan T.G., Longman R.S., Honda K., Littman D.R., Choi G.B., et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017;549:528–532. doi: 10.1038/nature23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirabella F., Desiato G., Mancinelli S., Fossati G., Rasile M., Morini R., Markicevic M., Grimm C., Amegandjin C., Termanini A., et al. Prenatal interleukin 6 elevation increases glutamatergic synapse density and disrupts hippocampal connectivity in offspring. Immunity. 2021;54:2611–2631.e8. doi: 10.1016/j.immuni.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed M.D., Yim Y.S., Wimmer R.D., Kim H., Ryu C., Welch G.M., Andina M., King H.O., Waisman A., Halassa M.M., et al. IL-17a promotes sociability in mouse models of neurodevelopmental disorders. Nature. 2020;577:249–253. doi: 10.1038/s41586-019-1843-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraguas D., Díaz-Caneja C.M., Ayora M., Hernández-Álvarez F., Rodríguez-Quiroga A., Recio S., Leza J.C., Arango C. Oxidative Stress and Inflammation in First-Episode Psychosis: A Systematic Review and Meta-analysis. Schizophr. Bull. 2019;45:742–751. doi: 10.1093/schbul/sby125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iakunchykova O., Leonardsen E.H., Wang Y. Genetic evidence for causal effects of immune dysfunction in psychiatric disorders: where are we? Transl. Psychiatry. 2024;14:63. doi: 10.1038/s41398-024-02778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berriat F., Lobsiger C.S., Boillée S. The contribution of the peripheral immune system to neurodegeneration. Nat. Neurosci. 2023;26:942–954. doi: 10.1038/s41593-023-01323-6. [DOI] [PubMed] [Google Scholar]
- 13.Michaud M., Balardy L., Moulis G., Gaudin C., Peyrot C., Vellas B., Cesari M., Nourhashemi F. Proinflammatory cytokines, aging, and age-related diseases. J. Am. Med. Dir. Assoc. 2013;14:877–882. doi: 10.1016/j.jamda.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Mildner A., Schlevogt B., Kierdorf K., Böttcher C., Erny D., Kummer M.P., Quinn M., Brück W., Bechmann I., Heneka M.T., et al. Distinct and non-redundant roles of microglia and myeloid subsets in mouse models of Alzheimer’s disease. J. Neurosci. 2011;31:11159–11171. doi: 10.1523/JNEUROSCI.6209-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peter I., Dubinsky M., Bressman S., Park A., Lu C., Chen N., Wang A. Anti-Tumor Necrosis Factor Therapy and Incidence of Parkinson Disease Among Patients With Inflammatory Bowel Disease. JAMA Neurol. 2018;75:939–946. doi: 10.1001/jamaneurol.2018.0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bannaga A.S., Selinger C.P. Inflammatory bowel disease and anxiety: links, risks, and challenges faced. Clin. Exp. Gastroenterol. 2015;8:111–117. doi: 10.2147/CEG.S57982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carloni S., Bertocchi A., Mancinelli S., Bellini M., Erreni M., Borreca A., Braga D., Giugliano S., Mozzarelli A.M., Manganaro D., et al. Identification of a choroid plexus vascular barrier closing during intestinal inflammation. Science. 2021;374:439–448. doi: 10.1126/science.abc6108. [DOI] [PubMed] [Google Scholar]
- 18.Hedemann T.L., Liu X., Kang C.N., Husain M.I. Associations between psoriasis and mental illness: an update for clinicians. Gen. Hosp. Psychiatry. 2022;75:30–37. doi: 10.1016/j.genhosppsych.2022.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Ortega J.M., Nogueras P., Muñoz-Negro J.E., Gutiérrez-Rojas L., González-Domenech P., Gurpegui M. Quality of life, anxiety and depressive symptoms in patients with psoriasis: A case-control study. J. Psychosom. Res. 2019;124 doi: 10.1016/j.jpsychores.2019.109780. [DOI] [PubMed] [Google Scholar]
- 20.Jones Amaowei E.E., Anwar S., Kavanoor Sridhar K., Shabbir K., Mohammed E.H., Bahar A.R., Talpur A.S., Bhat S., Zafar S., Qadar L.T. Correlation of Depression and Anxiety With Rheumatoid Arthritis. Cureus. 2022;14 doi: 10.7759/cureus.23137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machin A.R., Babatunde O., Haththotuwa R., Scott I., Blagojevic-Bucknall M., Corp N., Chew-Graham C.A., Hider S.L. The association between anxiety and disease activity and quality of life in rheumatoid arthritis: a systematic review and meta-analysis. Clin. Rheumatol. 2020;39:1471–1482. doi: 10.1007/s10067-019-04900-y. [DOI] [PubMed] [Google Scholar]
- 22.Marrie R.A., Hitchon C.A., Walld R., Patten S.B., Bolton J.M., Sareen J., Walker J.R., Singer A., Lix L.M., El-Gabalawy R., et al. Increased Burden of Psychiatric Disorders in Rheumatoid Arthritis. Arthritis Care Res. 2018;70:970–978. doi: 10.1002/acr.23539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uda M., Hashimoto M., Uozumi R., Torii M., Fujii T., Tanaka M., Furu M., Ito H., Terao C., Yamamoto W., et al. Factors associated with anxiety and depression in rheumatoid arthritis patients: a cross-sectional study. Adv. Rheumatol. 2021;61:65. doi: 10.1186/s42358-021-00223-2. [DOI] [PubMed] [Google Scholar]
- 24.Amaya-Uribe L., Rojas M., Azizi G., Anaya J.-M., Gershwin M.E. Primary immunodeficiency and autoimmunity: A comprehensive review. J. Autoimmun. 2019;99:52–72. doi: 10.1016/j.jaut.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 25.McCusker C., Upton J., Warrington R. Primary immunodeficiency. Allergy Asthma Clin. Immunol. 2018;14:61. doi: 10.1186/s13223-018-0290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picard C., Bobby Gaspar H., Al-Herz W., Bousfiha A., Casanova J.-L., Chatila T., Crow Y.J., Cunningham-Rundles C., Etzioni A., Franco J.L., et al. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J. Clin. Immunol. 2018;38:96–128. doi: 10.1007/s10875-017-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tangye S.G., Al-Herz W., Bousfiha A., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J.L., Holland S.M., Klein C., Morio T., et al. Human Inborn Errors of Immunity: 2019 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 2020;40:24–64. doi: 10.1007/s10875-019-00737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tangye S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., Klein C., Morio T., Oksenhendler E., Picard C., et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 2022;42:1473–1507. doi: 10.1007/s10875-022-01289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isung J., Williams K., Isomura K., Gromark C., Hesselmark E., Lichtenstein P., Larsson H., Fernández de la Cruz L.F., Sidorchuk A., Mataix-Cols D. Association of Primary Humoral Immunodeficiencies With Psychiatric Disorders and Suicidal Behavior and the Role of Autoimmune Diseases. JAMA Psychiatry. 2020;77:1147–1154. doi: 10.1001/jamapsychiatry.2020.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rider N.L., Kutac C., Hajjar J., Scalchunes C., Seeborg F.O., Boyle M., Orange J.S. Health-Related Quality of Life in Adult Patients with Common Variable Immunodeficiency Disorders and Impact of Treatment. J. Clin. Immunol. 2017;37:461–475. doi: 10.1007/s10875-017-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Titman P., Allwood Z., Gilmour C., Malcolmson C., Duran-Persson C., Cale C., Davies G., Gaspar H., Jones A. Quality of life in children with primary antibody deficiency. J. Clin. Immunol. 2014;34:844–852. doi: 10.1007/s10875-014-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manusama O.R., van Beveren N.J.M., van Hagen P.M., Drexhage H.A., Dalm V.A.S.H. Psychological Symptoms in Primary Immunodeficiencies: a Common Comorbidity? J. Clin. Immunol. 2022;42:695–698. doi: 10.1007/s10875-022-01207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anginot A., Nguyen J., Abou Nader Z., Rondeau V., Bonaud A., Kalogeraki M., Boutin A., Lemos J.P., Bisio V., Koenen J., et al. WHIM Syndrome-linked CXCR4 mutations drive osteoporosis. Nat. Commun. 2023;14 doi: 10.1038/s41467-023-37791-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dotta L., Notarangelo L.D., Moratto D., Kumar R., Porta F., Soresina A., Lougaris V., Plebani A., Smith C.I.E., Norlin A.-C., et al. Long-Term Outcome of WHIM Syndrome in 18 Patients: High Risk of Lung Disease and HPV-Related Malignancies. J. Allergy Clin. Immunol. Pract. 2019;7:1568–1577. doi: 10.1016/j.jaip.2019.01.045. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez P.A., Gorlin R.J., Lukens J.N., Taniuchi S., Bohinjec J., Francois F., Klotman M.E., Diaz G.A. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat. Genet. 2003;34:70–74. doi: 10.1038/ng1149. [DOI] [PubMed] [Google Scholar]
- 36.Kallikourdis M., Trovato A.E., Anselmi F., Sarukhan A., Roselli G., Tassone L., Badolato R., Viola A. The CXCR4 mutations in WHIM syndrome impair the stability of the T-cell immunologic synapse. Blood. 2013;122:666–673. doi: 10.1182/blood-2012-10-461830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawai T., Malech H.L. WHIM syndrome: congenital immune deficiency disease. Curr. Opin. Hematol. 2009;16:20–26. doi: 10.1097/MOH.0b013e32831ac557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balabanian K., Brotin E., Biajoux V., Bouchet-Delbos L., Lainey E., Fenneteau O., Bonnet D., Fiette L., Emilie D., Bachelerie F. Proper desensitization of CXCR4 is required for lymphocyte development and peripheral compartmentalization in mice. Blood. 2012;119:5722–5730. doi: 10.1182/blood-2012-01-403378. [DOI] [PubMed] [Google Scholar]
- 39.Balabanian K., Lagane B., Pablos J.L., Laurent L., Planchenault T., Verola O., Lebbe C., Kerob D., Dupuy A., Hermine O., et al. WHIM syndromes with different genetic anomalies are accounted for by impaired CXCR4 desensitization to CXCL12. Blood. 2005;105:2449–2457. doi: 10.1182/blood-2004-06-2289. [DOI] [PubMed] [Google Scholar]
- 40.Galli J., Pinelli L., Micheletti S., Palumbo G., Notarangelo L.D., Lougaris V., Dotta L., Fazzi E., Badolato R. Cerebellar involvement in warts Hypogammaglobulinemia immunodeficiency myelokathexis patients: neuroimaging and clinical findings. Orphanet J. Rare Dis. 2019;14:61. doi: 10.1186/s13023-019-1030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heusinkveld L.E., Yim E., Yang A., Azani A.B., Liu Q., Gao J.-L., McDermott D.H., Murphy P.M. Pathogenesis, diagnosis and therapeutic strategies in WHIM syndrome immunodeficiency. Expert Opin. Orphan Drugs. 2017;5:813–825. doi: 10.1080/21678707.2017.1375403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milanesi S., Locati M., Borroni E.M. Aberrant CXCR4 Signaling at Crossroad of WHIM Syndrome and Waldenstrom’s Macroglobulinemia. Int. J. Mol. Sci. 2020;21:5696. doi: 10.3390/ijms21165696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bianchi M.E., Mezzapelle R. The Chemokine Receptor CXCR4 in Cell Proliferation and Tissue Regeneration. Front. Immunol. 2020;11:2109. doi: 10.3389/fimmu.2020.02109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDermott D.H., Liu Q., Velez D., Lopez L., Anaya-O’Brien S., Ulrick J., Kwatemaa N., Starling J., Fleisher T.A., Priel D.A.L., et al. A phase 1 clinical trial of long-term, low-dose treatment of WHIM syndrome with the CXCR4 antagonist plerixafor. Blood. 2014;123:2308–2316. doi: 10.1182/blood-2013-09-527226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDermott D.H., Velez D., Cho E., Cowen E.W., DiGiovanna J.J., Pastrana D.V., Buck C.B., Calvo K.R., Gardner P.J., Rosenzweig S.D., et al. A phase III randomized crossover trial of plerixafor versus G-CSF for treatment of WHIM syndrome. J. Clin. Invest. 2023;133 doi: 10.1172/JCI164918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy A., Mondini M., Deutsch E. Plerixafor for the Treatment of WHIM Syndrome. N. Engl. J. Med. 2019;380 doi: 10.1056/NEJMc1901646. [DOI] [PubMed] [Google Scholar]
- 47.Özdemir E. Retrospective evaluation of adults with primary immunodeficiency disease. Postepy Dermatol. Alergol. 2022;39:976–979. doi: 10.5114/ada.2022.120887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dehkordy S.F., Aghamohammadi A., Ochs H.D., Rezaei N. Primary immunodeficiency diseases associated with neurologic manifestations. J. Clin. Immunol. 2012;32:1–24. doi: 10.1007/s10875-011-9593-8. [DOI] [PubMed] [Google Scholar]
- 49.Sowers K.L., Gayda-Chelder C.A., Galantino M.L. Self-reported cognitive impairment in individuals with Primary Immunodeficiency Disease. Brain Behav. Immun Health. 2020;9 doi: 10.1016/j.bbih.2020.100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yildirim M., Ayvaz D.C., Konuskan B., Gocmen R., Tezcan I., Topcu M., Topaloglu H., Anlar B. Neurologic Involvement in Primary Immunodeficiency Disorders. J. Child Neurol. 2018;33:320–328. doi: 10.1177/0883073817754176. [DOI] [PubMed] [Google Scholar]
- 51.Grossfeld P.D., Mattina T., Lai Z., Favier R., Jones K.L., Cotter F., Jones C. The 11q terminal deletion disorder: a prospective study of 110 cases. Am. J. Med. Genet. A. 2004;129A:51–61. doi: 10.1002/ajmg.a.30090. [DOI] [PubMed] [Google Scholar]
- 52.Sauer A.V., Hernandez R.J., Fumagalli F., Bianchi V., Poliani P.L., Dallatomasina C., Riboni E., Politi L.S., Tabucchi A., Carlucci F., et al. Alterations in the brain adenosine metabolism cause behavioral and neurological impairment in ADA-deficient mice and patients. Sci. Rep. 2017;7 doi: 10.1038/srep40136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Yu X., Ichikawa M., Lyons J.J., Datta S., Lamborn I.T., Jing H., Kim E.S., Biancalana M., Wolfe L.A., et al. Autosomal recessive phosphoglucomutase 3 (PGM3) mutations link glycosylation defects to atopy, immune deficiency, autoimmunity, and neurocognitive impairment J. Allergy Clin. Immunol. 2014;133:1400–1409. doi: 10.1016/j.jaci.2014.02.013. , 1409.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ammann S., Schulz A., Krägeloh-Mann I., Dieckmann N.M.G., Niethammer K., Fuchs S., Eckl K.M., Plank R., Werner R., Altmüller J., et al. Mutations in AP3D1 associated with immunodeficiency and seizures define a new type of Hermansky-Pudlak syndrome. Blood. 2016;127:997–1006. doi: 10.1182/blood-2015-09-671636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhala S., Best A.F., Giri N., Alter B.P., Pao M., Gropman A., Baker E.H., Savage S.A. CNS manifestations in patients with telomere biology disorders. Neurol. Genet. 2019;5:370. doi: 10.1212/NXG.0000000000000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oud M.M., Tuijnenburg P., Hempel M., van Vlies N., Ren Z., Ferdinandusse S., Jansen M.H., Santer R., Johannsen J., Bacchelli C., et al. Mutations in EXTL3 Cause Neuro-immuno-skeletal Dysplasia Syndrome. Am. J. Hum. Genet. 2017;100:281–296. doi: 10.1016/j.ajhg.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Versteegh J.J., Dulfer K., Stuvel K., Pasmans S.G., Utens E.M. Netherton syndrome; neuropsychological and psychosocial functioning of child and adult patients and their parents. J. Health Psychol. 2020;25:2296–2316. doi: 10.1177/1359105318790052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fageeh H.N. Papillon-Lefèvre Syndrome: A Rare Case Report of Two Brothers and Review of the Literature. Int. J. Clin. Pediatr. Dent. 2018;11:352–355. doi: 10.5005/jp-journals-10005-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phull T., Jyoti D., Malhotra R., Nayak S., Modi H., Singla I., P A. Diagnosis and Management of Papillon-Lefevre Syndrome: A Rare Case Report and a Brief Review of Literature. Cureus. 2023;15 doi: 10.7759/cureus.43335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mazar I., Stokes J., Ollis S., Love E., Espensen A., Barth P.G., Powers J.H., 3rd, Shields A.L. Understanding the life experience of Barth syndrome from the perspective of adults: a qualitative one-on-one interview study. Orphanet J. Rare Dis. 2019;14:243. doi: 10.1186/s13023-019-1200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolze A., Byun M., McDonald D., Morgan N.V., Abhyankar A., Premkumar L., Puel A., Bacon C.M., Rieux-Laucat F., Pang K., et al. Whole-exome-sequencing-based discovery of human FADD deficiency. Am. J. Hum. Genet. 2010;87:873–881. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kotlarz D., Marquardt B., Barøy T., Lee W.S., Konnikova L., Hollizeck S., Magg T., Lehle A.S., Walz C., Borggraefe I., et al. Human TGF-β1 deficiency causes severe inflammatory bowel disease and encephalopathy. Nat. Genet. 2018;50:344–348. doi: 10.1038/s41588-018-0063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Russ A., Mack J., Green-Murphy A., Occidental M., Mian A. Griscelli Type 2 Syndrome and Hemophagocytic Lymphohistiocytosis: Sisters With the Same Mutation but Different Presentations. J. Pediatr. Hematol. Oncol. 2019;41:473–477. doi: 10.1097/MPH.0000000000001522. [DOI] [PubMed] [Google Scholar]
- 64.Savic S., Parry D., Carter C., Johnson C., Logan C., Gutierrez B.M., Thomas J.E., Bacon C.M., Cant A., Hambleton S. A new case of Fas-associated death domain protein deficiency and update on treatment outcomes. J. Allergy Clin. Immunol. 2015;136:502–505.e4. doi: 10.1016/j.jaci.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 65.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M.M., Pletikos M., Meyer K.A., Sedmak G., et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sepp M., Leiss K., Murat F., Okonechnikov K., Joshi P., Leushkin E., Spänig L., Mbengue N., Schneider C., Schmidt J., et al. Cellular development and evolution of the mammalian cerebellum. Nature. 2024;625:788–796. doi: 10.1038/s41586-023-06884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hull C., Regehr W.G. The cerebellar cortex. Annu. Rev. Neurosci. 2022;45:151–175. doi: 10.1146/annurev-neuro-091421-125115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kozareva V., Martin C., Osorno T., Rudolph S., Guo C., Vanderburg C., Nadaf N., Regev A., Regehr W.G., Macosko E. A transcriptomic atlas of mouse cerebellar cortex comprehensively defines cell types. Nature. 2021;598:214–219. doi: 10.1038/s41586-021-03220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leto K., Arancillo M., Becker E.B.E., Buffo A., Chiang C., Ding B., Dobyns W.B., Dusart I., Haldipur P., Hatten M.E., et al. Consensus Paper: Cerebellar Development. Cerebellum. 2016;15:789–828. doi: 10.1007/s12311-015-0724-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voogd J., Glickstein M. The anatomy of the cerebellum. Trends Neurosci. 1998;21:370–375. doi: 10.1016/s0166-2236(98)01318-6. [DOI] [PubMed] [Google Scholar]
- 71.Dresselhaus E.C., Meffert M.K. Cellular Specificity of NF-κB Function in the Nervous System. Front. Immunol. 2019;10:1043. doi: 10.3389/fimmu.2019.01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaltschmidt B., Kaltschmidt C. NF-kappaB in the nervous system. Cold Spring Harb. Perspect. Biol. 2009;1 doi: 10.1101/cshperspect.a001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du H., Wang Z., Guo R., Yang L., Liu G., Zhang Z., Xu Z., Tian Y., Yang Z., Li X., et al. Transcription factors Bcl11a and Bcl11b are required for the production and differentiation of cortical projection neurons. Cereb. Cortex. 2022;32:3611–3632. doi: 10.1093/cercor/bhab437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simon R., Wiegreffe C., Britsch S. Bcl11 Transcription Factors Regulate Cortical Development and Function. Front. Mol. Neurosci. 2020;13:51. doi: 10.3389/fnmol.2020.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Molyneaux B.J., Goff L.A., Brettler A.C., Chen H.H., Hrvatin S., Rinn J.L., Arlotta P. DeCoN: Genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozawa P.M.M., Ariza C.B., Ishibashi C.M., Fujita T.C., Banin-Hirata B.K., Oda J.M.M., Watanabe M.A.E. Role of CXCL12 and CXCR4 in normal cerebellar development and medulloblastoma. Int. J. Cancer. 2016;138:10–13. doi: 10.1002/ijc.29333. [DOI] [PubMed] [Google Scholar]
- 77.Wang Y., Li G., Stanco A., Long J.E., Crawford D., Potter G.B., Pleasure S.J., Behrens T., Rubenstein J.L.R. CXCR4 and CXCR7 Have Distinct Functions in Regulating Interneuron Migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arpi M.N.T., Simpson T.I. SFARI genes and where to find them; modelling Autism Spectrum Disorder specific gene expression dysregulation with RNA-seq data. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-14077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alouche N., Bonaud A., Rondeau V., Hussein-Agha R., Nguyen J., Bisio V., Khamyath M., Crickx E., Setterblad N., Dulphy N., et al. Hematologic disorder-associated Cxcr4 gain-of-function mutation leads to uncontrolled extrafollicular immune response. Blood. 2021;137:3050–3063. doi: 10.1182/blood.2020007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Freitas C., Wittner M., Nguyen J., Rondeau V., Biajoux V., Aknin M.-L., Gaudin F., Beaussant-Cohen S., Bertrand Y., Bellanné-Chantelot C., et al. Lymphoid differentiation of hematopoietic stem cells requires efficient Cxcr4 desensitization. J. Exp. Med. 2017;214:2023–2040. doi: 10.1084/jem.20160806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mayol K., Biajoux V., Marvel J., Balabanian K., Walzer T. Sequential desensitization of CXCR4 and S1P5 controls natural killer cell trafficking. Blood. 2011;118:4863–4871. doi: 10.1182/blood-2011-06-362574. [DOI] [PubMed] [Google Scholar]
- 82.Biajoux V., Natt J., Freitas C., Alouche N., Sacquin A., Hemon P., Gaudin F., Fazilleau N., Espéli M., Balabanian K. Efficient Plasma Cell Differentiation and Trafficking Require Cxcr4 Desensitization. Cell Rep. 2016;17:193–205. doi: 10.1016/j.celrep.2016.08.068. [DOI] [PubMed] [Google Scholar]
- 83.Apps R., Hawkes R. Cerebellar cortical organization: a one-map hypothesis. Nat. Rev. Neurosci. 2009;10:670–681. doi: 10.1038/nrn2698. [DOI] [PubMed] [Google Scholar]
- 84.Anyan J., Amir S. Too Depressed to Swim or Too Afraid to Stop? A Reinterpretation of the Forced Swim Test as a Measure of Anxiety-Like Behavior. Neuropsychopharmacology. 2018;43:931–933. doi: 10.1038/npp.2017.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fox W.M. Reflex-ontogeny and behavioural development of the mouse. Anim. Behav. 1965;13:234–241. doi: 10.1016/0003-3472(65)90041-2. [DOI] [PubMed] [Google Scholar]
- 86.Feather-Schussler D.N., Ferguson T.S. A Battery of Motor Tests in a Neonatal Mouse Model of Cerebral Palsy. J. Vis. Exp. 2016;117 doi: 10.3791/53569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baumann O., Borra R.J., Bower J.M., Cullen K.E., Habas C., Ivry R.B., Leggio M., Mattingley J.B., Molinari M., Moulton E.A., et al. Consensus paper: the role of the cerebellum in perceptual processes. Cerebellum. 2015;14:197–220. doi: 10.1007/s12311-014-0627-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.MacNeilage P.R., Glasauer S. Gravity Perception: The Role of the Cerebellum. Curr. Biol. 2018;28:R1296–R1298. doi: 10.1016/j.cub.2018.09.053. [DOI] [PubMed] [Google Scholar]
- 89.Therrien A.S., Bastian A.J. Cerebellar damage impairs internal predictions for sensory and motor function. Curr. Opin. Neurobiol. 2015;33:127–133. doi: 10.1016/j.conb.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van der Heijden M.E., Lackey E.P., Perez R., Ișleyen F.S., Brown A.M., Donofrio S.G., Lin T., Zoghbi H.Y., Sillitoe R.V. Maturation of Purkinje cell firing properties relies on neurogenesis of excitatory neurons. eLife. 2021;10 doi: 10.7554/eLife.68045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lawton A.K., Engstrom T., Rohrbach D., Omura M., Turnbull D.H., Mamou J., Zhang T., Schwarz J.M., Joyner A.L. Cerebellar folding is initiated by mechanical constraints on a fluid-like layer without a cellular pre-pattern. eLife. 2019;8 doi: 10.7554/eLife.45019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Consalez G.G., Goldowitz D., Casoni F., Hawkes R. Origins, Development, and Compartmentation of the Granule Cells of the Cerebellum. Front. Neural Circuits. 2020;14 doi: 10.3389/fncir.2020.611841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yeung J., Ha T.J., Swanson D.J., Goldowitz D. A Novel and Multivalent Role of Pax6 in Cerebellar Development. J. Neurosci. 2016;36:9057–9069. doi: 10.1523/JNEUROSCI.4385-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sudarov A., Joyner A.L. Cerebellum morphogenesis: the foliation pattern is orchestrated by multi-cellular anchoring centers. Neural Dev. 2007;2:26. doi: 10.1186/1749-8104-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pellegrino L., Khodaparast S., Cabral J.T. Orthogonal wave superposition of wrinkled, plasma-oxidised, polydimethylsiloxane surfaces. Soft Matter. 2020;16:595–603. doi: 10.1039/C9SM02124H. [DOI] [PubMed] [Google Scholar]
- 96.Oomman S., Strahlendorf H., Finckbone V., Strahlendorf J. Non-lethal active caspase-3 expression in Bergmann glia of postnatal rat cerebellum. Brain Res. Dev. Brain Res. 2005;160:130–145. doi: 10.1016/j.devbrainres.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 97.Gulati G.S., Sikandar S.S., Wesche D.J., Manjunath A., Bharadwaj A., Berger M.J., Ilagan F., Kuo A.H., Hsieh R.W., Cai S., et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science. 2020;367:405–411. doi: 10.1126/science.aax0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sato M., Suzuki K., Nakanishi S. Expression profile of BDNF-responsive genes during cerebellar granule cell development. Biochem. Biophys. Res. Commun. 2006;341:304–309. doi: 10.1016/j.bbrc.2005.12.184. [DOI] [PubMed] [Google Scholar]
- 99.Limatola C., Giovannelli A., Maggi L., Ragozzino D., Castellani L., Ciotti M.T., Vacca F., Mercanti D., Santoni A., Eusebi F. SDF-1alpha-mediated modulation of synaptic transmission in rat cerebellum. Eur. J. Neurosci. 2000;12:2497–2504. doi: 10.1046/j.1460-9568.2000.00139.x. [DOI] [PubMed] [Google Scholar]
- 100.Witter L., Rudolph S., Pressler R.T., Lahlaf S.I., Regehr W.G. Purkinje Cell Collaterals Enable Output Signals from the Cerebellar Cortex to Feed Back to Purkinje Cells and Interneurons. Neuron. 2016;91:312–319. doi: 10.1016/j.neuron.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mapelli L., Dubochet O., Tedesco M., Sciacca G., Ottaviani A., Monteverdi A., Battaglia C., Tritto S., Cardot F., Surbled P., et al. Design, implementation, and functional validation of a new generation of microneedle 3D high-density CMOS multi-electrode array for brain tissue and spheroids. bioRxiv. 2022 doi: 10.1101/2022.08.11.503595. Preprint at. [DOI] [Google Scholar]
- 102.Geier C.B., Ellison M., Cruz R., Pawar S., Leiss-Piller A., Zmajkovicova K., McNulty S.M., Yilmaz M., Evans M.O., 2nd, Gordon S., et al. Disease Progression of WHIM Syndrome in an International Cohort of 66 Pediatric and Adult Patients. J. Clin. Immunol. 2022;42:1748–1765. doi: 10.1007/s10875-022-01312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Evans M.O., 2nd, McDermott D.H., Murphy P.M., Petersen M.M. Abnormal Newborn Screen in a WHIM Syndrome Infant. J. Clin. Immunol. 2019;39:839–841. doi: 10.1007/s10875-019-00686-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ara T., Tokoyoda K., Sugiyama T., Egawa T., Kawabata K., Nagasawa T. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity. 2003;19:257–267. doi: 10.1016/s1074-7613(03)00201-2. [DOI] [PubMed] [Google Scholar]
- 105.Christensen J.L., Wright D.E., Wagers A.J., Weissman I.L. Circulation and chemotaxis of fetal hematopoietic stem cells. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zou Y.R., Kottmann A.H., Kuroda M., Taniuchi I., Littman D.R. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 107.Alves de Lima K., Rustenhoven J., Da Mesquita S., Wall M., Salvador A.F., Smirnov I., Martelossi Cebinelli G., Mamuladze T., Baker W., Papadopoulos Z., et al. Meningeal γδ T cells regulate anxiety-like behavior via IL-17a signaling in neurons. Nat. Immunol. 2020;21:1421–1429. doi: 10.1038/s41590-020-0776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goyal M., Goyal R., Sanguinetti J.L. Ultrasound stimulation of the vagus nerve as a treatment modality for anxiety. Front. Psychiatry. 2024;15 doi: 10.3389/fpsyt.2024.1376140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fernandes D.J., Spring S., Corre C., Tu A., Qiu L.R., Hammill C., Vousden D.A., Spencer Noakes T.L., Nieman B.J., Bowdish D.M.E., et al. Mouse models of immune dysfunction: their neuroanatomical differences reflect their anxiety-behavioural phenotype. Mol. Psychiatry. 2022;27:3047–3055. doi: 10.1038/s41380-022-01535-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sawtell N.B. Multimodal integration in granule cells as a basis for associative plasticity and sensory prediction in a cerebellum-like circuit. Neuron. 2010;66:573–584. doi: 10.1016/j.neuron.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 111.Wang W.X., Lefebvre J.L. Morphological pseudotime ordering and fate mapping reveal diversification of cerebellar inhibitory interneurons. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-30977-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Silva C.G., Peyre E., Nguyen L. Cell migration promotes dynamic cellular interactions to control cerebral cortex morphogenesis. Nat. Rev. Neurosci. 2019;20:318–329. doi: 10.1038/s41583-019-0148-y. [DOI] [PubMed] [Google Scholar]
- 113.Komuro H., Yacubova E. Recent advances in cerebellar granule cell migration. Cell. Mol. Life Sci. 2003;60:1084–1098. doi: 10.1007/s00018-003-2248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chin P.W., Augustine G.J. The cerebellum and anxiety. Front. Cell. Neurosci. 2023;17 doi: 10.3389/fncel.2023.1130505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moreno-Rius J. The cerebellum in fear and anxiety-related disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2018;85:23–32. doi: 10.1016/j.pnpbp.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 116.Wester J.C., Mahadevan V., Rhodes C.T., Calvigioni D., Venkatesh S., Maric D., Hunt S., Yuan X., Zhang Y., Petros T.J., et al. Neocortical Projection Neurons Instruct Inhibitory Interneuron Circuit Development in a Lineage-Dependent Manner. Neuron. 2019;102:960–975.e6. doi: 10.1016/j.neuron.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lodato S., Rouaux C., Quast K.B., Jantrachotechatchawan C., Studer M., Hensch T.K., Arlotta P. Excitatory Projection Neuron Subtypes Control the Distribution of Local Inhibitory Interneurons in the Cerebral Cortex. Neuron. 2011;69:763–779. doi: 10.1016/j.neuron.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang S.S.-H., Kloth A.D., Badura A. The cerebellum, sensitive periods, and autism. Neuron. 2014;83:518–532. doi: 10.1016/j.neuron.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Morland B., Kepak T., Dallorso S., Sevilla J., Murphy D., Luksch R., Yaniv I., Bader P., Rößler J., Bisogno G., et al. Plerixafor combined with standard regimens for hematopoietic stem cell mobilization in pediatric patients with solid tumors eligible for autologous transplants: two-arm phase I/II study (MOZAIC) Bone Marrow Transplant. 2020;55:1744–1753. doi: 10.1038/s41409-020-0836-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Karres D., Ali S., van Hennik P.B., Straus S., Josephson F., Thole G., Glerum P.J., Herberts C., Babae N., Herold R., et al. EMA recommendation for the pediatric indications of plerixafor (mozobil) to enhance mobilization of hematopoietic stem cells for collection and subsequent autologous transplantation in children with lymphoma or malignant solid tumors. Oncologist. 2020;25:e976–e981. doi: 10.1634/theoncologist.2019-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.López-Bendito G., Sánchez-Alcañiz J.A., Pla R., Borrell V., Picó E., Valdeolmillos M., Marín O. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 2008;28:1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen T.W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee H.Y., Greene L.A., Mason C.A., Manzini M.C. Isolation and culture of post-natal mouse cerebellar granule neuron progenitor cells and neurons. J. Vis. Exp. 2009;23:990. doi: 10.3791/990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Krämer D., Minichiello L. Cell culture of primary cerebellar granule cells. Methods Mol. Biol. 2010;633:233–239. doi: 10.1007/978-1-59745-019-5_17. [DOI] [PubMed] [Google Scholar]
- 126.Losi G., Prybylowski K., Fu Z., Luo J.H., Vicini S. Silent synapses in developing cerebellar granule neurons. J. Neurophysiol. 2002;87:1263–1270. doi: 10.1152/jn.00633.2001. [DOI] [PubMed] [Google Scholar]
- 127.C Wood T. QUIT: QUantitative Imaging Tools. J. Open Source Softw. 2018;3:656. doi: 10.21105/joss.00656. [DOI] [Google Scholar]
- 128.Avants B.B., Tustison N.J., Song G., Cook P.A., Klein A., Gee J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Avants B.B., Epstein C.L., Grossman M., Gee J.C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Winkler A.M., Ridgway G.R., Webster M.A., Smith S.M., Nichols T.E. Permutation inference for the general linear model. Neuroimage. 2014;92:381–397. doi: 10.1016/j.neuroimage.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van der Walt S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T., scikit-image contributors scikit-image: image processing in Python. PeerJ. 2014;2 doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nunez-Iglesias J., Blanch A.J., Looker O., Dixon M.W., Tilley L. A new Python library to analyse skeleton images confirms malaria parasite remodelling of the red blood cell membrane skeleton. PeerJ. 2018;6 doi: 10.7717/peerj.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Boitnott A., Garcia-Forn M., Ung D.C., Niblo K., Mendonca D., Park Y., Flores M., Maxwell S., Ellegood J., Qiu L.R., et al. Developmental and Behavioral Phenotypes in a Mouse Model of DDX3X Syndrome. Biol. Psychiatry. 2021;90:742–755. doi: 10.1016/j.biopsych.2021.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Heyser C.J. Assessment of developmental milestones in rodents. Curr. Protoc. Neurosci. 2004;Chapter 8:Unit 8.18. doi: 10.1002/0471142301.ns0818s25. [DOI] [PubMed] [Google Scholar]
- 135.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 136.Luo W., Lin G.N., Song W., Zhang Y., Lai H., Zhang M., Miao J., Cheng X., Wang Y., Li W., et al. Single-cell spatial transcriptomic analysis reveals common and divergent features of developing postnatal granule cerebellar cells and medulloblastoma. BMC Biol. 2021;19:135. doi: 10.1186/s12915-021-01071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sun Z., Südhof T.C. A simple Ca2+-imaging approach to neural network analyses in cultured neurons. J. Neurosci. Methods. 2021;349 doi: 10.1016/j.jneumeth.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pilpel N., Landeck N., Klugmann M., Seeburg P.H., Schwarz M.K. Rapid, reproducible transduction of select forebrain regions by targeted recombinant virus injection into the neonatal mouse brain. J. Neurosci. Methods. 2009;182:55–63. doi: 10.1016/j.jneumeth.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 139.Kaneko M., Yamaguchi K., Eiraku M., Sato M., Takata N., Kiyohara Y., Mishina M., Hirase H., Hashikawa T., Kengaku M. Remodeling of monoplanar Purkinje cell dendrites during cerebellar circuit formation. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fujishima K., Horie R., Mochizuki A., Kengaku M. Principles of branch dynamics governing shape characteristics of cerebellar Purkinje cell dendrites. Development. 2012;139:3442–3455. doi: 10.1242/dev.081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gibson D.A., Ma L. Mosaic analysis of gene function in postnatal mouse brain development by using virus-based Cre recombination. J. Vis. Exp. 2011;54:2823. doi: 10.3791/2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Arganda-Carreras I., Fernández-González R., Muñoz-Barrutia A., Ortiz-de-Solorzano C. 3D reconstruction of histological sections: Application to mammary gland tissue. Microsc. Res. Tech. 2010;73:1019–1029. doi: 10.1002/jemt.20829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell gene expression datasets of P1 WHIM and control cerebella have been deposited at GEO and are publicly available as of the date of publication. GEO: GSE266671.
-
•
MRI data of P45 WHIM and control cerebella have been deposited on OpenNeuro and are publicly available as of the date of publication. OpenNeuro: ds005137.
-
•
The BrainSpan Atlas is available at https://www.brainspan.org/.
-
•
The human single-nuclei dataset is available at https://cellxgene.cziscience.com/collections/72d37bc9-76cc-442d-9131-da0e273862db/.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.