Abstract
Allelic loss of chromosome 1p is frequently observed in oligodendroglioma. We screened 177 oligodendroglial tumors for 1p deletions and found 6 tumors with localized 1p36 deletions. Several apoptosis regulation genes have been mapped to this region, including Tumor Protein 73 (p73), DNA Fragmentation Factor subunits alpha (DFFA) and beta (DFFB), and Tumor Necrosis Factor Receptor Superfamily Members 9 and 25 (TNFRSF9, TNFRSF25). We compared expression levels of these 5 genes in pairs of 1p-loss and 1p-intact tumors using quantitative reverse-transcriptase PCR (QRTPCR) to test if 1p deletions had an effect on expression. Only the DFFB gene demonstrated decreased expression in all tumor pairs tested. Mutational analysis did not reveal DFFB mutations in 12 tested samples. However, it is possible that DFFB haploinsufficiency from 1p allelic loss is a contributing factor in oligodendroglioma development.
Introduction
Oligodendroglial tumors with allelic losses on 1p usually display loss of relatively long regions, a phenomenon that has made the identification of putative 1p tumor suppressor genes difficult [1-6]. However, the vast majority of reported oligodendroglioma cases with 1p-loss have involved the 1p36 region, with several breakpoints within the region observed [5-9]. It is important to note that several apoptotic genes have been mapped to 1p36. Diminished apoptosis has been recognized as one of the hallmarks of most types of cancer, representing one of the major ways known for a tumor-cell population to expand [10]. Therefore, we tested TP73, TNFRSF9, TNFRSF25, DFFA, and DFFB, all of which are 1p genes involved in apoptosis, for differential expression in 1p-status subsets of oligodendroglioma. QRTPCR analysis of match-paired samples demonstrated that levels of DFFB were decreased in all 1p-allelic loss cases. In contrast, the other tested genes showed heterogeneous patterns of expression. This result suggests DFFB to be a key molecule affected by 1p-deletion in oligodendroglioma.
Materials and methods
Samples
The records of 177 patients who underwent treatment for oligodendroglial tumors at the University of Texas M.D. Anderson Cancer Center (UTMDACC) between 1981 and 2002 were collected and reviewed. These patients were initially diagnosed as having low-grade oligodendroglioma or mixed oligoastrocytoma, anaplastic oligodendroglioma or mixed oligoastrocytoma, or glioblastoma multiforme with significant oligodendroglial component by neuropathologists from UTMDACC and later confirmed by two of the authors (KA and GF). Mixed tumors were included in this study since clear pathologic discrimination between glioma subtypes is sometimes difficult, and as a group, oligoastrocytomas often have 1p deletions [4]. In fact, both the oligodendroglial and astrocytic components of mixed tumors have been observed to have this genetic signature [4].
Tissue for DNA isolation was obtained from paraffin-embedded samples. Each tissue block was histologically assessed for tumor by a neuropathologist (KA). Sections were directly cut from the block for DNA isolation if at least 90% of the tissue was determined to be tumor. If the proportion of tumor was <90%, 10 to 20 unstained slides were prepared from the block and tumor tissue was dissected from normal tissue. DNA was isolated by digesting deparaffinized tumor sections for 3 to 5 days with proteinase K at 55°C (0.5 mg/ml in 100 mmol/L NaCl, 10 mmol/L Tris-HCl, pH 8.0, 25 mmol/L ethylenediaminetetraacetic acid, 0.5% sodium dodecyl sulfate), followed by a phenol:chloroform:isoamyl alcohol extraction and isopropanol precipitation.
Tissue for RNA isolation was obtained from fresh/frozen samples. Each frozen section was histologically assessed for tumor by a neuropathologist (GF or KA) and used only if at least 90% of the tissue was determined to be tumor. For RNA isolation, up to 50 mg of tissue was frozen in liquid nitrogen, crushed into powder using a mortar and pestle, and dissolved in 1 ml of Trizol® Reagent. 200 μl chloroform was added to the sample, vortexed at high speed for 15 seconds, and centrifuged at 12,000 × g for 15 minutes at 4°C. After transfer of the aqueous phase to fresh 1.5 ml Eppendorf tube, an equal volume of 70% ethanol was added and mixed by tube inversion. The sample was then loaded onto a QIAGEN RNeasy® mini column and centrifuged at 16,000 × g for 20 seconds (QIAGEN Sciences, Germantown, MD). The column was washed twice with 500 μl of QIAGEN's RPE buffer. RNA was eluated off the column in 50 μl of nuclease-free water. RNA was quantified using a spectrophotometer and qualitated with an Agilent BioAnalyzer 2100 (Agilent Technologies, Palo Alto, CA).
Detection of 1p allelic loss
Quantitative Microsatellite Analysis (QuMA) was used as previously described to determine 1p allelic loss in 177 tumors in this study [11-13].
Quantitative Reverse Transcription-Polymerase Chain Reaction (QRTPCR) Analysis
Initial experiments were performed to determine the valid range of RNA concentrations and to demonstrate the similarity of PCR efficiencies for each gene of interest compared to the endogenous control gene cyclophilin. To determine fold-changes in each gene, QRTPCR was performed on the ABI Prism 7700 using the commercially available gene expression assay for p73, DFFA, and DFFB (Hs00232088_m1, Hs00189336_m1, Hs00237077_m1, respectively) and the cylophilin Vic-labeled Pre-Developed Assay Reagent (Applied Biosystems, Foster City, CA) without multiplexing. In triplicate, we amplified 50 ng cDNA for each sample for each assay in a reaction containing 1× TaqMan® Universal PCR Master Mix without AmpErase UNG and 1× gene expression assay with the following cycling conditions: 10 minutes at 95°C, then 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. Calculations were performed using the δδCt method to determine fold-difference in 1p-loss cases relative to the matched 1p-intact cases. Fold changes for TNFSF5, TNFRSF9, TNFRSF11a, and TNFRSF25 were determined in a similar fashion, using commercially available gene expression assays (Hs00374176_m1, Hs00155512_m1, Hs00187189_m1, and Hs00237054_m1, respectively) and the 18S rRNA TaqMan® Endogenous Control (Hs99999901_s1).
Mutation screening
PCR amplifications of exons 1–6 were carried out using 100-μL reaction volumes with 1.5 mmol/L MgCl2; 200 μmol/L each of deoxy (d)-ATP, dGTP, dTTP, and dCTP; 2 pmol of each primer; 100 ng template DNA; and 1 U AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA). Amplifications of exon 7 were the same with the exception that the reaction mix had a concentration of 7% dimethylsulphoxide (DMSO). PCR cycling conditions were 10 min at 94°C, followed by 40 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min, followed by 15 min at 72°C. Sequencing reactions were setup using the BigDye Terminator Cycle Sequencing Reaction Kit with AmpliTaq DNA polymerase FS (Applied Biosystems, Foster City, CA) according to the manufacturer's specifications, and were subjected to gel electrophoresis on an ABI PRISM 3700 (Applied Biosystems, Foster City, CA). Sequencing data were aligned with the Sequencer program using DFFB sequence as reported by the Human Genome Database [14]. Forward and reverse PCR primer sequences are listed in Table 1.
Table 1.
Primer sets used for amplifying and sequencing the coding regions of DFFB.
| Name | F primer seq | R primer seq |
| DFFBamp1 | gcttgcagagctcaccaggtgc | cggctgaggcgaacgaaaactacc |
| DFFBseq1 | acggatctgagcagctgg | ctcctattctccccacacgc |
| DFFBamp2 | aagcacagctcattccggtcg | tgatgggcacctggagctaagc |
| DFFBseq2 | gccctcgtcttgagacc | aggacctcggagagtgc |
| DFFBamp3 | gggggaagatgtggtcagaggctc | ccacctgagtccttgctgggtacc |
| DFFBseq3 | cttgtgaccggggcag | atccaacttcttctggcacc |
| DFFBamp4 | gctgtagtaagctgtgttcgtgccactg | gcgctagcttccctcaccagagc |
| DFFBseq4 | ggaggacagagcaagacc | ccagatccacgcaagc |
| DFFBamp5 | gggtctcagagggccatggag | cctgtgtgcactgcagcttgagag |
| DFFBseq5 | atggatcgagagccagtg | ggcaagggctgaaggtc |
| DFFBamp6 | cgggaggcggaggttgtagtaagc | ctgggctgtaacacgggtgcag |
| DFFBseq6 | gccactgcactccagc | ccatggcagggacagg |
| DFFBamp7 | gggaatttgtgaagagctgtgactgc | ccccaacaattcagaaatgtaatgaaatcag |
| DFFBseq7 | gctatgacctgttgcctgtg | ggcacctgttaaaatgatgc |
Results and Discussions
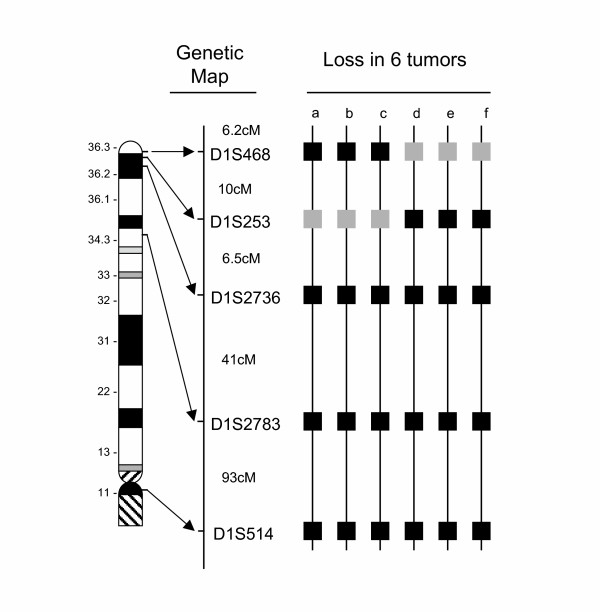
We evaluated 177 oligodendroglial tumors using QuMA for 1p-allelic loss in an attempt to determine a consensus region of deletion [11-13]. Loss was observed in 92 tumors, which in most cases involved the entire chromosomal arm. However, six tumors demonstrated localized loss involving the 1p36 region, defining a consensus region of deletion (Figure 1). These results were similar to those observed for other oligodendroglial 1p-deletion mapping studies, in which consensus regions of deletion involved 1p36 [5,6,8,9].
Figure 1.
Common region of allelic loss on the short arm of chromosome 1 in oligodendrogliomas. Markers used for screening 1p-allelic loss and their placement on the genetic and cytogenetic maps of 1p. Black squares indicate where tumors retained allelic balance, whereas gray squares indicate allelic loss.
The second part of our strategy included the identification of abrogated cellular pathways in oligodendrogliomas with 1p/19q allelic loss. The transcriptsomes of eight pairs of gender- and age-matched tumors were measured using a Pathway microarray consisting of 1,500 functionally characterized genes constructed in our Cancer Genomics Core Laboratory. We used paired sample tests (the Sign Rank test and the paired t-test) to identify differentially expressed genes. While the sign rank test uses the null hypothesis that the medians for the two classes are same, the paired t-test uses the null hypothesis that the means of the two classes are same. We recognize a gene as significant when the sample data for the gene gives a p-value less than 0.01 (99% confidence). This analysis revealed a number of genes that demonstrated robust differential expression (McDonald and Zhang, unpublished results).
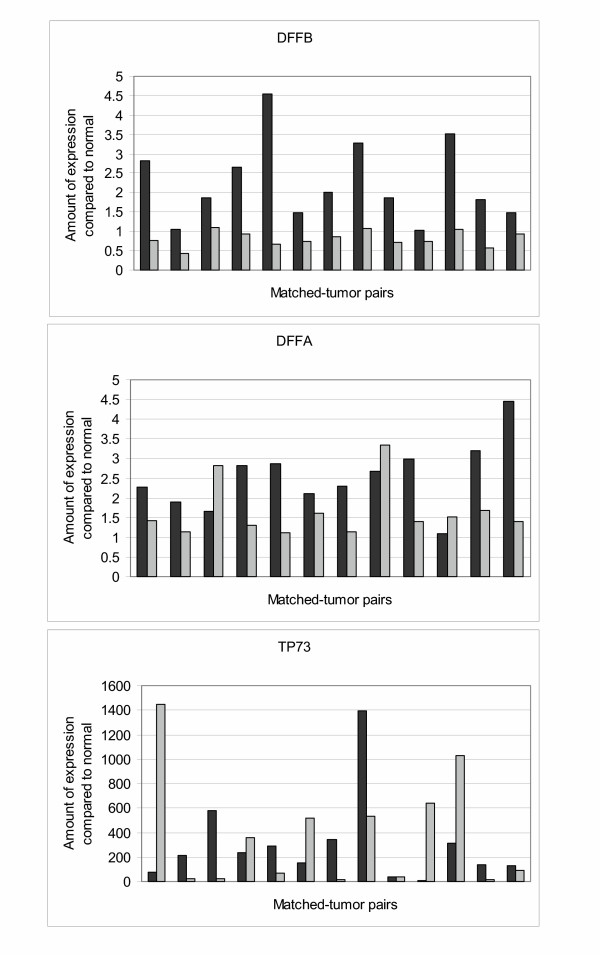
We evaluated three of these genes with QRTPCR either because of location in our consensus region of deletion (p73), or due to their relationship to genes in our region of interest (Tumor Necrosis Factor Super Family Ligand 5 [TNFSF5] and Tumor Necrosis Factor Receptor Super Family 11a [TNFRSF11a]). Both TNFSF5 and TNFRSF11a are involved in apoptotic pathways that include several genes located in our region of interest: TNFRSF9, TNFRSF25, DFFA, and DFFB. Therefore, these genes were also tested via QRTPCR to determine if they had differential gene expression. Based on fresh/frozen tissue availability of the original 170 cases, total RNA samples from thirteen age- and gender-matched pairs of 1p/19q loss and intact tumors were evaluated for differential gene expression of p73 and the six TNF pathway genes. Of the seven genes, only DFFB was differentially expressed in all 13 pairs of tumor samples (Figure 2). Figure 2 also displays the differential expression levels for DFFA and TP73 for the tested tumor pairs. In contrast to DFFB, there were 3 pairs (25%) in which the 1p/19q loss tumors had higher DFFA expression. Likewise, 5 of the 13 pairs (38%) had higher TP73 expression in the 1p/19q loss tumors. Similarly, 33%, 50%, 50%, and 83% of tested pairs had higher expression of TNFSF5, TNFRSF9, TNFRSF11a, and TNFRSF25 in the 1p/19q loss tumors, respectively (data not shown). Since DFFB was the only tested gene that was differentially expressed in the same direction by all 13 pairs of tumors, we viewed DFFB as the best tumor suppressor gene candidate in our study.
Figure 2.
QRTPCR results in oligodendroglioma subsets. Black bar, 1p-intact samples; light gray bars, 1p-loss samples. A) Expression of DFFB was lower in all 1p-loss samples as compared to their matched 1p-intact samples. Ten of the thirteen 1p-loss tumors had lower expression of DFFB compared to normal brain, with three tumors demonstrating 1–2× the amount of normal brain DFFB expression. In contrast, all 1p-intact tumors had DFFB expression greater than or equal to that seen in normal brain. B) Expression of DFFA was lower in most 1p-loss samples as compared to their matched 1p-intact samples. Only three 1p-loss tumors had higher DFFA expression. Differential expression was detected to a degree; most 1p-loss tumors (10 of 12) demonstrated 1–2× the amount of normal brain DFFA expression, whereas most 1p-intact tumors (9of 12) demonstrated ≥ 2× the amount of normal brain DFFA expression. C). Differential p73 expression was not detected. In five of the pairs, 1p-loss tumors had higher expression, whereas in seven other pairs, 1p-intact tumors had higher expression.
We addressed the candidacy of DFFB as an oligodendroglioma tumor suppressor gene by mutation analysis. Twelve tumors with 1p-allelic loss were screened for mutations by sequencing the 1.2 kb coding region of DFFB. No coding region mutations were detected in any of the samples, which may indicate that haploinsufficiency of DFFB is enough of a genetic insult to contribute to tumorigenesis. In order to thoroughly test this hypothesis, it will be necessary to further investigate DFFB, perhaps by determining if the DFFB promoter has been hypermethylaed and/or if intronic sequence has been mutated in tumor samples.
DFFB-null mouse lines have been established via gene targeting [15]. Resultant mice developed normally but their lymphocytes were more susceptible to DNA damage. These animal model experiments suggest that DFFB is a weak tumor suppressor, which may only manifest its function in the presence of stress and DNA damage. Brain tissue samples from three six-month-old specimens revealed neuropil spongiosis, but no tumor development was observed (data not shown). We are not clear at present the implication of the neuropil spongiosis phenotype. Consistent with our data, a sequencing effort in a neuroblastoma study did not reveal a tumor-specific mutation in DFFB [16]. The gene expression level of DFFB was not analyzed in that study.
Thus, this study revealed that attenuated expression of the DFFB gene is a signature of oligodendrogliomas with 1p-allelic loss. Since DFFB contributes to both chromosomal condensation and DNA degradation during apoptosis, decreased expression of DFFB may subject cells to DNA damage stresses, which in turn may contribute to both tumorigenesis and better response to DNA damaging chemotherapy. Further studies are needed to investigate the role of the DFFB gene in the etiology of oligodendroglioma.
Acknowledgments
Acknowledgements
This work was partially supported by a gift from the "Anthony Bullock III Research fund".
Contributor Information
J Matthew McDonald, Email: jonmatth@mdanderson.org.
Valerie Dunmire, Email: VDunmire@Houston.rr.com.
Ellen Taylor, Email: eetaylor@mdanderson.org.
Raymond Sawaya, Email: rsawaya@mdanderson.org.
Janet Bruner, Email: jbruner@mdanderson.org.
Gregory N Fuller, Email: gfuller@mdanderson.org.
Kenneth Aldape, Email: kaldape@mdanderson.org.
Wei Zhang, Email: wzhang@mdanderson.org.
References
- Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145:1175–1190. [PMC free article] [PubMed] [Google Scholar]
- Bello MJ, Vaquero J, de Campos JM, Kusak ME, Sarasa JL, Saez-Castresana J, Pestana A, Rey JA. Molecular analysis of chromosome 1 abnormalities in human gliomas reveals frequent loss of 1p in oligodendroglial tumors. Int J Cancer. 1994;57:172–175. doi: 10.1002/ijc.2910570207. [DOI] [PubMed] [Google Scholar]
- Bello MJ, Leone PE, Vaquero J, de Campos JM, Kusak ME, Sarasa JL, Pestana A, Rey JA. Allelic loss at 1p and 19q frequently occurs in association and may represent early oncogenic events in oligodendroglial tumors. Int J Cancer. 1995;64:207–210. doi: 10.1002/ijc.2910640311. [DOI] [PubMed] [Google Scholar]
- Kraus JA, Koopmann J, Kaskel P, Maintz D, Brandner S, Schramm J, Louis DN, Wiestler OD, von Deimling A. Shared allelic losses on chromosomes 1p and 19q suggest a common origin of oligodendroglioma and oligoastrocytoma. J Neuropathol Exp Neurol. 1995;54:91–95. doi: 10.1097/00005072-199501000-00011. [DOI] [PubMed] [Google Scholar]
- Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G, Hosek SM, Kimmel D, O'Fallon J, Yates A, Feuerstein BG, Burger PC, Scheithauer BW, Jenkins RB. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18:4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- Husemann K, Wolter M, Buschges R, Bostrom J, Sabel M, Reifenberger G. Identification of two distinct deleted regions on the short arm of chromosome 1 and rare mutation of the CDKN2C gene from 1p32 in oligodendroglial tumors. J Neuropathol Exp Neurol. 1999;58:1041–1050. doi: 10.1097/00005072-199910000-00002. [DOI] [PubMed] [Google Scholar]
- Dong SM, Pang JC, Poon WS, Hu J, To KF, Chang AR, Ng HK. Concurrent hypermethylation of multiple genes is associated with grade of oligodendroglial tumors. J Neuropathol Exp Neurol. 2001;60:808–816. doi: 10.1093/jnen/60.8.808. [DOI] [PubMed] [Google Scholar]
- Dong Z, Pang JS, Ng MH, Poon WS, Zhou L, Ng HK. Identification of two contiguous minimally deleted regions on chromosome 1p36.31-p36.32 in oligodendroglial tumours. Br J Cancer. 2004;91:1105–1111. doi: 10.1038/sj.bjc.6602093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res. 2005;11:1119–1128. [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Moore DH, Ginzinger DG, Godfrey TE, Barclay J, Powell B, Pinkel D, Zaloudek C, Lu K, Mills G, Berchuck A, Gray JW. An approach to analysis of large-scale correlations between genome changes and clinical endpoints in ovarian cancer. Cancer Res. 2000;60:5382–5385. [PubMed] [Google Scholar]
- Ginzinger DG, Godfrey TE, Nigro J, Moore DH, Suzuki S, Pallavicini MG, Gray JW, Jensen RH. Measurement of DNA copy number at microsatellite loci using quantitative PCR analysis. Cancer Res. 2000;60:5405–5409. [PubMed] [Google Scholar]
- Nigro JM, Takahashi MA, Ginzinger DG, Law M, Passe S, Jenkins RB, Aldape K. Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay. Am J Pathol. 2001;158:1253–1262. doi: 10.1016/S0002-9440(10)64076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Edgar R, Federhen S, Helmberg W, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Suzek TO, Tatusova TA, Wagner L. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004 doi: 10.1093/nar/gkh073. 32 (Database issue):D35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawane K, Fukuyama H, Yoshida H, Nagase H, Ohsawa Y, Uchiyama Y, Okada K, Iida T, Nagata S. Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nat Immunol. 2003;4:138–144. doi: 10.1038/ni881. [DOI] [PubMed] [Google Scholar]
- Judson H, van Roy N, Strain L, Vandesompele J, Van Gele M, Speleman F, Bonthron DT. Structure and mutation analysis of the gene encoding DNA fragmentation factor 40 (caspase-activated nuclease), a candidate neuroblastoma tumour suppressor gene. Hum Genet. 2000;106:406–413. doi: 10.1007/s004390000257. [DOI] [PubMed] [Google Scholar]