Abstract
The transition to flowering involves major changes in the shoot apical meristem and in the fate of existing leaf primordia. Transcripts of the Arabidopsis thaliana flowering-promoting gene FLOWERING LOCUS T (FT) are present in leaf tissue but can also promote flowering when artificially introduced into the meristem. FT may normally act in the leaf and/or the meristem, initiating or constituting a mobile flower-promoting signal. We studied FT-dependent events in the rosette leaf, some of which might precede or mimic events in the meristem and its primordia. We show FT-dependent transcript accumulation of the MADS box transcription factors FRUITFULL (FUL) and SEPALLATA3 (SEP3) in leaves. Abnormally high levels of FT further increase the expression of these genes, leading to morphological changes in the leaves. Loss of the flowering-time gene FD, as well as environmental conditions that delay flowering, reduce FT's effect on leaves via reduced activation of its targets. FUL, SEP3, and APETALA1 accumulation in the meristem is associated with and contributes to the transition to flowering. We propose that FT functions through partner-dependent transcriptional activation of these and as-yet-unknown genes and that this occurs at several sites. Organ fate may depend on both degree of activation and the developmental stage reached by the organ before activation occurs.
INTRODUCTION
Many plants time their reproductive stage to a season with favorable climate. Correct timing of this transition is an important adaptive trait to adverse environmental conditions. One environmental cue is the gradual change in daylength (photoperiodism) that occurs in nonequatorial regions. Some plants flower as days become shorter, whereas others respond to increasing photoperiods. Daylength is perceived by the leaves (Knott, 1934), which when induced produce an unknown flowering stimulus (florigen; Chailakhyan, 1936). This graft-transmissible systemic signal is thought to move from the induced leaf to the meristem (Zeevaart, 1976). Much of our current molecular understanding of the control of flowering time was obtained in the model plant Arabidopsis thaliana (Boss et al., 2004; Hayama and Coupland, 2004; Sung and Amasino, 2004). Arabidopsis grown under long-day conditions flowers earlier and with fewer leaves compared with short-day conditions. In Arabidopsis, increasing daylength leads to the accumulation of the transcription factor CONSTANS (CO; Putterill et al., 1995). The circadian clock produces daily expression rhythms of CO transcript that are modified by changes in daylength (Suarez-Lopez et al., 2001). CO protein is stabilized by blue and far-red light and is degraded in red light and in the dark (Valverde et al., 2004). High levels of transcript during the light hours occur only under long days, allowing accumulation of sufficient levels of CO protein required for flowering. CO directly activates the floral integrators SUPPRESSOR OF OVEREXPRESSION OF CO 1 and FLOWERING LOCUS T (FT) (Samach et al., 2000). CO misexpression from phloem-specific promoters caused early flowering, and this induction was graft-transmissible (An et al., 2004; Ayre and Turgeon, 2004). On the other hand, CO misexpression from meristem-specific promoters did not trigger flowering, while its target FT acted in both the phloem and the meristem to trigger flowering (An et al., 2004). These experiments pinpointed the focus on the transmissible signal on events downstream of CO expression, one of them being FT transcript accumulation. It is still unclear whether FT normally acts in the leaf, the apical meristem, or both. FT transcript or protein may move or promote the formation of a downstream mobile signal. FT encodes an ∼20-kD protein (Kardailsky et al., 1999; Kobayashi et al., 1999) belonging to the CETS (CEN, TFL1, SP) family (Pnueli et al., 2001). Overexpression of FT causes early flowering (Kardailsky et al., 1999; Kobayashi et al., 1999), and loss-of-function alleles are late-flowering (Koornneef et al., 1991). The precise mode of action of CETS proteins is still unknown. Interestingly, they share similarities in sequence and protein folding structure with the mammalian RAF-kinase-inhibitor protein (RKIP) (Banfield and Brady, 2000). RKIP has been shown to play a pivotal modulatory role in several protein kinase signaling cascades (Yeung et al., 1999; Lorenz et al., 2003; Keller et al., 2004). A putative ligand binding domain and neighboring effector sites are also conserved between RKIP and CETS, suggesting that their mode of action might be similar. RKIP is also a precursor of the hippocampal cholinergic neurostimulating peptide (Tohdoh et al., 1995), although this region in RKIP does not share strong homology with CETS. Several potential protein interactors of CETS proteins have been identified using the yeast two-hybrid system (Pnueli et al., 2001). Once the decision to form flowers has been made, the development of the complex flower involves many genes in several tiers, which are switched on or off, both spatially and temporally (Zik and Irish, 2003). Indeed, using global expression analysis, hundreds of genes were shown to go through a twofold or higher change in expression in the apex during the FT-dependent transition to flowering (Schmid et al., 2003). Genetic evidence suggests that FT function is closely associated with activation of the APETALA1 (AP1) gene. FT acts redundantly with the floral integrator LEAFY (LFY) to activate AP1 transcription (Ruiz-Garcia et al., 1997), and a plant containing mutations in both FT and LFY completely lacked floral structures and AP1 expression. However, FT must have additional targets because flowering of Pro35S:FT plants is not delayed by loss of AP1 function (Kardailsky et al., 1999), and ap1 mutants are not late flowering. In Arabidopsis and most other plants, the transition to flowering also affects the fate of leaf primordia (Poethig, 2003). The last leaves produced before flower formation (cauline leaves as opposed to the earlier formed rosette leaves) acquire a different fate. Cauline leaves have certain morphological characteristics, such as reduced size and changes in trichome distribution. The meristem in the axil of a cauline leaf differentiates relatively earlier as a secondary inflorescence. Enhanced internode elongation between cauline leaves eventually leads to their appearance on the inflorescence stem. In early-flowering ecotypes exposed to long photoperiods, the fate of existing leaf primordia changes with the transition to flowering (Hempel and Feldman, 1994). Although FT is involved in promoting the decision to make flowers, FT transcript is detected in leaves and the vasculature (Takada and Goto, 2003) but not in the apical meristem (Schmid et al., 2005). FT mRNA or protein might move to the meristem to affect its targets. Alternatively, FT might act completely or partially within the leaf.
Here, we looked for FT-dependent events within the leaf. Using loss-of-function and gain-of-function approaches, we show FT-dependent accumulation of the MADS box transcription factors FRUITFULL (FUL) and SEPALLATA3 (SEP3) transcripts in mature wild-type rosette leaves. The FUL gene is highly expressed in the inflorescence meristem and in cauline leaves (Mandel and Yanofsky, 1995; Gu et al., 1998; Ferrandiz et al., 2000b; Schmid et al., 2005), and the null ful-1 allele delays flowering and increases cauline leaf size (Gu et al., 1998; Ferrandiz et al., 2000b). We show that ful-1 suppresses both flowering and cauline leaf phenotypes of transgenic plants expressing high levels of FT. We show that full FT action in the leaf is dependent on the flowering-time gene FD and that the environment can change the timing of the transition to flowering not only by modifying FT transcript levels but also by modifying FT activity. The implications of our findings on understanding the mode and site of FT action are discussed.
RESULTS
High Levels of FT Cause Growth Condition–Dependent Vegetative Phenotypes
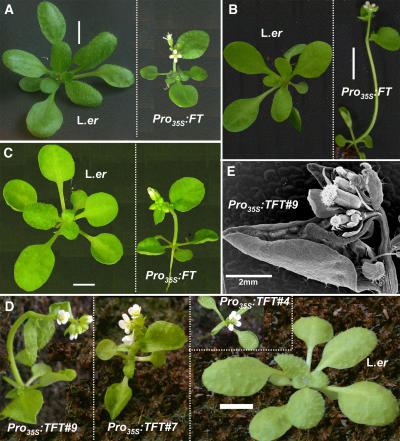
We began our study of FT-dependent events within the leaf by examining vegetative phenotypes of plants overexpressing FT. We used a Pro35S:FT transgene originally introduced into Columbia (Col) but introgressed into the Landsberg erecta (Ler) background by five backcrosses (YK#1-5L, kindly provided by T. Araki, Kyoto University, Kyoto, Japan). These plants flower extremely early after producing, on average, two rosette leaves and two cauline leaves in all photoperiods tested (see Supplemental Table 1 online). Flowering time is also reduced compared with similar genotype lines in the Col background (Kardailsky et al., 1999; Kobayashi et al., 1999; Abe et al., 2005). We noticed that in addition to early flowering and early termination of the inflorescence meristem, under certain growth conditions the rosette leaves were reduced in size and folded inwards and upwards (curling) along the axis of the major leaf vein (Figure 1). The degree of the phenotype was highly variable between growth conditions but fairly uniform within identical growth conditions. For example, plants grown under long days in a growth room (treatment A1 in Table 1) were not curled (Figure 1A). Substantial leaf curling occurred under blue fluorescent long days in a growth chamber (treatment A3 in Table 1, Figure 1B), and intermediate leaf curling occurred in a short-day growth chamber (treatment A4 in Table 1, Figure 1C). Once a leaf became curled, changing environmental conditions did not reverse the phenotype.
Figure 1.
Leaf Phenotypes of Pro35S:FT/TFT Plants.
(A) to (C) Growth conditions affect leaf phenotypes of Pro35S:FT plants. Wild-type and Pro35S:FT plants were grown in a long-day growth room (A1 in Table 1; 23-d-old plants; [A]), a blue long-day chamber (A3 in Table 1; 18-d-old plants; [B]), and a short-day growth chamber (A4 in Table 1; 23-d-old plants; [C]).
(D) Phenotypes of independent 22-d-old transgenic plants homozygous for Pro35S:TFT. All lines are early-flowering and small with very curled leaves.
(E) A cauline leaf and the terminal inflorescence meristem of one of the transgenic Pro35S:TFT plants viewed by a scanning electron microscope. Note the severe leaf curling and the termination of the meristem.
Dashed lines within panels border between different genotypes, grown together under identical conditions, and photographed separately at the same age. Bars = 1 cm in (A) to (D) and 2 mm in (E).
Table 1.
Degree of Leaf Curling and Different Growth Conditions
| Temperature (C°)a
|
Relative Humidity (%)a
|
Lighting
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experiment | Treatment Name | Min. | Max. | Avg. | Min. | Max. | Avg. | Daylengthb | Color | Typec | Irradianced | Leaf Curlinge | Chamberf |
| General | A1 LD Rm1 | 19.1 | 20.2 | 19 | 29.5 | 63.0 | 50 | LDs | White | F+inc | 88 | – | 1 |
| A2 LD Rm2 | 16.6 | 19.5 | 18 | 56.6 | 91.4 | 69 | LDs | White | 10F+6inc | 88 | * | 2 | |
| A3 LD blue | 21.5 | 24.5 | 23 | 56.0 | 94.6 | 72 | LDs | Blue | F | 102 | *** | 3 | |
| A4 SD | 21.1 | 22.6 | 21 | 59.3 | 89.9 | 79 | SDs | White | F | 180 | ** | 4 | |
| A5 SD blue | 22.2 | 24.7 | 23 | 68.7 | 97.3 | 85 | SDs | Blue | F | 102 | *** | 3 | |
| Temperature | B1 SD white | 22.0 | 23.6 | 23 | 73.9 | 97.0 | 88 | SDs | White | F | 133 | ** | 3 |
| B2 SD white cold | 11.7 | 13.4 | 12 | 68.3 | 94.5 | 82 | SDs | White | F | 127 | – | ||
Temperature and relative humidity values shown are minimum (min.), maximum (max.), and average (avg.).
Daylength terms: LDs are long days (16/8 day/night photoperiods). SDs are short days (8/16 day/night photoperiods).
Lighting type is either fluorescent (F), flourescent light supplemented by 8 h of incandescent light in the end of the day (F+inc), or 10 h of fluorescent light followed by 6 h of incandescent light in the end of the day(10F+6inc).
Irradiance was measured in microEinsteins per second per square meter (see Methods).
Degree of leaf curling of Pro35S:FT was given a semiquantitative measure using asterisks, with highest curling receiving three asterisks, medium curling receiving two asterisks, low curling receiving one asterisk, and no curling receiving –.
Chambers: 1, growth room; 2, growth room 2; 3, TC16 Conviron chamber; 4, AR95L Percival Arabidopsis growth chamber.
Functional analysis in Arabidopsis of the FT-like tomato (Lycopersicon esculentum) gene SP3D (Carmel-Goren et al., 2003) provided similar phenotypes. SP3D encodes a protein homologous to FT (see Supplemental Figure 1 online), which causes early flowering when overexpressed in tomato and was renamed Tomato FT (TFT; E. Lifschitz, personal communication). We transformed Arabidopsis (Ler) with a Pro35S:TFT construct (generously provided by E. Lifschitz; see Methods). Nine independent transgenic lines all showing very early, day-length-insensitive flowering (Table 2), severe leaf curling, and reduced leaf size (Figures 1D and 1E) were obtained. Rosette and cauline leaf curling in different Pro35S:TFT lines was generally more severe than in Pro35S:FT plants but was still dependent on growth conditions. Line Pro35S:TFT#7 was chosen for further genetic analysis, since it showed the highest expression of the TFT transgene (see Supplemental Figure 2 online). These results indicate that leaf curling and leaf size reduction are common responses to abnormally high levels of FT/TFT activity in leaves.
Table 2.
Flowering Time of Transgenic Lines Overexpressing FT or TFT (SP3D)
| Rosette
|
Cauline
|
Total
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Daylength | Genotype | Avg.a | se | Rng.b | Avg. | se | Rng. | Avg. | se | Rng. | No. of Rpts.c |
| Long daysd | Ler | 8.43 | 0.20 | 8–9 | 3.57 | 0.30 | 3–5 | 12.00 | 0.22 | 11–13 | 7 |
| Pro35S:TFT#7 | 2.00 | 0.00 | 2 | 4.00 | 0.26 | 3–5 | 6.00 | 0.26 | 5–7 | 10 | |
| Pro35S:TFT#9 | 2.33 | 0.14 | 2–3 | 3.17 | 0.19 | 2–4 | 5.50 | 0.17 | 4–7 | 18 | |
| Pro35S:TFT#4 | 3.00 | 0.23 | 2–4 | 0.92 | 0.14 | 0–2 | 3.92 | 0.24 | 3–5 | 13 | |
| Pro35S:FT | 2.00 | 0.00 | 2 | 2.00 | 0.00 | 2 | 4.00 | 0.00 | 4 | 9 | |
| Short dayse | Ler | 30.44 | 0.56 | 29–33 | 8.89 | 0.45 | 6–10 | 39.33 | 0.65 | 37–43 | 9 |
| Pro35S:TFT#7 | 3.50 | 0.17 | 2–4 | 1.86 | 0.10 | 1–2 | 5.36 | 0.17 | 4–6 | 14 | |
| Pro35S:TFT#9 | 3.06 | 0.10 | 2–4 | 1.76 | 0.14 | 1–3 | 4.82 | 0.13 | 4–6 | 17 | |
| Pro35S:TFT#4 | 3.00 | 0.00 | 3 | 0.36 | 0.13 | 0–1 | 3.36 | 0.13 | 3–4 | 14 | |
| Pro35S:FT | 2.00 | 0.00 | 2 | 1.70 | 0.15 | 1–2 | 3.70 | 0.15 | 3–4 | 10 | |
Avg., average.
Rng., range.
No. of rpts., number of plants.
Long days: 16/8 day/night photoperiods.
Short days: 8/16 day/night photoperiods.
Expression of SEP3 and FUL in Leaves Is Dependent on FT Levels
We looked for genes that are regulated by FT within rosette-leaf tissue. Here, we took a candidate gene approach by studying the expression of specific genes encoding MADS box transcription factors that had been shown (using loss-of-function approaches) to play a role in inflorescence and flower meristem and organ identity. The genes tested were AGAMOUS (AG), AP1, AP3, CAULIFLOWER (CAL), FUL, and SEP3. CAL and FUL plays a redundant role with AP1 in flower meristem identity (Ferrandiz et al., 2000b), while AG, AP3, and SEP3 are highly expressed in TERMINAL FLOWER 2 mutants that misexpress FT (Kotake et al., 2003). All these genes are upregulated in the apical meristem upon the transition to flowering, and this increase is delayed in an ft mutant (Schmid et al., 2003).
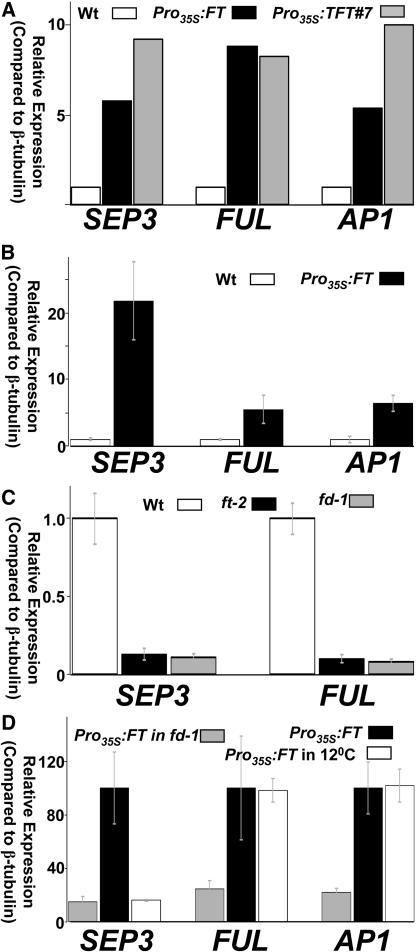
Initially, we compared expression in the seedlings of different transgenic and wild-type lines grown under conditions that promote strong curling (treatment A3 in Table 1). We avoided collecting inflorescence tissue (in early-flowering transgenics) by sampling 6-d-old seedlings. In these seedlings, the AP1, FUL, and SEP3 genes were highly misexpressed in the transgenic lines (Figure 2A; biological repeat in Supplemental Figure 3A online), whereas no significant misexpression of CAL, AP3, or AG could be detected (data not shown). We then compared expression of AP1, FUL, and SEP3 in mature rosette leaves of wild-type plants or those overexpressing FT (Figure 2B) or TFT (see Supplemental Figure 3B online). Plants were grown for 30 d under conditions that enhance curling (conditions A2 and A4 in Table 1, respectively). All three genes were highly misexpressed in the rosette leaves of both transgenic lines. Fold induction ranged between 5.4 (FUL) and 21 (SEP3) for Pro35S:FT plants grown under long days.
Figure 2.
SEP3 and FUL Are Activated by FT in Leaves.
Expression was measured by quantitative real-time RT-PCR (see Methods). Expression of each gene was calculated relative to β-TUBULIN (AT5G62690). Bars in (B) to (D) show standard error of the mean of three biological repeats. The experiment in (A) was performed at least twice, with similar results (see Supplemental Figure 3 online).
(A) Comparison of 6-d-old seedlings from different genotypes grown under blue long days (A3 in Table 1).
(B) Expression in rosette leaves of 30-d-old wild-type and Pro35S:FT plants grown under long days (A2 in Table 1).
(C) Expression in rosette leaves of 30-d-old wild-type, ft-2, or fd-1 plants grown under long days (A2 in Table 1). Expression of AP1 is not shown due to low levels and high variance among samples.
(D) Expression in rosette leaves of 30-d-old Pro35S:FT plants with or without a mutation in FD, grown under long days at 18°C (A2 in Table 1) or Pro35S:FT plants grown at 12°C.
FT is normally expressed in rosette leaves (Takada and Goto, 2003) as well as in modified leaves such as cauline leaves and sepals (Schmid et al., 2005). Here, we introduced abnormally higher levels of FT in the leaves and perhaps into cells within the leaf in which FT is not normally expressed. FT activity under these conditions might not reflect its normal functions. FUL and SEP3, although predominantly expressed in the meristem and floral tissue, are also detected above baseline levels at specific stages of rosette/cauline leaf development. In fact, the organs showing the highest levels of FUL expression are cauline leaves (Schmid et al., 2005). To determine whether FT is normally involved in the regulation of these genes in rosette leaves, we compared their expression in rosette leaves of wild-type and ft-2 mutant plants grown under long days (condition A2 in Table 1). Using real-time PCR, we detected clear above-background signals in wild-type leaves of both SEP3 and FUL. In rosette leaves from 19-d-old plants, we detected a slight reduction in the gene expression of SEP3 and FUL in ft-2 mutants (data not shown). In 30-d-old rosette leaves, we detected a consistent 10-fold decrease in the expression of SEP3 and FUL in ft-2 mutants (Figure 2C). Thus, FT is normally required for SEP3 and FUL accumulation in mature rosette leaves.
FT-Dependent Changes in Gene Expression and Morphology of Leaves Require the Flowering-Time Gene, FD
The flowering time gene FD was assigned to the same group as FT, since plants carrying mutations in either gene are late-flowering under long days and do not exhibit an increased response to vernalization (Koornneef et al., 1991). FD was recently shown to encode a basic region/leucine zipper transcription factor that interacts with FT (Abe et al., 2005; Wigge et al., 2005). We therefore asked whether FD is required for FT-dependent accumulation of SEP3 and FUL in mature rosette leaves. Indeed, the fd-1 mutation had an effect similar to that of ft-2 in reducing the expression of these genes in mature rosette leaves (Figure 2C).
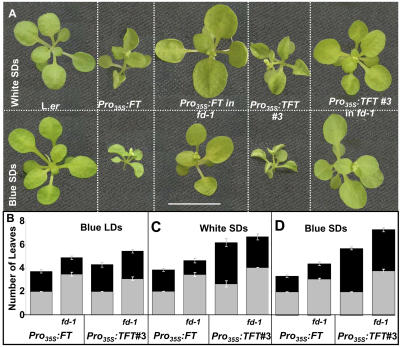
Suppression of Pro35S:FT early flowering by fd-1 has been reported by others (Abe et al., 2005; Wigge et al., 2005). We screened an array of different levels of TFT in an fd-1 mutant (see Methods) and introduced the Pro35S:FT line into an fd-1 background. In all cases, loss of FD fully suppressed leaf curling and leaf-size reduction (Figure 3A), yet caused a relatively slight delay in flowering time of Pro35S:FT/TFT transgenics (Figures 3B to 3D).
Figure 3.
Interaction of Pro35S:FT/TFT Plants with fd-1.
(A) The fd-1 mutation suppresses leaf phenotypes of Pro35S:FT/TFT plants. Phenotypes of 21-d-old plants grown under white (top row, A4 in Table 1) or blue (bottom row, A5 in Table 1) short days (SDs). Dashed lines within panels border between different genotypes, grown together under identical conditions, and photographed separately at the same age. Bar = 1 cm.
(B) to (D) Flowering time of Pro35S:FT plants and Pro35S:TFT#3 plants in a wild-type or fd-1 homozygous mutant background. The different growth conditions, blue long days (LDs; A3), white short days (A4), and blue short days (A5), are depicted in each panel. Total leaf number of wild-type plants (data not shown) is 7 in A3 (see Figure 6C) and >30 in A4 (Table 2; see Supplemental Table 1 online) and A5. Under all conditions, a mutation in FD suppresses early flowering by 0.5 to 1.5 leaves. The major effect of FD is on rosette leaf number. The difference between the Pro35S:FT and Pro35S:TFT#3 plants becomes more clear under white short days, perhaps due to lack of internal FT under these conditions. Flowering time is measured by counting rosette (gray) and cauline (black) leaves. Mean leaf number is shown ± se (n = 5 to 22).
Pro35S:FT/TFT in fd-1 plants showed early flowering with abnormally large leaves, a unique phenotype for early-flowering plants. This suggested that with the loss of FD, leaf fate is no longer affected by high levels of FT (see Discussion). We then asked whether the changes in leaf phenotype correlate with reduced FT-dependent gene activation in rosette leaves. A mutation in FD caused a significant reduction in the expression of all three genes (Figure 2D; see Supplemental Figure 3C online). Loss of FD in the Pro35S:FT background reduced target gene expression at least fourfold. Thus, a mutation in FD reduced target gene expression in rosette leaves of both wild-type and Pro35S:FT plants. In the wild-type background, this change in gene expression within the leaf could be interpreted as an aftereffect of the delayed transition to flowering caused by fd-1. In the Pro35S:FT background, the reduction in gene expression suggests a more direct involvement of FD in the regulation of these genes in leaves. The early-flowering Pro35S:FT/TFT in fd-1 plants still express abnormally high levels of all three target genes in leaves (Figure 2D; see Supplemental Figure 3C online) and young seedlings (data not shown).
Temperature-Dependent FT Activation of SEP3
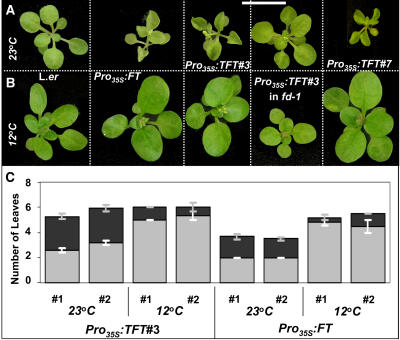
Loss of FD reduced target gene expression and leaf curling, suggesting a possible correlation and perhaps even a cause-and-effect relationship between the two phenomena. To further test this correlation, we grew Pro35S:TFT#3 plants under conditions that either enhance (short days, A4 in Table 1) or reduce (long days, A1 in Table 1) curling; indeed, we detected a significant reduction in SEP3, FUL, and AP1 expression in leaves from plants grown under noncurling conditions (see Supplemental Figure 3D online). We noticed reduced curling in growth chambers with lower ambient temperatures. Reduced ambient temperatures delay flowering time in Arabidopsis (Blazquez et al., 2003), although this delay was not attributed to reduced FT function. To test the effect of temperature, we grew Pro35S:FT/TFT plants at 12 or 23°C under otherwise identical short-day growth conditions (Table 1, B treatments; Figure 4). Significantly, low temperature influenced four parameters of FT overexpression: leaf curling, reduction in leaf size (Figures 4A and 4B), and early termination of the meristem and flowering time (Figure 4C). Pro35S:FT/TFT plants grown at 12°C had much larger leaves with no sign of curling (Figures 4A and 4B). At 23°C, termination of the inflorescence meristem occurred, on average, after 6.7 and 7.5 flowers in Pro35S:TFT#3 and Pro35S:FT plants, respectively. At 12°C, termination was severely delayed with >14 and 36 flowers in Pro35S:TFT#3 and Pro35S:FT plants, respectively. Both Pro35S:TFT#3 and Pro35S:FT plants produced significantly more rosette leaves at 12°C (Figure 4C). When measuring total leaf number, the Pro35S:FT plants were significantly late-flowering at 12°C, suggesting that temperature affects a common mechanism.
Figure 4.
Effect of Growth Temperature on FT/TFT Function.
(A) and (B) Plants of different genotypes were grown at 23°C (A) or at 12°C (B) under otherwise similar short-day growth conditions (Table 1, B treatments). While low temperatures slightly increased leaf size of wild-type (Ler) plants, it severely increased leaf size and eliminated leaf curling of Pro35S:FT, Pro35S:TFT#3, and Pro35S:TFT#7. Note that cold temperatures had no noticeable effect on Pro35S:TFT#3 in fd-1 plants, besides a reduction in petiole length. Plants in (A) are 21 d old, and in (B), they are 26 d old. Dashed lines within panels border between different genotypes, grown together under identical conditions, and photographed separately at the same age. Bar = 1 cm.
(C) Flowering time of Pro35S:FT and Pro35S:TFT#3 plants at different temperatures. Flowering time is measured by counting rosette (gray) and cauline (black) leaves. Both genotypes responded to lower temperatures by an increase in rosette leaf number. Since no corresponding reduction in cauline leaf number occurred in Pro35S:FT plants, flowering was significantly delayed. #, repeat number of experiment. Mean leaf number is shown ± se (n = 5 to 22).
We asked whether changes in ambient temperature affect target gene expression. Here, for the first time in our experiments, we detected a major difference in the responses of SEP3 and FUL. Reducing the temperature had a specific effect on SEP3 expression, without any noticeable reduction in that of AP1 or FUL (Figure 2D). Thus, temperature might affect the ability of FT to activate SEP3 and seemingly not the two other genes we identified. Of course, activation of other, as-yet-unknown targets of FT may also be regulated by temperature.
A Mutation in SEP3 Suppresses Pro35S:FT/TFT Leaf Phenotypes
Our results highlighted the SEP3 gene as a likely target of FT action in the leaf. SEP3 expression also showed the highest correlation with leaf curling in transgenic Pro35S:FT/TFT plants. We asked whether a mutation in SEP3 would suppress leaf curling in Pro35S:FT/TFT plants. In fact, curling of Pro35S:FT/TFT leaves was completely suppressed by replacing even one wild-type allele with the null sep3-2 allele (Col-0; Pelaz et al., 2001). Leaves of Pro35S:FT/+; sep3-2/+ and Pro35S:TFT#7/+; sep3-2/+ plants were completely uncurled and larger under conditions that promote curling (Figures 5A and 5B).
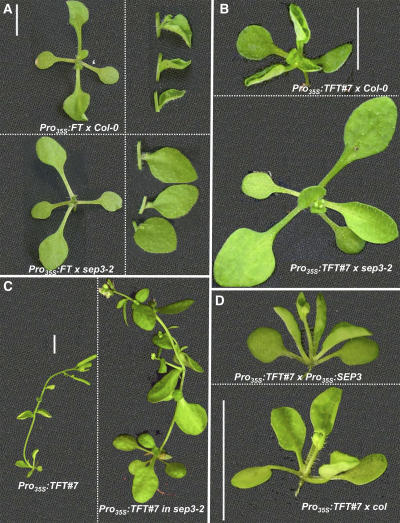
Figure 5.
SEP3 Is the Rate-Limiting Factor in FT/TFT-Dependent Leaf Curling.
(A) Phenotypes of 14-d-old F1 plants grown under blue long days (A3 in Table 1) from a cross between Pro35S:FT and sep3-2 (in Col-0) or Col-0. Loss of leaf phenotypes are noticed in both rosette (left) and cauline (right) leaves.
(B) Phenotypes of 14-d-old F1 plants grown under the same conditions as in (A), from a cross between Pro35S:TFT#7 and sep3-2 or Col-1. Note the complete absence of leaf curling by losing one allele of SEP3 under conditions that normally cause strong curling.
(C) Phenotypes of 35-d-old plants grown under short days (A4 in Table 1) homozygous for Pro35S:TFT#7 or Pro35S:TFT#7 in sep3-2.
(D) Phenotypes of 15-d-old F1 plants grown under short days (A4 in Table 1). Note the increase in curling in the cross to Pro35S:SEP3.
Dashed lines within panels border between different genotypes, grown together under identical conditions, and photographed separately at the same age. All bars = 1 cm.
In the F2 generation of the cross between Pro35S:TFT#7 and sep3-2, only 19.4% of the plants had curled leaves, proving that leaf curling requires two wild-type copies of SEP3 (see Supplemental Table 2 online). As expected, a line homozygous for Pro35S:TFT#7 in sep3-2 also had uncurled leaves (Figure 5C).
High levels of SEP3 alone (Pro35S:SEP3 plants) are not sufficient for severe curling under our growth conditions (data not shown), suggesting that SEP3 is not the only component required for leaf curling. Introducing even higher levels of SEP3 into Pro35S:TFT#7 plants increased the degree of curling (Figure 5D), suggesting that SEP3 levels are rate-limiting in leaf curling.
While no late-flowering phenotype was described for the sep3-2 mutant in a wild-type background, loss of SEP3 caused a slight yet significant delay in the Pro35S:TFT#7 background (Figures 5C, 6A, and 6B). This delay required the absence of both SEP3 copies: it was first noticed in a segregating F2 population (Figure 6A) and later confirmed in a homozygous line for Pro35S:TFT#7 in sep3-2 (Figure 6B). The effect of sep3-2 was partially masked by the fact that Col-0 seems to carry a recessive suppressor of Pro35S:TFT early flowering (Figure 6A; see Supplemental Table 2 online). A suppressor of Pro35S:FT in Col has also been found by others (Y. Kobayashi and T. Araki, personal communication). Under the growth conditions tested, the sep3-2 allele did not significantly suppress Pro35S:FT early flowering (data not shown).
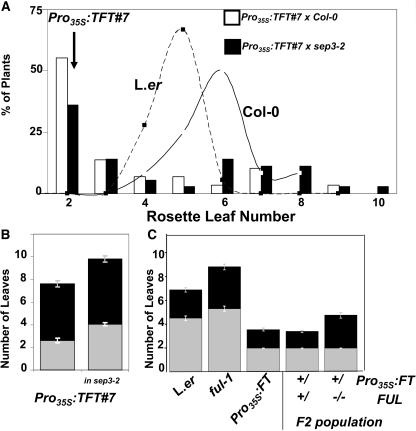
Figure 6.
Flowering-Time Phenotypes Caused by Mutations in SEP3 and FUL.
(A) Flowering time under blue long days (A3 in Table 1) of segregating F2 populations from a cross of Pro35S:TFT#7 to Col-0 or to sep3-2. Graph shows percentage of plants flowering with different amounts of rosette leaves. Segregation of both Ler and Col-0 is shown as a line graph. Arrow shows that all Pro35S:TFT#7 (in Ler) plants flowered with two rosette leaves. In the segregating F2 population of the control cross to Col-0, 55%, rather than the expected 75%, of the plants flowered with less than three rosette leaves (see Supplemental Table 3 online). A recessive suppressor should reduce the ratio of early-flowering plants to 56% [0.75(Pro35S:TFT+/+;+/−) × 0.75(Ler +/+;+/−)]. Above the noise created by the Col-0 background, loss of SEP3 clearly affected flowering time of the segregating Pro35S:TFT population. The percentage of plants that flowered with less than three rosette leaves was reduced from 55 to 36%. The average number of rosette leaves increased by 1.5 leaves in the whole population and from 2.36 to 3.37 in the earliest 75th percentile (likely including at least one copy of Pro35S:TFT).
(B) Flowering time of a line homozygous for both Pro35S:TFT#7 and sep3-2 under short days (A4 in Table 1). Plants are late-flowering compared with Pro35S:TFT#7. See Figure 5C for picture of plants.
(C) A mutation in FUL delays flowering in a wild-type and Pro35S:FT background. Flowering time of wild-type, ful-1, Pro35S:FT, or F2 plants from a cross of Pro35S:FT to ful-1. Early-flowering plants containing at least one copy of Pro35S:FT (+/) were separated to those containing (+/) or not containing (−/−) an intact FUL allele. The latter were identified by their distinct ful-1 silique phenotype and verified by PCR. Plants were grown under the same conditions as in (A). See Figure 7D for additional phenotypes of these plants. Mean leaf number is shown ± se (n = 5 to 46).
Higher Levels of AP1 Enhance Flowering and Meristem Termination of Pro35S:TFT
Unlike mutations in SEP3, loss of AP1 did not suppress leaf curling of the Pro35S:TFT#7 line (Figure 7A). Introducing even higher levels of AP1 (Pro35S:AP1 line #563.CI1.5; Liljegren et al., 1999) into the Pro35S:TFT#7 background severely increased leaf curling (Figure 7B). AP1 is therefore not essential for leaf curling but can synergistically contribute to its severity.
Figure 7.
Interactions with AP1 and FUL.
(A) Phenotypes of 35-d-old F3 plants grown under short days (A4 in Table 1) from a cross between Pro35S:TFT#7 and ap1-7. Both plants are early-flowering, and the plant on the left is homozygous for ap1-7 (see floral phenotype). Leaves of both plants are curled.
(B) Phenotypes of 23-d-old plants grown under similar conditions. The inflorescence meristem in Pro35S:TFT#7 × Pro35S:AP1 F1 plants is completely transformed into a single flower, and leaves are severely curled.
(C) GUS staining of first rosette leaves from 19-d-old plants (containing the ful-1 allele) grown under blue long days (A3 in Table 1). Overexpression of FT causes higher expression of the FUL promoter in leaves.
(D) Phenotypes of 18-d-old plants grown under blue long days. The ful-1 mutation causes late flowering and abnormally large cauline leaves in a wild-type and Pro35S:FT background.
Dashed lines within panels border between different genotypes, grown together under identical conditions, and photographed separately at the same age. Bars = 1 cm.
There is likely to be a degree of functional redundancy among FT targets, since a mutation in AP1 does not suppress early flowering of Pro35S:FT plants (Kardailsky et al., 1999). We found that the strong ap1-7 (Bowman et al., 1993) Col allele did not cause a significant delay in flowering in the Pro35S:TFT#7 line (Figure 7A). On the other hand, higher levels of AP1 accelerated Pro35S:TFT#7 flowering time (Figure 7B). The amount of rosette leaves decreased, partially due to elongation between the cotyledon and the first leaf, in many of the plants. In addition, meristem termination was strongly enhanced by introducing Pro35S:AP1. Inflorescences were replaced by a single flower (Figure 7B). Thus, AP1 might be a rate-limiting factor in FT-dependent early meristem termination.
A Mutation in FUL Suppresses Pro35S:FT Leaf and Flowering-Time Phenotypes
The ful-1 loss-of-function allele is caused by the insertion of a DsE transposable enhancer trap element into the 5′ untranslated leader of the FUL gene (Gu et al., 1998). The transposable element contains a β-glucuronidase (GUS) reporter gene so that GUS activity is found in cells that normally transcribe FUL (Mandel and Yanofsky, 1995; Gu et al., 1998). Plants homozygous or heterozygous for the ful-1 allele showed clear GUS expression in the vasculature of rosette leaves grown under blue long days (Figure 7C). A similar pattern, under other growth conditions, has been previously shown (Gu et al., 1998). We crossed Pro35S:FT into the ful-1 background, and in the F1 generation, we could observe a severe FT-dependent increase in GUS expression in the youngest rosette leaves (Figure 7C), confirming our direct analysis of FUL expression.
A mutation in FUL delays flowering under continuous light (Ferrandiz et al., 2000b) and, as we show here, under blue long days (Figures 6C and 7D). Flowering time of Pro35S:FT was clearly delayed by loss of FUL. In a segregating F2 population, plants containing the Pro35S:FT construct that were homozygous for ful-1 (see Methods) flowered after producing an additional cauline leaf (Figures 6C and 7D). A mutation in FUL increases cauline leaf size (Gu et al., 1998). The ful-1 allele clearly suppressed the reduction in cauline leaf size caused by Pro35S:FT. Pro35S:FT in ful-1 cauline leaves were abnormally large, though curling of rosette leaves was not significantly reduced (Figure 7D). These results suggest that FT, through FD, may promote flowering and changes in leaf fate via upregulation of FUL.
DISCUSSION
FT is a potent promoter of the transition to flowering, as demonstrated by the rapid transition to flowering of Pro35S:FT plants under noninductive environmental conditions. FT, acting through transcription factors, is likely to affect transcription of select genes, which together initiate a cascade of events leading to FT-dependent transcriptional changes in hundreds of genes within the apex (Schmid et al., 2003). Here, we provide evidence suggesting that FT promotes flowering through transcriptional activation of FUL, SEP3, and AP1. Although the expression of these genes has been previously shown to rise in the meristem with the transition to flowering, FT transcript has only been detected in the vasculature of leaves. Here, we report an FT-dependent rise in expression of FUL and SEP3 in rosette leaves. This raises the possibility that FT promotes flowering by regulating the expression of genes within leaves. The FUL- and SEP3-dependent reduction in leaf size, caused by FT and FD, suggests that these genes may also take part in changing the fate of leaf primordia during the transition to flowering.
Where Does FT Promote Flowering?
We detected FT-dependent accumulation of SEP3 and FUL in mature rosette leaves. FT and FUL expression has been detected in rosette-leaf vascular tissues (Gu et al., 1998; Takada and Goto, 2003). Introducing abnormally high levels of FT severely increases the levels of SEP3, FUL, and AP1 genes in rosette leaves. Using a gain-of-function approach, we magnified events that normally happen in certain leaf cells (SEP3 and FUL induction) and other events that may not (AP1 induction). If FT normally causes a local change in transcripts in cells within the leaf, how does this affect flowering time? FT might move to the meristem and cause similar changes there or might remain in the phloem cells, while one of its first or downstream targets moves. This remains a fascinating puzzle that is definitely worth solving. Although it is now clear that FT is expressed and acts in leaves, we cannot rule out a precedent action within the meristem.
A Role for FT and FD in Phase Change?
Normally, early flowering is associated with small leaves. An association between ontogenetic changes in vegetative metamers (heteroblasty) during plant development and the transition to the reproductive stage was noted and documented a century ago, but the link between these two processes is still unclear (Goebel, 1900; Jones, 1999). In Arabidopsis, long days reduce both flowering time and rosette leaf size. Most late-flowering mutants, including ft and fd loss-of-function mutants, have larger rosette leaves. In early-flowering ecotypes exposed to long photoperiods, the fate of existing leaf primordia is changed with the transition to flowering (Hempel and Feldman, 1994). It appears as if a signal is sent downward from the shoot apical meristem to already-existing leaf primordia, and this message gets diluted with distance or with the developmental stage of the leaf, so that it only affects some of them. As a result, one can observe basipetal (top to base) initiation of inflorescence shoots from the axils of these leaves that have acquired a cauline fate (Hempel and Feldman, 1994).
Pro35S:FT in fd-1 plants present a unique phenotype, since they are early-flowering with relatively large rosette and cauline leaves. It appears that the leaf primordia in this genotype lack the competence to respond to the previously suggested downward signal. Mutations in SEP3 significantly increased rosette leaf size of Pro35S:FT plants, and mutations in FUL significantly increased cauline leaf size in those plants. Since FUL normally accumulates in cauline leaves, and mutations in FUL increase cauline leaf size in wild-type plants (Gu et al., 1998), our results suggest that FT might act via FD to reduce cauline leaf size by increasing FUL expression.
Recently, several examples of a two-phase transition, with the production of cauline leaves preceding any production of flowers, have been demonstrated in certain Arabidopsis genotypes and environmental conditions (Suh et al., 2003). We noticed that cases described in that report were associated with reduced levels of FT activity. A two-phase transition was found in short-day-grown plants and in long-day-grown plants containing the fwa-2 allele (a dominant inhibitor of FT function; Kardailsky et al., 1999; Kobayashi et al., 1999) or overexpressing the TFL1 protein (Ratcliffe et al., 1998), a CETS protein with an antagonistic affect on FT (Kobayashi et al., 1999). It is therefore possible that the ability to change the fate of existing leaf primordia depends on the joint action of FT and FD.
The Dependence of FT on FD in Flowering
The recently cloned FD gene encodes a basic region/leucine zipper transcription factor that interacts with FT (Abe et al., 2005; Wigge et al., 2005). In a wild-type background, the fd-1 allele causes late flowering under long days (Koornneef et al., 1991). In a Pro35S:FT/TFT background, the fd-1 allele causes a major change in leaf morphology and a slight delay in flowering time. How can FD be essential for FT activation of flowering in a wild-type background yet partially redundant for FT action in Pro35S:FT/TFT plants?
The transition to flowering in wild-type plants can be considered a gradual, step-dependent transformation of primordia on the meristem from rosette leaf to cauline leaf to flower. FD may be required for all steps in the transition, its action on rosette/cauline leaves being less redundant than its action in the meristem (which can be replaced by other proteins). In a wild-type background, loss of FD will cause a delay in the first transition, leading to a delay in flowering time. By introducing high levels of FT directly into most cells, including the meristem, we have short circuited the normal transition. As a result, ectopically expressed FT, in the absence of FD, leads to a unique phenotype-a direct transition from very large leaves to flowers.
FUL Is a Regulator of Flowering Time
The FUL (AGL8) gene is required in several developmental processes, silique development being the most obvious one. Its role in the transition to flowering is based on loss-of-function phenotypes in a wild-type (Ferrandiz et al., 2000b) and a Pro35S:FT background (our results). Overexpression of FUL causes early flowering (Ferrandiz et al., 2000a). A role in the suppression of cauline leaf size can also be shown in a wild-type (Gu et al., 1998) and Pro35S:FT (our results) background.
FUL is expressed in the vasculature of rosette leaves (Gu et al., 1998), similar to FT. Here, we show that FUL accumulation in the leaf is dependent on FT and FD. FUL expression is much higher in cauline leaves, where relatively high levels of FT transcript are detected as well (Mandel and Yanofsky, 1995; Gu et al., 1998; Schmid et al., 2003; Zimmermann et al., 2004).
These results clearly present FUL as a target of FT function in the transition to flowering. As with FT, we still do not know where FUL acts to promote flowering. Unlike FT, FUL transcript has been reported to appear in the shoot apical meristem shortly after the transition to flowering in a broad range of cells (Mandel and Yanofsky, 1995; Hempel et al., 1997; Schmid et al., 2003). How FT regulates this accumulation remains unknown.
FT Regulates SEP3 Accumulation in Rosette Leaves
Expression of the SEP3 gene in mature rosette leaves is regulated by FT and FD. In a Pro35S:FT background, loss of FD reduced SEP3 expression, and regulation of SEP3 seems to be affected by an additional factor that is sensitive to temperature changes. In a sep3-2 loss-of-function background, early flowering of Pro35S:TFT plants was slightly, albeit significantly suppressed, proving that TFT can work partially through SEP3 to promote flowering. We could not detect a delay in Pro35S:FT flowering, probably due to higher levels of other targets that play a redundant role with SEP3. SEP3 can physically interact with AP1, and when both genes are overexpressed, flowering time is severely reduced (Pelaz et al., 2001). A combination of ap1 and ful mutations has been shown to cause a delay in flowering time in the cal mutant background (Ferrandiz et al., 2000b). Perhaps the role of SEP3 in flowering time of wild-type plants would be revealed in an ap1/ful background. SEP3's role in organ identity was revealed in a sep1 sep2 double mutant background (Pelaz et al., 2000). It would be interesting to test whether SEP1 and SEP2 are also under FT regulation.
Here, we show that SEP3, a gene that is normally controlled by FT, is a rate-limiting factor in Pro35S:FT/TFT leaf curling. Losing one allele of SEP3 has a dramatic affect on flowers of a sep1 sep2 sep4 triple mutant as well (Ditta et al., 2004). SEP3 has been shown to form higher-order complexes with other MADS box proteins and provides the complex with an activation domain and possibly target specificity (Egea-Cortines et al., 1999; Honma and Goto, 2001; Pelaz et al., 2001). There are several reports in the literature on leaf-curling phenotypes caused by ectopically expressing different MADS box proteins. It seems that many members of this large family of transcription factors are capable of interfering in the normal processes of leaf development, when introduced into new tissue or when accumulated to abnormally high levels. We do not regard leaf curling as a modified floral response but as a very sensitive assay of FT's activation of SEP3. We exploited this assay to follow the effect of FD and the environment on FT activity.
The Relationship between FT and AP1
FT acts redundantly with the floral integrator LFY to activate AP1 transcription in the meristem (Ruiz-Garcia et al., 1997), and a plant containing mutations in both FT and LFY completely lacks floral structures and AP1 expression. FT-dependent activation of AP1 seems to be specific to the meristem. We could not detect a reduction in AP1 expression within the ft-2 mature leaf, although we did detect a reduction in the expression of FUL and SEP3. We show that AP1 can accumulate in leaves of Pro35S:FT/TFT plants but, again, less dramatically than FUL and SEP3. A mutation in FD or conditions that reduce curling affected FUL and SEP3 expression more than AP1 expression in leaves. Such differences in response, also reflected by the unique patterns of expression of these three genes outside and within the inflorescence and flower (Zimmermann et al., 2004; Schmid et al., 2005), suggest that FT's action on them is not uniform and is likely dependent on partners located in the different sites of FT action and/or on additional positive and negative gene-specific transcriptional regulators. It has also been suggested that these genes regulate each other. The presence of functional AP1 seems to reduce FUL expression in stage 1 floral primordia (Mandel and Yanofsky, 1995). Whether leaves of Pro35S:FT/TFT plants have distinct regions of AP1 and FUL expression remains to be examined.
A mutation in AP1 does not delay flowering time in a wild-type or Pro35S:FT/TFT background. AP1 might not be required for the FT effect on flowering, although it is more likely to play an important, though redundant, role in the control of flowering time. Based on our expression and genetic analyses, the FUL and SEP3 genes are likely candidate targets of FT with such possible redundant roles. It is also reasonable to assume that other genes, not identified here, play such a role.
FT Function Is Reduced at Low Ambient Temperatures
Environmental conditions affect flowering time of most plant species, and there are several examples of how the environment modifies events upstream of FT transcription (Boss et al., 2004). Does the environment regulate FT function as well? Low ambient temperatures delay flowering time of Arabidopsis plants (Blazquez et al., 2003; Thingnaes et al., 2003). Here, we show that low temperatures reduce the function of FT/TFT. Pro35S:FT/TFT plants, in which FT transcript is no longer controlled by the environment, produce additional rosette leaves and show significantly reduced meristem termination and leaf curling when grown at 12°C. To the best of our knowledge, there has been no previous evidence of events downstream of FT transcription being controlled by the environment. Cold temperatures might affect the FT protein or the accumulation or function of one of its partners. Our results, showing a specific effect of cold temperature on FT-dependent SEP3 accumulation, suggest that temperature is controlling a gene (possibly encoding an FT partner) involved in SEP3 regulation. Exposing plants that were previously grown at 12°C to 96 h of 22°C did not affect curling of mature leaves or SEP3 expression within those leaves (data not shown). This suggests that the temperature-dependent factor is no longer present in these mature leaves. It is unlikely that cold temperatures work entirely on FD, since loss of FD affects all targets. It is also unlikely that cold temperatures work entirely through SEP3, since the sep3-2 phenotype seems less dramatic than growing plants at low temperatures. Other unknown targets are worth identifying. Lastly, it is likely that other environmental stimuli affect FT action, since the degree of leaf curling is not always correlated with growth temperature. The leaf-curling assay should be helpful in this regard, since it provides a simple tool to monitor FT-dependent activation of SEP3 within the leaf.
Conclusions
In this study, we provide evidence that FT acts in organs in which it is normally expressed, we highlight FUL and SEP3 genes as targets of FT and FD function in flowering and possibly in phase change, and we reveal the environmental regulation of FT activity. While we still need to identify the site in which FT activates the floral transition, the cells that express FD and other partners of FT, and additional targets of FT, our work provides a basis for understanding the mode of action of this universal regulator of flowering time.
METHODS
Constructs, Transformation, and Selection for Transformants
The Pro35S:TFT construct was provided by E. Lifschitz and contains the cDNA of the SP3D gene (Carmel-Goren et al., 2003) under the cauliflower mosaic virus 35S promoter in the PJD330 vector (Gallie et al., 1987) moved to the binary vector PCGN1548 (McBride and Summerfelt, 1990). Arabidopsis thaliana ecotype Ler was transformed using the floral-dip method (Clough and Bent, 1998). Selection for kanamycin resistance was performed by spraying T1 seedlings with 500 μg/mL kanamycin after 1 week, and then every 3 d thereafter.
Plant Genotypes
The Arabidopsis FD, FWA, FT, and CO mutant alleles fd-1, fwa-1, ft-2, and co-2 (Koornneef et al., 1991) and the FUL allele ful-1 (Gu et al., 1998) are all in the Ler background. The Arabidopsis SEP3 null allele sep3-2 (Pelaz et al., 2000) and ectopic expression transgenic line Pro35S:SEP3 (Pelaz et al., 2001) are in the Col-0 background. The AP1 allele ap1-7 (Bowman et al., 1993) is in Col.
Genetic Analysis
Identification of the TFT transgene in plants was performed by PCR using primers that recognize the cauliflower mosaic virus 35S promoter (5′-GCCATCATTGCGATAAAGGAAAG-3′) and NOS terminator (5′-GATAATCATCGCAAGACCGGC-3′) from vector PJD330 (Gallie et al., 1987).
An array of different levels of TFT in an fd-1 mutant was obtained using two approaches: one consisted of introducing the fd-1 mutation into Pro35S:TFT#7 and the other of transforming Pro35S:TFT directly into an fd-1 mutant. In all cases, the vegetative leaf-curling phenotype was lost in the absence of FD. Once established, new homozygous Pro35S:TFT fd-1 lines (from direct transformation) were backcrossed to a wild-type Ler plant. A functional FD allele was sufficient to regain leaf curling in all lines.
A line homozygous for Pro35S:TFT#7 in sep3-2 was identified in the following way. We collected seeds from individual plants (early and uncurled) from an F2 population segregating for sep3-2, Pro35S:TFT#7, Col-0, and Ler. We selected for homozygous Pro35S:TFT#7/sep3-2 plants by screening F3 populations under short-day conditions, which enhance curling (condition A4 in Table 1). Under these conditions, it was easy to score for the presence of Pro35S:TFT#7 (early in short days) and sep3-2 (no curling). A line producing uniform early-flowering plants with no leaf curling was chosen and shown by PCR to be Pro35S:TFT#7/sep3-2.
Pro35S:FT plants homozygous for the ful-1 allele were identified by PCR using primers that recognize the FUL wild-type allele (Gu et al., 1998), and the genotype was verified by following the unique ful-1 silique phenotype (Gu et al., 1998).
Plant Growth Conditions
Seeds were sown in soil and placed in the dark at 4°C for 2 to 3 d before moving them into lighted growth cabinets or greenhouses (see Table 1 for different growth conditions). Flowering time (rosette and cauline leaf number), leaf curling, and meristem termination of different genotypes were measured under a large range of growth conditions. Specific growth conditions are provided for each experiment. Different light qualities were obtained using cool white fluorescent lamps, blue (Philips TLD 18W/18 blue) fluorescent lights, and incandescent bulbs. Light spectrum and intensity were measured using an LI-1800 portable spectroradiometer (LI-COR) and an LI-250 light meter (LI-COR), respectively. Relative humidity and temperature were measured using a TESTO data logger.
Histochemical Detection of GUS
Histochemical analysis of GUS was performed as described by Ori et al. (2000). The first rosette leaves of 19-d-old plants grown under blue-lit long days (A3 in Table 1) were treated for 6 h.
RNA Analysis
Seedlings or rosette leaves (depending on the experiment) were harvested at a similar age and time of day in all compared treatments. Total RNA was extracted (Logemann et al., 1987), and poly(A)+ RNA was purified using the DYNAL Dynabeads mRNA purification kit. cDNA was synthesized from poly(A)+ RNA using Invitrogen Superscript II RNase H-Reverse Transcriptase and oligo (dT)12-18 primers. Reactions for quantitative real-time RT-PCR on the cDNA were performed using kits from Abgene: ABsolute QPCR SYBR Green Mix kit (AB-1162) for reactions with Syber-Green and ABsolute QPCR Mix (AB-1138) for reactions with Taqman probes. Reactions were run on a Corbett Research Rotor-Gene 2000 cycler. For each gene tested, at least one of the primers used spanned an exon-exon border so that only cDNA could be amplified. A list of primer and probe sequences is given in Table 3. As a housekeeping gene, we used β-TUBULIN (AT5G62690). A standard curve was obtained for each gene using a plasmid or fragment containing its amplified region. Reactions for each gene in each cDNA sample were repeated independently at least four times. Quantification of each gene was performed using Corbett Research Rotor-Gene software. The expression of each gene was an average of at least three repeats. We only used repeats in which the standard deviation of a population was <1.5% of the average. Relative expression of a gene in a certain sample was initially obtained by dividing the gene level (in arbitrary units) by the β-TUBULIN level (in arbitrary units). Relative expression units are shown by setting the sample with the lowest expression at a value of 1 or the control treatment at a value of 100.
Table 3.
Primers and Probes Used for Real-Time RT-PCR
| Gene | Forward Primer (5′/3′) | Reverse Primer (5′/3′) | Probe (5′/3′) |
|---|---|---|---|
| CAL | TGTACCACAGCCACAACCATT | CTTGGTACAAACCACCCATATTTAGA | AAGGAGAAGTCTGATGAGCGATCATGTAAAGAT |
| SEP3 | CTAAGACTAAGGTTAGCTGATGGGTA | ATGATGACGACCGTAGTGATCAA | ATGCCACTCCAGCTGAACCCTAACCAA |
| FUL | TGCGTAACCTCCTCCAGAGAT | GTTCTACTCGTTCGTAGTGGTAGGAC | AGAGAACGGTGGTGCATCGTCGTT |
| AP1 | ACATCCGCACTAGAAAAAACCAAC | CTTGGTTCTGCTGATCCCACA | ATGCTGTTTTGCTCCTGTATGGCCTT |
| TFT | CCAAGTCCGAGTGATCCAAATC | TTGATGGTCTTGGACTTTCATAGC | CTACCACAGGTTCAAGTTTTGGGCAAGAAA |
| β-TUBULIN | AAACTCACTACCCCCAGCTTTG | CACCAGACATAGTAGCAGAAATCAAGT |
Accession Numbers
The Arabidopsis Genome Initiative locus numbers for the major genes discussed in this article are At1g65480 for FT, At1g69120 for AP1, At1g24260 for SEP3, and At5g60910 for FUL.
Supplementary Material
Acknowledgments
We are grateful to E. Lifschitz (Technion, Haifa, Israel) for the Pro35S:TFT construct, to T. Araki (Kyoto University, Kyoto, Japan) for the pro35S:FT introgressed into Ler seeds and for sharing results prior to publication, and to P. Wigge (John Innes Centre, Norwich, UK) for sharing results prior to publication. We are also grateful to P. Wigge, D. Weiss, N. Ori (Hebrew University), G. Coupland (Max Planck Institute for Plant Breeding, Koln, Germany), and T. Araki for their very helpful comments on the manuscript and to D. Zamir (Hebrew University) for his encouragement throughout the project. We thank M. Yanofsky (University of California at San Diego, La Jolla, CA) for sep3-2 and pro35S:SEP3 seeds and the Nottingham Arabidopsis Stock Centre for other seeds used in this research. This research was supported by a Charles H. Revson Foundation grant (436/00-1) from the Israel Science Foundation and equipment from the Wolfson Advanced Research Center for Plant Genomics and Biotechnology in Semi-Arid Climates.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alon Samach (samach@agri.huji.ac.il).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035766.
References
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615–3626. [DOI] [PubMed] [Google Scholar]
- Ayre, B.G., and Turgeon, R. (2004). Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiol. 135, 2271–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield, M.J., and Brady, R.L. (2000). The structure of Antirrhinum CENTRORADIALIS protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 297, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Blazquez, M.A., Ahn, J.H., and Weigel, D. (2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33, 168–171. [DOI] [PubMed] [Google Scholar]
- Boss, P.K., Bastow, R.M., Mylne, J.S., and Dean, C. (2004). Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 16 (suppl.), S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Alvarez, J., Weigel, D., Meyerowitz, E.M., and Smyth, D.R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743. [Google Scholar]
- Carmel-Goren, L., Liu, Y.S., Lifschitz, E., and Zamir, D. (2003). The SELF-PRUNING gene family in tomato. Plant Mol. Biol. 52, 1215–1222. [DOI] [PubMed] [Google Scholar]
- Chailakhyan, M. (1936). New facts in support of the hormonal theory of plant development. CR (Doklady) Acad. Sci. URSS 13, 77. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., and Yanofsky, M.F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz, C., Gu, Q., Martienssen, R., and Yanofsky, M.F. (2000. b). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734. [DOI] [PubMed] [Google Scholar]
- Ferrandiz, C., Liljegren, S.J., and Yanofsky, M.F. (2000. a). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289, 436–438. [DOI] [PubMed] [Google Scholar]
- Gallie, D., Sleat, D., Watts, J., Turner, P., and Wilson, T. (1987). The 5′-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res. 15, 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel, K. (1900). Organography of Plants I. General Organography (English translation by I.B. Balfour). (New York: Hafner).
- Gu, Q., Ferrandiz, C., Yanofsky, M., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517. [DOI] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel, F., and Feldman, L. (1994). Bi-directional inflorescence development in Arabidopsis thaliana: Acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192, 276–286. [Google Scholar]
- Hempel, F.D., Weigel, D., Mandel, M.A., Ditta, G., Zambryski, P.C., Feldman, L.J., and Yanofsky, M.F. (1997). Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124, 3845–3853. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. [DOI] [PubMed] [Google Scholar]
- Jones, C.S. (1999). An essay on juvenility, phase change, and heteroblasty in seed plants. Int. J. Plant Sci. 160, S105–S111. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Keller, E.T., Fu, Z., and Brennan, M. (2004). The role of Raf kinase inhibitor protein (RKIP) in health and disease. Biochem. Pharmacol. 68, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Knott, J.E. (1934). Effect of localized photoperiod on spinach. Proc. Soc. Hort. Sci. 31, 152–154. [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Kotake, T., Takada, S., Nakahigashi, K., Ohto, M., and Goto, K. (2003). Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 44, 555–564. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Gustafson-Brown, C., Pinyopich, A., Ditta, G.S., and Yanofsky, M.F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann, J., Schell, J., and Willmitzer, L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163, 16–20. [DOI] [PubMed] [Google Scholar]
- Lorenz, K., Lohse, M.J., and Quitterer, U. (2003). Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature 426, 574–579. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7, 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, K.E., and Summerfelt, K.R. (1990). Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14, 269–276. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control KNOX gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Gustafson-Brown, C., Kohalmi, S.E., Crosby, W.L., and Yanofsky, M.F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26, 385–394. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Gutfinger, T., Hareven, D., Ben-Naim, O., Ron, N., Adir, N., and Lifschitz, E. (2001). Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13, 2687–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig, R.S. (2003). Phase change and the regulation of developmental timing in plants. Science 301, 334–336. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Amaya, I., Vincent, C.A., Rothstein, S., Carpenter, R., Coen, E.S., and Bradley, D.J. (1998). A common mechanism controls the life cycle and architecture of plants. Development 125, 1609–1615. [DOI] [PubMed] [Google Scholar]
- Ruiz-Garcia, L., Madueno, F., Wilkinson, M., Haughn, G., Salinas, J., and Martinez-Zapater, J.M. (1997). Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9, 1921–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Uhlenhaut, N.H., Godard, F., Demar, M., Bressan, R., Weigel, D., and Lohmann, J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130, 6001–6012. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Suh, S.S., Choi, K.R., and Lee, I. (2003). Revisiting phase transition during flowering in Arabidopsis. Plant Cell Physiol. 44, 836–843. [DOI] [PubMed] [Google Scholar]
- Sung, S., and Amasino, R.M. (2004). Vernalization and epigenetics: How plants remember winter. Curr. Opin. Plant Biol. 7, 4–10. [DOI] [PubMed] [Google Scholar]
- Takada, S., and Goto, K. (2003). TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingnaes, E., Torre, S., Ernstsen, A., and Moe, R. (2003). Day and night temperature responses in Arabidopsis: Effects on gibberellin and auxin content, cell size, morphology and flowering time. Ann. Bot. (Lond.) 92, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohdoh, N., Tojo, S., Agui, H., and Ojika, K. (1995). Sequence homology of rat and human HCNP precursor proteins, bovine phosphatidylethanolamine-binding protein and rat 23-kDa protein associated with the opioid-binding protein. Brain Res. Mol. Brain Res. 30, 381–384. [DOI] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Yeung, K., Seitz, T., Li, S., Janosch, P., McFerran, B., Kaiser, C., Fee, F., Katsanakis, K.D., Rose, D.W., Mischak, H., Sedivy, J.M., and Kolch, W. (1999). Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401, 173–177. [DOI] [PubMed] [Google Scholar]
- Zeevaart, J.A.D. (1976). Physiology of flower formation. Annu. Rev. Plant Physiol. 27, 321–348. [Google Scholar]
- Zik, M., and Irish, V.F. (2003). Flower development: Initiation, differentiation, and diversification. Annu. Rev. Cell Dev. Biol. 19, 119–140. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.