Abstract
Auxin is central to many aspects of plant development; accordingly, plants have evolved several mechanisms to regulate auxin levels, including de novo auxin biosynthesis, degradation, and conjugation to sugars and amino acids. Here, we report the characterization of an Arabidopsis thaliana mutant, IAA carboxyl methyltransferase1-dominant (iamt1-D), which displayed dramatic hyponastic leaf phenotypes caused by increased expression levels of the IAMT1 gene. IAMT1 encodes an indole-3-acetic acid (IAA) carboxyl methyltransferase that converts IAA to methyl-IAA ester (MeIAA) in vitro, suggesting that methylation of IAA plays an important role in regulating plant development and auxin homeostasis. Whereas both exogenous IAA and MeIAA inhibited primary root and hypocotyl elongation, MeIAA was much more potent than IAA in a hypocotyl elongation assay, indicating that IAA activities could be effectively regulated by methylation. IAMT1 was spatially and temporally regulated during the development of both rosette and cauline leaves. Changing expression patterns and/or levels of IAMT1 often led to dramatic leaf curvature phenotypes. In iamt1-D, the decreased expression levels of TCP genes, which are known to regulate leaf curvature, may partially account for the curly leaf phenotype. The identification of IAMT1 and the elucidation of its role in Arabidopsis leaf development have broad implications for auxin-regulated developmental process.
INTRODUCTION
Indole-3-acetic acid (IAA), the main auxin in plants, is known to be involved in various plant growth and developmental processes, and plants have evolved a complicated network to precisely regulate auxin activities (for recent reviews, see Leyser, 2002; Ljung et al., 2002; Berleth et al., 2004; Dharmasiri and Estelle, 2004). IAA activity is regulated at three distinct but interdependent levels: homeostasis, polar transport, and auxin responses. The auxin concentration in plants can be regulated through de novo biosynthesis of IAA, degradation of IAA, and conjugation/deconjugation of IAA with sugars or amino acids. Although several key genes in auxin biosynthesis have been identified, there is still no clear picture of how auxin is synthesized in plants (Bartel, 1997; Zhao et al., 2001, 2002; Cohen et al., 2003). IAA conjugates have also been subjected to intensive studies for decades (Ljung et al., 2002). It is generally believed that free IAA is the active form of auxin, whereas IAA–sugar and IAA–amino acid conjugates are not active per se but can be reversibly converted to free IAA. Therefore, IAA conjugates are part of a biochemical system for the homeostasis of IAA levels in higher plants (e.g., IAA storage or transport, protection of IAA against peroxidative degradation, and compartmentalization or detoxification of excess IAA) (Cohen and Bandurski, 1982). The biosynthesis of IAA–amino acid conjugates was first characterized in bacteria systems in which the enzyme iaaL conjugates IAA to the amino acid Lys (Hutzinger and Kosuge, 1968; Glass and Kosuge, 1986; Romano et al., 1991; Spena et al., 1991). Recently, a family of GH3 proteins in Arabidopsis thaliana has been shown to be involved in synthesizing IAA–amino acid conjugates (Staswick et al., 2005). IAA–amino acid conjugates can be converted back to free IAA. Several enzymes that hydrolyze certain IAA–amino acid conjugates in vivo to release free IAA have been identified in Arabidopsis by genetic screens for mutants insensitive to the amino acid conjugates (Bartel and Fink, 1995; Davies et al., 1999; Lasswell et al., 2000; LeClere et al., 2002; Rampey et al., 2004).
IAA conjugates with a monosaccharide or disaccharide via an ester linkage have also been studied extensively (Ljung et al., 2002). The first IAA conjugate biosynthesis gene, IAGlu, was isolated from maize (Zea mays), and its product catalyzes the formation of IAA glucose ester from IAA and glucose (Szerszen et al., 1994). Overexpression of the Arabidopsis homolog of IAGlu, UGT84B1, resulted in alteration of the homeostatic level of IAA in transgenic plants and consequently produced rounded, wrinkled, and curly leaves, suggesting that the homeostatic level of IAA might play an important role in the regulation of adaxial/abaxial cell growth in leaves (Jackson et al., 2002). Here, we present evidence that the conversion of IAA to methyl IAA ester (MeIAA) by an IAA carboxyl methyltransferase also plays an important role in regulating auxin homeostasis and plant development.
Unlike IAA–sugar and IAA–amino acid conjugates that are charged or that contain many polar groups, MeIAA is essentially nonpolar. Thus, formation of an IAA methyl ester offers a distinctive way to regulate IAA activities. Small-molecule methyl esters are secondary metabolites that have been known to play important roles in many plant processes, including plant defense responses, insect pollination, and regulation of cell growth (Shulaev et al., 1997; Dudareva et al., 2000; Murfitt et al., 2000; Chen et al., 2003). For example, methyl salicylic acid is a basic fragrance component in flowers (Ross et al., 1999) and accumulates at wound sites, serving as an airborne signal to induce the defense response in unwounded organs and adjacent plants (Shulaev et al., 1997). Methyl jasmonates are important cellular regulators mediating diverse developmental processes, including seed germination, flower and fruit development, leaf abscission, senescence, and disease resistance (Creelman and Mullet, 1995; Seo et al., 2001). These small-molecule methyl esters are synthesized in planta via a reaction catalyzed by carboxyl methyltransferases whereby a methyl group is transferred from S-adenosyl-l-Met to the carboxyl group of the small molecular compounds, such as salicylic acid, benzoic acid, and jasmonic acid (Zubieta et al., 2003). In Arabidopsis, there are at least 24 confirmed or putative carboxyl methyltransferase genes, and one of them, At5g55250, appears to encode an enzyme that specifically converts IAA to MeIAA in vitro (D'Auria et al., 2003; Zubieta et al., 2003).
Here, we present the analysis of IAMT1, an Arabidopsis gene that we originally identified in a screen for mutants in leaf development but that when molecularly characterized proved to be the IAA carboxyl methyltransferase gene At5g55250. We demonstrate that the spatial expression pattern of IAMT1 is developmentally regulated in leaves and that overexpression of IAMT1 resulted in a curly leaf phenotype in transgenic Arabidopsis, suggesting that conversion of IAA to MeIAA has a profound effect on auxin homeostasis and plant development. In addition to the curly leaf phenotypes, overexpression of IAMT1 led to various auxin-related phenotypes, including agravitropic growth in both hypocotyls and roots. Furthermore, we show that, like free IAA, exogenous MeIAA inhibits primary root growth and hypocotyl elongation. However, exogenously applied MeIAA appears to be much more potent than free IAA, indicating that IAA methylation could serve as an effective means to regulate IAA activities. This work thus adds a new layer of complexity to auxin regulation and has profound implications for auxin-regulated developmental processes.
RESULTS
Isolation of the IAA Carboxyl Methyltransferase Mutant iamt1-D
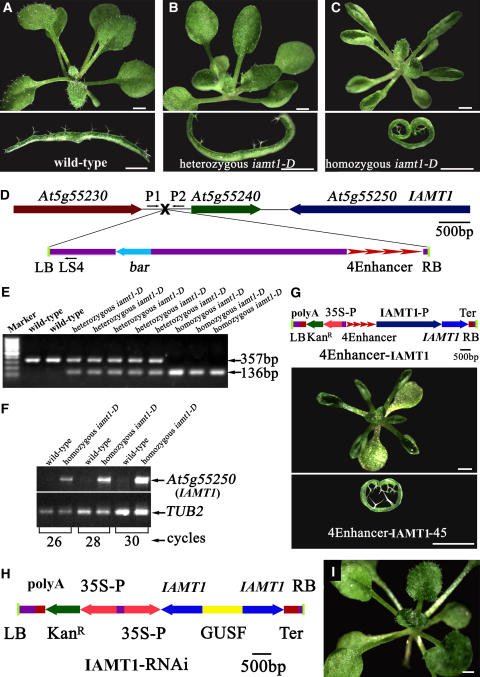
We screened ∼25,000 activation-tagged Arabidopsis lines for mutants with defects in leaf development. Among the eight putative mutants from the initial screen, one mutant had a dramatic upward-curling leaf phenotype and was first designated curly1 and later renamed iamt1-D for the reasons discussed below (Figures 1A to 1C). The iamt1-D leaves appeared indistinguishable from those of the wild type when they first emerged. However, the mature leaves all curled upward and resulted in a cylinder-like structure (Figure 1C). The iamt1-D mutant is semidominant, as the heterozygous iamt1-D displayed partially curly and crinkled leaves (Figure 1B). Although iamt1-D leaves were curled, they appeared similar in size to wild-type leaves when flattened (Figures 1A to 1C).
Figure 1.
Characterization of the IAA Carboxyl Methyltransferase Gene IAMT1.
(A) to (C) Top, 26-d-old wild-type, heterozygous, and homozygous IAA carboxyl methyltransferase1-dominant (iamt1-D) mutant Arabidopsis plants. Bottom, leaf transverse sections from the corresponding plants.
(D) Scheme of the genomic region flanking the T-DNA insertion site in iamt1-D. Genes are represented by different colored arrows, intergenic regions by lines, and the T-DNA insertion site by a black X. The arrow direction represents the transcriptional orientation of the genes. The four red arrowheads represent the four 35S enhancers from pSKI015. The thin black arrows represent the primers used in cosegregation analysis. LB, T-DNA left border; bar, Basta resistance gene; 4Enhancer, CaMV 35S enhancer tetrad; RB, T-DNA right border.
(E) Linkage analysis of the T-DNA insertion and hyponastic phenotypes. P1 and P2 will amplify a 357-bp fragment from the wild type, and P1 and LS4 will amplify a 136-bp fragment from the homozygous iamt1-D mutant. Marker, 100-bp ladder.
(F) Expression of IAMT1 in the wild type and the homozygous iamt1-D mutant by RT-PCR with the primer pair IAMT-1 and IAMT-2. TUB2, β-tubulin gene as an internal control.
(G) Recapitulation of iamt1-D phenotypes by overexpression of IAMT1 cDNA. The IAMT1 cDNA (coding region) was overexpressed under the control of four copies of the 35S enhancers and the IAMT1 promoter. The transgenic plants displayed hyponastic leaf phenotypes. LB, T-DNA left border; polyA, CaMV 35S poly(A); KanR, kanamycin resistance gene NPTII; 35S-P, CaMV 35S promoter; 4Enhancer, CaMV 35S enhancer tetrad; IAMT1-P, promoter of IAMT1; IAMT1, open reading frame of IAMT1; Ter, nopaline synthase terminator; RB, T-DNA right border.
(H) Construct of the IAMT1-RNAi plasmid. LB, polyA, KanR, 35S-P, IAMT1, Ter, and RB are as in (G). GUSF, 1-kb fragment of GUS.
(I) Suppression of iamt1-D phenotypes by the IAMT1-RNAi construct. A 26-d-old iamt1-D homozygous mutant plant transformed with the IAMT1-RNAi construct is shown. Bar = 1 mm.
iamt1-D was isolated as a semidominant mutant from a collection of activation-tagged Arabidopsis T-DNA lines (Qin et al., 2003); therefore, it is likely that iamt1-D is a gain-of-function mutant caused by a T-DNA insertion. We identified a single T-DNA insertion in the intergenic region between genes At5g55230 and At5g55240 (Figure 1D). To examine whether the T-DNA insertion cosegregates with the observed leaf phenotypes, we genotyped a T3 population of the iamt1-D mutant. Among 399 T3 plants, 82 were wild type without the T-DNA insertion, 98 were homozygous for the T-DNA insertion, and 219 were heterozygous for the T-DNA insertion (Figure 1E). All of the plants that were homozygous for the T-DNA insertion displayed the curly leaf phenotype, whereas all of the plants without the T-DNA insertion did not display the leaf phenotypes, suggesting that the curly leaf phenotype likely is caused by this single T-DNA insertion.
The T-DNA insert contains four copies of the 35S enhancer in the right border (Figure 1D); therefore, we examined whether the transcripts of the genes near the T-DNA insertion site were affected. We analyzed the expression levels of 10 genes upstream and 10 genes downstream of the insertion site by RT-PCR and found that the expression levels of two downstream genes (At5g55240 and At5g55250) were greatly increased (Figure 1F shows results for At5g55250). We next overexpressed either At5g55240 or At5g55250 in wild-type Arabidopsis to identify which gene was responsible for the observed leaf phenotypes. As shown in Figure 1G, overexpression of At5g55250, but not At5g55240, under its own promoter with four copies of the 35S enhancer recapitulated the curly leaf phenotype, indicating that At5g55250 is the IAMT1 gene. RT-PCR analysis also confirmed that IAMT1 was indeed overexpressed in these iamt1-D–like transgenic plants (Figure 1G).
To further confirm that overexpression of At5g55250 caused the iamt1-D phenotypes, we transformed iamt1-D with an RNA interference (RNAi) construct of At5g55250 driven by a cauliflower mosaic virus (CaMV) 35S promoter (Figure 1H). The majority (75 of 92) of the transgenic plants obtained had a wild-type leaf phenotype in the iamt1-D background (Figure 1I), indicating that the curly leaf mutant phenotype was caused by overexpression of the IAMT1 gene.
At5g55250 belongs to a recently defined novel family of carboxyl methyltransferases in plants (Zubieta et al., 2003). Members of this methyltransferase family catalyze the transfer of the methyl group from S-adenosyl-l-Met to carboxylic acid–containing substrates to form small-molecule methyl esters (Zubieta et al., 2003). Two members of this methyltransferase family, salicylic acid carboxyl methyltransferase and jasmonic acid methyltransferase, have been demonstrated to methylate, both in vitro and in vivo, two important plant organic acids, the hormones salicylic acid and jasmonic acid, respectively (Ross et al., 1999; Seo et al., 2001; Chen et al., 2003; D'Auria et al., 2003; Zubieta et al., 2003). Recently, At5g55250 was shown to encode an enzyme that specifically methylates the plant hormone IAA in vitro with a much higher value of the kinetic specificity constant (Kcat/Km) for IAA than for salicylic acid or other compounds. Additional structural and biochemical experiments further indicate that At5g55250-encoded enzyme is highly specific for IAA (Y. Yang, J.R. Ross, E. Pichersky, and J.P. Noel, unpublished data). These data provide the basis for the designation of At5g55250 as IAMT1 (for IAA CARBOXYL METHYLTRANSFERASE1) and for the designation of the mutant as iamt1-D.
Overexpression of IAMT1 in Arabidopsis Affects Auxin Responses
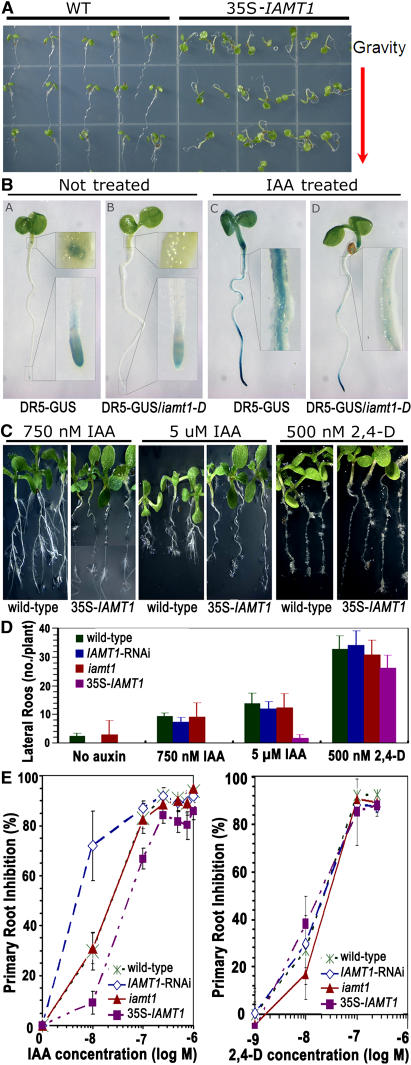
We expressed IAMT1 in Escherichia coli and confirmed that the recombinant IAMT1 converts IAA to MeIAA in vitro (data not shown). The biochemical activities of IAMT1 prompted us to investigate whether IAMT1 affects other auxin-regulated processes. As shown in Figure 2A, overexpression of IAMT1 led to the disruption of proper gravitropic responses that are often associated with malfunctions in auxin homeostasis or signal transduction, indicating that overexpression of IAMT1 affects auxin-regulated processes. IAMT1 overexpression lines also showed decreased responses to IAA treatment, as indicated by the induction levels of the auxin-responsive DR5 promoter (Figure 2B) and the differences in root elongation compared with wild-type plants (Figures 2C and 2E). Furthermore, IAMT1 overexpression lines initiated fewer lateral roots in response to IAA treatment (Figures 2C and 2D). However, the overexpression lines did not alter the responses to 2,4-D treatment in either lateral root development or root elongation (Figures 2C to 2E). Moreover, the IAMT1-RNAi lines appeared to be more sensitive to IAA than the wild-type line and the overexpression lines (Figure 2E).
Figure 2.
Auxin Responses of IAMT1 Overexpression Lines.
(A) Overexpression of IAMT1 resulted in agravitropism.
(B) GUS staining of the auxin-responsive DR5-GUS reporter in roots of iamt1-D plants treated with or without 20 μM IAA for 5 h.
(C) and (D) Lateral root response of the iamt1 mutants to IAA and 2,4-D.
(E) Primary root elongation response of the iamt1-D mutants to IAA and 2,4-D. Error bars represent standard deviations.
IAMT1-RNAi Transgenic Plants Displayed Phenotypes Opposite to Those of iamt1-D
We identified a T-DNA insertion mutant of iamt1 (SALK_072125) from the Salk T-DNA collection (Alonso et al., 2003), but no obvious developmental phenotypes were observed for this T-DNA line. The T-DNA insertion occurs at the end of the third of the four exons of this gene, causing an insertion of nine nucleotides at the site of integration and interrupting the C-terminal third of the protein. However, a truncated transcript of IAMT1 was detected, which would encode a truncated protein with a deletion of 92 amino acids at the C terminus (see Supplemental Figure 1 online). It is possible that this altered protein still has IAMT activity. Alternatively, it is possible that other methyltransferase genes may be able to compensate for the loss of function of IAMT1, because there are additional, still uncharacterized carboxyl methyltransferase genes in the Arabidopsis genome (D'Auria et al., 2003; Zubieta et al., 2003).
We next constructed IAMT1-RNAi transgenic lines and found that ∼20% of the RNAi lines (11 of 65 total) developed an epinastic leaf phenotype that is opposite to that shown by iamt1-D (Figure 3A). RT-PCR analysis showed that, although transcripts of both endogenous and transgenic IAMT1 were detected in the RNAi lines with the primers IAMT1-3 and IAMT1-4, transcripts of the endogenous IAMT1 were barely detected with the primers IAMT1-5 and IAMT1-6, which would otherwise amplify the coding region and 3′ untranslated region of IAMT1 in wild-type plants (Figure 3B). On the other hand, transcripts of both endogenous and transgenic IAMT1 were detected in the overexpression lines with both sets of primers (Figure 3B). We also observed a good correlation between the leaf phenotypes and the expression levels of IAMT1 (Figure 3C). These data suggest that the epinastic leaf phenotype in the RNAi lines is attributable to the suppression of IAMT1 and, perhaps, of other related genes. Furthermore, the RNAi lines displayed additional phenotypes, including smaller leaves, dwarfism (Figure 3A), and low fertility (Figures 3D to 3F), suggesting that IAMT1 plays a role in the regulation of other aspects of plant development during both vegetative and reproductive growth.
Figure 3.
Analysis of IAMT1-RNAi Transgenic Plants.
(A) IAMT1-RNAi knockdown plants in the wild-type background displayed an epinastic leaf phenotype. A 35-d-old plant of a transgenic line transformed with the IAMT1-RNAi construct in Figure 1H (top) and a close-up view of the transverse section of its leaf (bottom). Bars = 1 mm.
(B) Analysis of IAMT1 expression levels in wild-type, 4Ehancer-IAMT1, and IAMT1-RNAi transgenic plants by RT-PCR. Top, primers used in the analysis. Bottom, expression with primers IAMT1-3 and IAMT1-4 (first row) and IAMT1-5 and IAMT1-6 (second row). TUB2 was used as an internal control (third row).
(C) Correlation between IAMT1 expression levels and the curly leaf phenotypes in different IAMT1-RNAi transgenic lines.
(D) Adult wild-type Arabidopsis plant.
(E) IAMT1-RNAi transgenic plant.
(F) Comparison of siliques from wild-type and IAMT1 RNAi plants.
Expression of IAMT1 Is Developmentally Regulated in Leaves
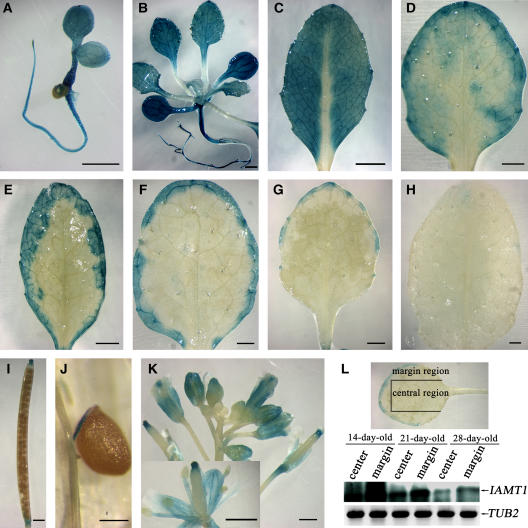
We constructed transgenic plants that express a β-glucuronidase (GUS) reporter gene under the control of a 2.7-kb fragment upstream of the IAMT1 gene. As shown in Figure 4, the GUS gene was expressed ubiquitously in rosette leaves before the eighth true leaves emerged (Figures 4A and 4B). After the eighth true leaves emerged, GUS expression gradually faded away from the center of the leaves and was detected primarily toward the edges of the leaves as leaf development proceeded (Figures 4C to 4G). After the leaves fully expanded, GUS expression was detectable only at the edge of the leaf blade (Figure 4H). A similar pattern was observed in cauline leaves (data not shown). RT-PCR analysis confirmed that the expression level of IAMT1 was indeed lower in the central region than it was in the margin region of the leaves (Figure 4L). Apart from leaves, IAMT1 expression was detected at a relatively high level in the stigma (Figure 4I), funiculus (Figure 4J), and vascular bundles in sepals, petals, and stamens (Figure 4K). The spatial and temporal regulation of IAMT1 expression revealed that the expression pattern of this gene in the leaf margin correlated with the curly leaf phenotype, in which the marginal region of the leaves curled upward. This finding further supports the hypothesis that IAMT1 plays an important role in leaf pattern formation.
Figure 4.
Spatial Expression Pattern of IAMT1.
The expression pattern of IAMT1 was determined using transgenic lines transformed with pIAMT1P-GUS, in which a GUS reporter gene was driven by the IAMT1 promoter.
(A) A 3-d-old seedling of the pIAMT1P-GUS reporter line.
(B) A 22-d-old seedling of the pIAMT1P-GUS reporter line.
(C) to (H) Rosette leaves of pIAMT1P-GUS reporter lines, from young to old. The expression of IAMT1 wanes from the midvein to the margin with the maturation of the leaf.
(I) Silique of the pIAMT1P-GUS reporter line.
(J) Close-up view of funiculus and seed from the pIAMT1P-GUS reporter line.
(K) Inflorescence and flower of the pIAMT1P-GUS reporter line.
(L) Analysis of IAMT1 expression level in leaf marginal and central sections by RT-PCR. RNA was extracted from the marginal or central section (dissected as shown at top) of leaves from 14-, 21-, and 28-d-old wild-type Arabidopsis. A DNA gel blot of RT-PCR products probed with cDNA of IAMT1 is shown. TUB2 was used as an internal control. The expression of IAMT1 is higher in the margin than in the central section and higher in young leaves than in old leaves.
Bars = 200 μm in (J) and 1 mm in all other panels.
Overexpression of IAMT1 in Abaxial Layers Results in Epinastic Leaf Phenotypes
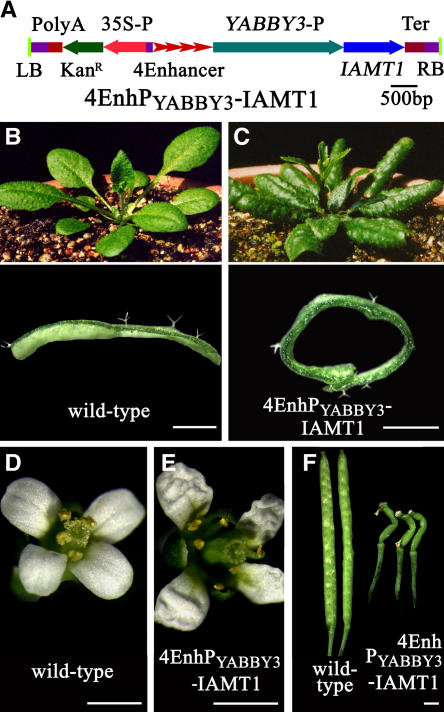
Because IAMT1 expression appeared developmentally regulated and important for leaf adaxial/abaxial deferential growth, we hypothesized that misexpression of the IAMT1 gene in Arabidopsis leaves may also cause differential growth problems. Therefore, we put the IAMT1 gene under the control of four copies of the 35S enhancer and the YABBY3 promoter (Figure 5A), which directs abaxial layer–specific gene expression (Siegfried et al., 1999). Among the 28 transgenic lines obtained, 9 lines displayed dramatic epinastic leaf phenotypes (Figures 5B and 5C). Furthermore, the petals of these transgenic plants were crinkled (Figures 5D and 5E) and the siliques were twisted (Figure 5F).
Figure 5.
Abaxial Overexpression of IAMT1 Results in the Epinastic Leaf Phenotype.
(A) 4EnhPYABBY3-IAMT1 construct. LB, polyA, KanR, 35S-P, 4Ehancer, IAMT1, Ter, and RB are as in Figure 1. YABBY3-P, promoter of the YABBY3 gene.
(B) Thirty-two-day-old wild-type Arabidopsis plant (top) with a transverse leaf section (bottom).
(C) Thirty-two-day-old 4EnhPYABBY3-IAMT1 transgenic plant (top) with a transverse leaf section (bottom).
(D) A wild-type Arabidopsis flower.
(E) A flower from a 4EnhPYABBY3-IAMT1 transgenic plant.
(F) Comparison of siliques from wild-type and 4EnhPYABBY3-IAMT1 transgenic plants.
Bars = 1 mm.
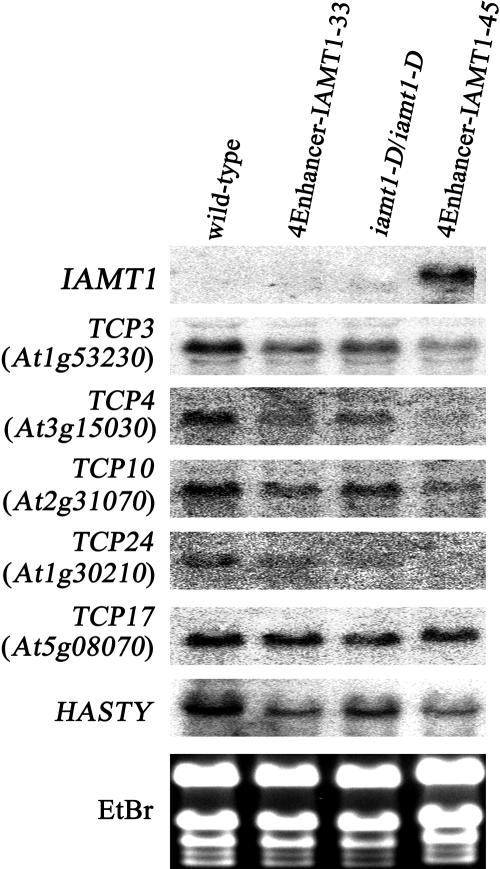
Expression of Several TCP Genes Is Downregulated in iamt1-D
We examined the expression of several genes known to be involved in regulating leaf curvature development. RNA gel blots showed that four TCP genes (i.e., TCP3, TCP4, TCP10, and TCP24) that are involved in the regulation of leaf development were downregulated in iamt1-D (Figure 6). Higher IAMT1 expression levels were correlated with lower expression levels of the TCP genes (Figure 5). These four TCP genes are homologous with CINCINNATA, a curvature regulation gene in snapdragon (Nath et al., 2003). Other TCP gene members (e.g., TCP17) showed no change in their expression in iamt1-D (Figure 6). HASTY, another gene that results in a curly leaf phenotype when mutated (Telfer and Poethig, 1998), was also downregulated in iamt1-D (Figure 6). The expression levels of AGAMOUS (Mizukami and Ma, 1992) and CLF (Goodrich et al., 1997), however, were not altered in iamt1-D plants (data not shown).
Figure 6.
Expression Analysis of Leaf Curvature–Related Genes by RNA Gel Blot.
TCP3, TCP4, TCP10, TCP17, and TCP24 are homologous with CINCINNATA, which regulates leaf curvature in snapdragon (Nath et al., 2003). HASTY is another gene that results in the curly leaf phenotype when mutated (Telfer and Poethig, 1998). Except for TCP17, these genes were downregulated in iamt1-D. Ethidium bromide (EtBr) staining was used as the loading control.
Effects of Exogenous MeIAA on Plant Growth and Development
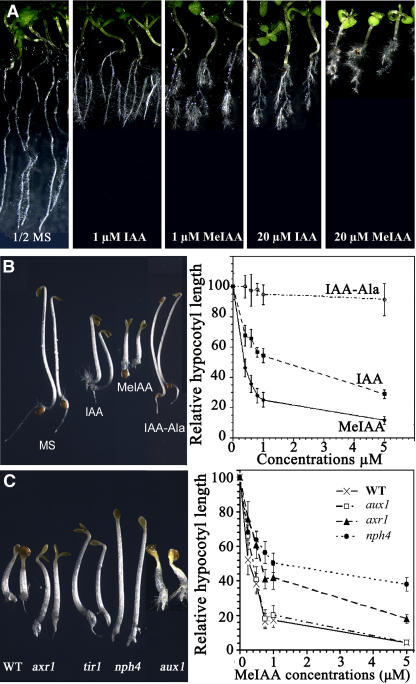
Although the leaf phenotypes of iamt1-D lines are consistent with perturbations of IAA concentration and/or gradient through the conversion of IAA to MeIAA, it is not clear what the physiological roles might be for MeIAA itself. To examine this question, we first tested the effects of exogenous MeIAA on plant growth and development. Like IAA, MeIAA also inhibits primary root elongation and stimulates adventitious root development of light-grown seedlings, and we found that MeIAA was more active than IAA (Figure 7A). In the dark, MeIAA inhibited hypocotyls and root elongation (Figure 7B). Interestingly, MeIAA was much more potent than IAA in inhibiting hypocotyl elongation of dark-grown seedlings (Figure 7B). We next investigated whether other IAA esters, including IAA ethyl ester (ethyl-3-indoleacetate) and IAA hexyl ester (hexyl-3-indoleacetate), also had auxin activities. Both the IAA ethyl and hexyl esters displayed activities similar to those of MeIAA, and both esters were much more potent than IAA (data not shown). By contrast, the IAA amino acid conjugate IAA–Ala [N-(3-indolylacetyl)-Ala] was much less active than IAA (Figure 7B).
Figure 7.
Effects of Exogenous MeIAA on Arabidopsis Seedling Growth and Development.
(A) Comparison of Arabidopsis responses to exogenous MeIAA or IAA. Seeds of Columbia ecotype Arabidopsis were germinated in a Petri dish with solid half-strength (1/2) MS medium for 3 d after stratification for 3 d. The seedlings with average growth were transferred to half-strength MS medium with or without different concentrations of MeIAA or IAA and grown vertically for 5 d.
(B) Inhibition of hypocotyl elongation by MeIAA. Wild-type Arabidopsis seeds were germinated on medium containing various levels of the indicated chemicals for 3 d before hypocotyl length was measured. The seedlings at left were 3-d-old dark-grown wild-type Arabidopsis grown on half-strength MS medium or half-strength MS medium containing MeIAA or the indicated compounds. IAA, MeIAA, and IAA–Ala concentrations were 800 nM. Right, hypocotyl elongation of dark-grown seedlings is more inhibited by MeIAA than by IAA or IAA–Ala.
(C) Effect of MeIAA on the growth of the auxin-resistant mutants axr1, tir1, and nph4 and the auxin influx carrier mutant aux1. Seedlings were grown on 1 μM MeIAA in the dark for 3 d. The graph at right shows the decreased sensitivity of auxin-resistant mutants to MeIAA. Wild-type Arabidopsis and mutants were germinated on medium containing various concentrations of MeIAA for 3 d before hypocotyl length was measured. Because the phenotype of tir1 on MeIAA is very similar to that of axr1, only the axr1 data are shown. Error bars represent standard deviations.
We then tested the responses of known auxin signaling mutants to exogenous MeIAA. As shown in Figure 7C, the auxin-resistant mutants axr1 (Lincoln et al., 1990), tir1 (Ruegger et al., 1998), and nph4 (Harper et al., 2000) all had decreased sensitivity to MeIAA, suggesting that these auxin signaling components are also critical for the auxin activities of MeIAA. By contrast, the auxin transport mutant aux1 had the same sensitivity to MeIAA as wild-type plants (Figure 7C), suggesting that MeIAA can bypass the auxin polar transport system.
DISCUSSION
In this work, we present evidence that IAMT1, an IAA carboxyl methyltransferase, plays critical roles in Arabidopsis leaf development. In addition, characterization of the iamt1-D phenotype provides a basis for further analysis of the role of IAA methylation in auxin homeostasis and plant development.
We showed that overexpression of IAMT1 resulted in hyponastic leaf phenotypes, whereas knockdown of IAMT1 expression produced an opposite leaf phenotype (Figures 1 and 3), indicating that IAMT1 can affect the differential growth of leaf cells. Both plant hormone biosynthetic/signaling genes and nonhormone genes have been reported to regulate Arabidopsis leaf development. Auxin overproduction mutants such as rooty (King et al., 1995), sur2 (Delarue et al., 1998; Barlier et al., 2000), yucca (Zhao et al., 2001), and iaaM overexpression lines (Romano et al., 1995) all have epinastic leaves similar to the phenotype we observed for IAMT1-RNAi transgenic plants. On the other hand, the auxin signaling mutant arf7/nph4 displayed a hyponastic leaf phenotype similar to those of iamt1-D and IAMT1 overexpression transgenic lines (Harper et al., 2000).
Many other genes, including some of the TCP genes, have been shown to participate in regulating leaf curvature development (Nath et al., 2003). Decreased mRNA levels of some TCP genes in the jaw1 overexpression mutant led to leaf defects (Palatnik et al., 2003). Interestingly, the expression levels of four Arabidopsis TCP genes (TCP3, TCP4, TCP10, and TCP24) that are highly similar to the snapdragon curvature regulation gene CINCINNATA (Nath et al., 2003) are also downregulated in iamt1-D (Figure 5). The higher the IAMT1 expression level, the lower the TCP mRNA steady state level (Figure 5). Another curly leaf gene, HASTY (Telfer and Poethig, 1998), was also downregulated in iamt1-D plants (Figure 5), but the expression levels of AGAMOUS (Mizukami and Ma, 1992) and CLF (Goodrich et al., 1997) were not altered (data not shown). Therefore, it appears that increased expression of IAMT1 leads to decreased expression of leaf developmental genes such as TCP and HASTY, which in turn causes the curly leaf phenotypes. Although the exact mechanism by which IAMT1 affects the expression of TCP genes and other genes is not clear, it appears that alteration of IAA homeostasis caused by changes of the expression levels of the IAA carboxyl methyltransferase gene IAMT1 may be involved.
The biological activities of IAA esters including MeIAA have long been studied in bioassays; however, the evidence that these esters play any physiological roles in plant growth and development has never been conclusive. Although early studies found that IAA esters displayed auxin activities similar to free IAA and that in some cases IAA esters appeared to be even more potent than free IAA, it is not clear whether plants even make MeIAA in vivo (Zimmerman and Hitchcock, 1937). The search for MeIAA in planta has a long history, but most of the early reports of MeIAA in plants were later described as artifacts generated during the extraction and purification processes with alcohol-containing buffers. In collaboration with Jerry Cohen, we developed an isotope dilution method with 13C-labeled MeIAA as the internal standard to analyze MeIAA in planta (J. Cohen and Y. Zhao, unpublished data), but we did not detect any MeIAA in Arabidopsis rosette leaves (Y. Zhao and J. Cohen, unpublished data). We suspect that the MeIAA levels in leaves could be below the detection limit of our method or that MeIAA undergoes rapid turnover.
Although we do not yet have direct evidence that IAMT1 produces MeIAA in plants, the data presented here strongly suggest that IAMT1 is involved in auxin homeostasis, likely through the methylation of IAA. First, like many known auxin mutants, such as aux1, eir1, and axr2, IAMT1 overexpression lines lacked proper responses to gravity in both hypocotyls and roots (Figure 2A), a phenotype consistent with our hypothesis that IAMT1 affects auxin homeostasis. Second, IAMT1 overexpression lines showed decreased responses to exogenous IAA but not to the synthetic auxin 2,4-D (Figure 2C), presumably because IAMT1 can methylate IAA but could not use 2,4-D as a substrate. In response to exogenous IAA treatment, the expression levels of an auxin reporter, DR5, in IAMT1 overexpression lines were much lower than in wild-type plants. Moreover, IAMT1 overexpression lines make fewer lateral roots than wild-type plants in response to IAA treatment (Figure 2C). Finally, IAMT1 has been shown to specifically methylate IAA in vitro with a much higher value of the kinetic specificity constant (Kcat/Km) for IAA than for salicylic acid or other tested compounds (Zubieta et al., 2003). Furthermore, the IAMT1 crystal structure and additional biochemical experiments indicate that IAMT1 is highly specific for IAA (Y. Yang, J.R. Ross, E. Pichersky, and J.P. Noel, unpublished data).
Because of the nonpolar nature of MeIAA, methylation of IAA could provide a regulatory mechanism for modulating auxin activities that previously known IAA modifications such as forming conjugates with amino acids and sugars could not achieve. A major difference between MeIAA and IAA conjugates with sugar or amino acid is that MeIAA is a nonpolar molecule, whereas the other conjugates contain many polar or charged groups, which could render different distribution/transport efficiency for these compounds. In general, a nonpolar molecule can diffuse through membranes, whereas a polar molecule often requires an active transport system. That MeIAA is more active than IAA in bioassays may result from a more efficient uptake of MeIAA than IAA, an explanation that is also consistent with our findings that all of the IAA esters tested in this work were more potent than IAA in bioassays. In addition, defects in auxin transport genes such as AUX1 led to decreased responses to IAA treatment but did not affect the potency of MeIAA, indicating that MeIAA does not require the auxin transport system for its uptake.
METHODS
Mutagenesis of Arabidopsis and Growth Conditions
Arabidopsis thaliana plants, ecotype Columbia, were grown at 22 ± 2°C under long-day conditions (16 h of light/8 h of darkness) and were transformed with Agrobacterium tumefaciens GV3101 harboring the activation-tagging plasmid pSKI015 (Weigel et al., 2000) by the floral dip method (Clough and Bent, 1998). Harvested seeds were placed on half-strength Murashige and Skoog (MS) medium containing 20 μg/mL dl-phosphinothricin and synchronized for 3 d at 4°C before being placed under long-day conditions for 1 week. Drug-resistant seedlings were transferred to soil and grown under the same conditions as described above.
For root elongation assay and lateral root initiation, surface-sterilized seeds were sowed on half-strength MS medium containing 1% sucrose at a density of 200 per plate and incubated in the dark for 48 h at 4°C. Plates were vertically placed in a long-day, 22°C incubator for 4 d. The seedlings were then transferred to assay plates containing various concentrations of either IAA or 2,4-D, and the initial positions of the root tips were marked to determine root elongation. The plates were photographed after 3 d of incubation, and root elongation was determined using SPOT software. The number of lateral roots was counted at day 5 after transferring.
Primers and PCR Conditions
The flanking sequence of the T-DNA insertion was determined by thermal asymmetric interlaced PCR (Liu and Huang, 1998). The specific and arbitrary degenerate primers used were as described previously (Qin et al., 2003). Three primers, P1, P2, and LS4, were designed for cosegregation analysis, with P1 and P2 corresponding to Arabidopsis genomic sequences flanking the T-DNA insertion and LS4 corresponding to the sequence of the T-DNA vector (Figure 1). The primers were as follows: P1, 5′-AAGTAGGCAAGGCATGATCT-3′; P2, 5′-TTACAACGAGTCAAGGACAC-3′; and LS4, 5′-TTGGTAATTACTCTTTCTTTTCCTCC-3′. For RT-PCR analysis and isolation of the IAMT1 gene, the primer pair IAMT1-1 (5′-ATGGGTTCTAAGGGAGACAACG-3′) and IAMT1-2 (5′-TTAAGTAAAAGACAAAGAAGCGACAATG-3′) was designed according to the cDNA sequence from the National Center for Biotechnology Information (accession number NM_124907). The TUB2 (for β-tubulin) gene was used as an internal control in RT-PCR, and the primers for TUB2 were 5′-GTTCTCGATGTTGTTCGTAAG-3′ and 5′-TGTAAGGCTCAACCACAGTAT-3′. Two primers, IAMT1-3 (5′-TGAAAGGTGGCAAAGGACAAGA-3′) and IAMT1-4 (5′-ATGCAAGGAGAAGGCAGAGTGG-3′) were designed based on the coding region sequences to detect both endogenous transcripts and transgene transcripts in RNAi lines. Two other primers, IAMT1-5 (5′-TTCTTCCACCACTTGTCTCTAA-3′) and IAMT1-6 (5′-CACACGAACATATTTCTTTTTC-3′) were designed based on the coding region and 3′ untranslated region sequence to detect only the endogenous transcripts of IAMT1 (Figure 2C). PCR was performed for 26 to 40 cycles (94°C for 30 s, 58°C for 30 s, and 72°C for 1.5 min) (Qu et al., 2003).
Gel Blot Hybridization
Total RNA was extracted from leaves of 35-d-old plants, and 12 μg of total RNA was used for RNA gel blot analysis as described previously (Qu et al., 2003). The probes for IAMT1, TCP3, TCP4, TCP10, TCP17, TCP24, and HASTY were amplified from wild-type Arabidopsis leaf mRNA. The primers used were as follows: for IAMT1, primers IAMT1-1 and IAMT1-2; for TCP3, 5′-ACCGTCACGAGGCAATACAC-3′ and 5′-AGAATCGGATGAAGCAGAGG-3′; for TCP4, 5′-GATGGTCCACCGTCGCTTCT-3′ and 5′-CCGTCGTGCTGCTCCTCTTC-3′; for TCP10, 5′-TACTAAACCGGAATCTCCCA-3′ and 5′-ATCCCAAGAACGAAACGAAT-3′; for TCP17, 5′-TTGACGCAACGATGGAATAA-3′ and 5′-GACCACCACCGAGAAACGAA-3′; for TCP24, 5′-TCGTCTCGTATCATTAGGGTTT-3′ and 5′-CGGTTACTCGGTTGTTGGTC-3′; for HASTY, 5′-ACAAGAGGGCAGAGCAAAGG-3′ and 5′-AGTGAGACACGGGAGCGAAA-3′. The PCR products were purified before being labeled with [32P]dCTP. Membrane hybridization and washes were performed as described (Qu et al., 2003). For RT-PCR, first-strand cDNA was synthesized in a 20-μL reaction containing 3 μg of total RNA, oligo-dT12–18, and SuperScript reverse transcriptase (Invitrogen). The reaction was allowed to proceed at 42°C for 50 min before being terminated by treatment at 70°C for 15 min. Subsequent RT-PCR was performed for 40 cycles with the IAMT1-1 and IAMT1-2 primers, and the products were transferred to membranes and hybridized as described above.
Overexpression and RNAi Constructs and Arabidopsis Transformation
The cDNA of IAMT1 was amplified from wild-type Arabidopsis by RT-PCR and cloned into the EcoRV site of pBluescript SK+. Recombinant plasmids with the IAMT1 gene in both sense (designated pBIAMT1) and antisense (designated pAIAMT1) orientations were identified and confirmed by sequencing. The IAMT1 promoter was amplified from Arabidopsis genomic DNA with primers 5′-TTAGGGAAACAAGAATGACAACA-3′ and 5′-TCTCTTTCTCTTTCTCTATGGATC-3′ and cloned (designated pBIAMT1P). The CaMV 35S enhancer tetrad was amplified from pSKI015 with the primers 5′-TAATACGACTCACTATAGGG-3′ and 5′-CTAGATCCGAAACTATCAGTGTT-3′ and cloned into the EcoRV site of pBluescript SK+ (designated pA4Ehancer). The YABBY3 promoter (a 2.3-kb fragment upstream from the start codon of YABBY3) was amplified from genomic DNA of wild-type Arabidopsis plants with the primers 5′-CGAGATCAATGGCTAGAAGAACCA-3′ and 5′-GGAGTAAGAGAGAGAGGAGGGCT-3′ and then cloned into the EcoRV site of pBluescript SK+ (designated pBYABBY3P). The 1-kb GUS fragment was amplified from pCAMBIA1381 with the primers GUS-1 (5′-GCTTCGCGTCGGCATCC-3′) and GUS-2 (5′-CACCGAAGTTCATGCCAGTC-3′) and cloned into the EcoRV site of pBluescript SK+ (designated pBGUS). For the preparation of plant expression vector, the HindIII-EcoRI fragment of yy43 (Yamamoto et al., 1998) was introduced into pPZP111 (Hajdukiewicz et al., 1994) to generate pQG110. 4Enhancer-IAMT1 was constructed by ligation of four DNA fragments: the HindIII-XbaI fragment from pQG110, the HindIII-EcoRI enhancer tetrad fragment from pA4Ehancer, the EcoRI-KpnI IAMT1 promoter fragment from pBIAMT1P, and the KpnI-XbaI IAMT1 fragment from pAIAMT1. IAMT1-RNAi was constructed by ligation of the following DNA fragments: the XbaI-KpnI fragment from pQG110, the XbaI-SalI IAMT1 fragment from pACUR, the SalI-EcoRI 1-kb GUS fragment from pBGUS, and the EcoRI-KpnI fragment from pBIAMT1. 4EnPYABBY3-IAMT1 was obtained by ligation of four DNA fragments: the HindIII-XbaI larger fragment from pQG110, the HindIII-ScaI enhancer tetrad fragment from pA4Ehancer, the ScaI-KpnI IAMT1 promoter fragment from pBYABBY3P, and the KpnI-XbaI IAMT1 fragment from pAIAMT1.
Wild-type or iamt1-D homozygous mutant plants were used as the recipients for Agrobacterium-mediated transformation by the floral dip method. The seeds of transgenic plants were screened on half-strength MS medium containing 50 μg/mL kanamycin, and the resistant seedlings were transferred to soil.
Construction of pIAMT1P-GUS and Histochemical GUS Assays
IAMT1 promoter (2.7 kb upstream from the start codon) was amplified from Arabidopsis genomic DNA with the primers 5′-TTAGGGAAACAAGAATGACAACA-3′ and 5′-TGTCTCCCTTAGAACCCATTCTC-3′ and cloned (designated pBIAMT1PA). The EcoRI-HindIII IAMT1 promoter fragment from pBIAMT1PA was cloned into pCAMBIA1381Xc to generate pIAMT1P-GUS. The histochemical GUS assay was performed in a staining solution containing 0.5 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide in 0.1 M Na2HPO4, pH 7.0, 10 mM Na2EDTA, 0.5 mM potassium ferricyanide/ferrocyanide, and 0.06% Triton X-100 (Jefferson et al., 1987). Samples were infiltrated under vacuum for 10 min and then incubated at 37°C overnight. The staining buffer was removed, and the samples were cleared in 70% ethanol.
The auxin-responsive DR5-GUS reporter line was crossed with iamt1-D, and the F3 seedlings homozygous for both the DR5-GUS reporter and iamt1-D were subjected to GUS staining as described above.
Bioassays for MeIAA Activities
For analysis of dark-grown seedlings, Arabidopsis seeds were put on half-strength MS medium containing various concentrations of MeIAA, IAA, IAA–Ala, or other chemicals. The plates were subjected to cold treatment for 2 d in total darkness at 4°C and then transferred to light at 23°C for 2 h before they were wrapped with aluminum foil. The seeds were germinated in total darkness at 23°C for 3 d, and the dark-grown seedlings were photographed and characterized. The hypocotyl length of the dark-grown seedlings was measured using NIH Image software.
For analysis of light-grown seedlings, Arabidopsis seeds (Columbia ecotype) were germinated on solid half-strength MS medium for 3 d after stratification for 3 d. The seedlings were then transferred to half-strength MS medium with or without different concentrations of IAA, MeIAA and 2,4-D and grown vertically for 5 d. The roots were photographed, and root elongation and lateral root initiation were analyzed with SPOT software.
Accession Numbers
Sequence data for IAMT1, TCP3, TCP4, TCP10, TCP17, TCP24, HASTY, and YABBY3 can be found in the GenBank/EMBL data libraries under accession numbers NM_124907 (At5g55250), NM_104201 (At1g53230), NM_180258 (At3g15030), NM_128662 (At2g31070), NM_120889 (At5g08070), NM_179399 (At1g30210), AY198396 (At3g05040), and NM_116235 (At4g00180), respectively.
Supplementary Material
Acknowledgments
We thank Xing Wang Deng and Tim Nelson (Yale University), Hongwei Guo and Hai Li (SALK Institute), Xiangdong Fu (John Innes Center), Matthew Terry (University of Southampton), Haruko Okamoto (University of Oxford), Hongwei Xue (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences), Jian Hua (Cornell University), and Lin Wang (Institute of Microbiology, Chinese Academy of Sciences) for helpful suggestions and valuable discussions regarding this article. We also thank the other members of the authors' group at Peking University, M. Liu, X.-H. Deng, J. Ma, and J. Wang, for technical assistance. This work was supported by the National Natural Science Foundation of China (Grants 30221120261 and 30321001), the National Special Projects for R&D of Transgenic Plants (J99-A-001), the Key Grant Project of the Chinese Ministry of Education (NO 00-08), and the Excellent Young Teachers Program of the Ministry of Education of China (to L.-J.Q.). Part of this work was also funded by National Institutes of Health Grant 1RO1GM68631 to Y.Z. and National Science Foundation Grant MCB-0312466 to E.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Li-Jia Qu (qulj@pku.edu.cn).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.034959.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 51–66. [DOI] [PubMed] [Google Scholar]
- Bartel, B., and Fink, G.R. (1995). ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748. [DOI] [PubMed] [Google Scholar]
- Berleth, T., Krogan, N.T., and Scarpella, E. (2004). Auxin signals—Turning genes on and turning cells around. Curr. Opin. Plant Biol. 7, 553–563. [DOI] [PubMed] [Google Scholar]
- Chen, F., D'Auria, J.C., Tholl, D., Ross, J.R., Gershenzon, J., Noel, J.P., and Pichersky, E. (2003). An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36, 577–588. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen, J.D., and Bandurski, R.S. (1982). Chemistry and physiology of the bound auxins. Annu. Rev. Plant Physiol. 33, 403–430. [Google Scholar]
- Cohen, J.D., Slovin, J.P., and Hendrickson, A.M. (2003). Two genetically discrete pathways convert tryptophan to auxin: More redundancy in auxin biosynthesis. Trends Plant Sci. 8, 197–199. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1995). Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 92, 4114–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria, J.C., Chen, F., and Pichersky, E. (2003). The SABATH family of methyltransferases in Arabidopsis thaliana and other plant species. Recent Adv. Phytochem. 37, 95–125. [Google Scholar]
- Davies, R.T., Goetz, D.H., Lasswell, J., Anderson, M.N., and Bartel, B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue, M., Prinsen, E., Onckelen, H.V., Caboche, M., and Bellini, C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14, 603–611. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., and Estelle, M. (2004). Auxin signaling and regulated protein degradation. Trends Plant Sci. 9, 302–308. [DOI] [PubMed] [Google Scholar]
- Dudareva, N., Murfitt, L.M., Mann, C.J., Gorenstein, N., Kolosova, N., Kish, C.M., Bonham, C., and Wood, K. (2000). Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell 12, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, N.L., and Kosuge, T. (1986). Cloning of the gene for indoleacetic acid-lysine synthetase from Pseudomonas syringae subsp. savastanoi. J. Bacteriol. 166, 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A polycomb-group gene regulated homeotic gene expression in Arabidopsis. Nature 386, 44–51. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Harper, R.M., Stowe-Evans, E.L., Luesse, D.R., Muto, H., Tatematsu, K., Watahiki, M.K., Yamamoto, K., and Liscum, E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutzinger, O., and Kosuge, T. (1968). Microbial synthesis and degradation of indole-3-acetic acid. III. The isolation and characterization of indole-3-acetyl-ɛ-l-lysine. Biochemistry 7, 601–605. [DOI] [PubMed] [Google Scholar]
- Jackson, R.G., Kowalczyk, M., Li, Y., Higgins, G., Ross, J., Sandberg, G., and Bowles, D.J. (2002). Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: Phenotypic characterisation of transgenic lines. Plant J. 32, 573–583. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J.J., Stimart, D.P., Fisher, R.H., and Bleecker, A.B. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7, 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasswell, J., Rogg, L.E., Nelson, D.C., Rongey, C., and Bartel, B. (2000). Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis. Plant Cell 12, 2395–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere, S., Tellez, R., Rampey, R.A., Matsuda, S.P., and Bartel, B. (2002). Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452. [DOI] [PubMed] [Google Scholar]
- Leyser, O. (2002). Molecular genetics of auxin signaling. Annu. Rev. Plant Biol. 53, 377–398. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., and Huang, N. (1998). Efficient amplification of insert end sequences from bacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol. Biol. Rep. 16, 175–181. [Google Scholar]
- Ljung, K., Hul, A.K., Kowalczyk, M., Marchant, A., Celenza, J., Cohen, J.D., and Sandberg, G. (2002). Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 50, 309–332. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131. [DOI] [PubMed] [Google Scholar]
- Murfitt, L.M., Kolosova, N., Mann, C.J., and Dudareva, N. (2000). Purification and characterization of S-adenosyl-L-methionine:benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methyl benzoate in flowers of Antirrhinum majus. Arch. Biochem. Biophys. 382, 145–151. [DOI] [PubMed] [Google Scholar]
- Nath, U., Crawford, B.C., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Qin, G., et al. (2003). Obtaining and analysis of flanking sequences from T-DNA transformants of Arabidopsis. Plant Sci. 165, 941–949. [Google Scholar]
- Qu, L.-J., et al. (2003). Molecular cloning and functional analysis of a novel type of Bowman-Birk inhibitor gene family in rice. Plant Physiol. 133, 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey, R.A., LeClere, S., Kowalczyk, M., Ljung, K., Sandberg, G., and Bartel, B. (2004). A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, C.P., Hein, M.B., and Klee, H.J. (1991). Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev. 5, 438–446. [DOI] [PubMed] [Google Scholar]
- Romano, C.P., Robson, P.R., Smith, H., Estelle, M., and Klee, H. (1995). Transgene-mediated auxin overproduction in Arabidopsis: Hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 27, 1071–1083. [DOI] [PubMed] [Google Scholar]
- Ross, J.R., Nam, K.H., D'Auria, J.C., and Pichersky, E. (1999). S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 367, 9–16. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Song, J.T., Cheong, J.J., Lee, Y.-H., Lee, Y.-W., Hwang, I., Lee, J.S., and Choi, Y.D. (2001). Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98, 4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev, V., Silverman, P., and Raskin, I. (1997). Airborne signaling by methyl salicylate in plant pathogen resistance. Nature 385, 718–721. [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Spena, A., Prinsen, E., Fladung, M., Schulze, S.C., and Van Onckelen, H. (1991). The indoleacetic acid-lysine synthetase gene of Pseudomonas syringae subsp. savastanoi induces developmental alterations in transgenic tobacco and potato plants. Mol. Gen. Genet. 227, 205–212. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M.T., Maldonado, M.C., and Suza, W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerszen, J.B., Szczyglowski, K., and Bandurski, R.S. (1994). iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265, 1699–1701. [DOI] [PubMed] [Google Scholar]
- Telfer, A., and Poethig, R.S. (1998). HASTY: A gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125, 1889–1898. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Y.Y., Matsui, M., Ang, L.H., and Deng, X.W. (1998). Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell 10, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Christensen, S.K., Fankhauser, C., Cashman, J.R., Cohen, J.D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Hull, A.K., Gupta, N.R., Goss, K.A., Alonso, J., Ecker, J.R., Normanly, J., Chory, J., and Celenza, J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16, 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, P.W., and Hitchcock, A.E. (1937). Comparative effectiveness of acids, esters, and salts as growth substances and methods of evaluating them. Contrib. Boyce Thompson Inst. 12, 321–343. [Google Scholar]
- Zubieta, C., Ross, J.R., Koscheski, P., Yang, Y., Pichersky, E., and Noel, J.P. (2003). Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15, 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.