Abstract
The two guard cells of a stoma are produced by a single symmetric division just before terminal differentiation. Recessive mutations in the FOUR LIPS (FLP) gene abnormally induce at least four guard cells in contact with one another. These pattern defects result from a persistence of precursor cell identity that leads to extra symmetric divisions at the end of the cell lineage. FLP is likely to be required for the correct timing of the transition from cell cycling to terminal differentiation. FLP encodes a two-repeat (R2R3) MYB protein whose expression accumulates just before the symmetric division. A paralogous gene, MYB88, overlaps with FLP function in generating normal stomatal patterning. Plants homozygous for mutations in both genes exhibit more severe defects than flp alone, and transformation of flp plants with a genomic MYB88 construct restores a wild-type phenotype. Both genes compose a distinct and relatively basal clade of atypical R2R3 MYB proteins that possess an unusual pattern of amino acid substitutions in their putative DNA binding domains. Our results suggest that two related transcription factors jointly restrict divisions late in the Arabidopsis thaliana stomatal cell lineage.
INTRODUCTION
Cell distribution and differentiation require a balance between proliferation and cell specification in time and space. This is especially evident in stem cell lineages where some progenitors divide a number of times, then stop dividing and undergo terminal differentiation (Tokumoto et al., 2002). Stomata are a terminally differentiated product of a specialized stem cell compartment in the developing epidermis of plant shoots (Nadeau and Sack, 2003). Each stoma is a hydrostatic valve made up of two guard cells around a pore, which restricts water loss and allows entry of carbon dioxide used in photosynthesis. Stomata are thus essential for plant survival and productivity, and the genes required for their formation were critical for the evolution of land plants.
Cell divisions are central to the production and spacing of Arabidopsis thaliana stomata. A stoma forms after at least one asymmetric division, followed by a single symmetric one (Nadeau and Sack, 2002b). Each asymmetric division produces a smaller cell, the meristemoid, that ultimately forms the stoma and a larger cell that can divide unequally to extend the stem cell lineage (Nadeau and Sack, 2003). Meristemoids acquire a guard mother cell (GMC) fate after asymmetric divisions cease. Terminal differentiation occurs after the GMC divides equally and the two daughter cells coordinately differentiate into the stoma (Geisler et al., 2000).
Stomata are separated by at least one intervening cell (Figure 1A), a pattern that optimizes gas exchange and guard cell ion transport. This spacing arises when a piggyback asymmetric division is oriented so that the new cell wall does not touch the preexisting stoma (GMC3 in Figure 1G), a process likely to require intercellular signaling (Geisler et al., 2000). Mutations in several genes disrupt these spacing divisions and induce clusters of stomata in contact with one another. These genes include TOO MANY MOUTHS (TMM), which encodes a receptor, STOMATAL DENSITY AND DISTRIBUTION1 (SDD1), which encodes a processing protease, and YODA, which encodes a mitogen-activated protein kinase kinase kinase, all of which are identities consistent with functions in signal transduction (Berger and Altmann, 2000; Nadeau and Sack, 2002a; Bergmann et al., 2004). In addition to stomatal clusters, mutations in each locus cause excess asymmetric divisions, suggesting that these genes also function as negative regulators of stem cell proliferation (Nadeau and Sack, 2003; Bergmann, 2004).
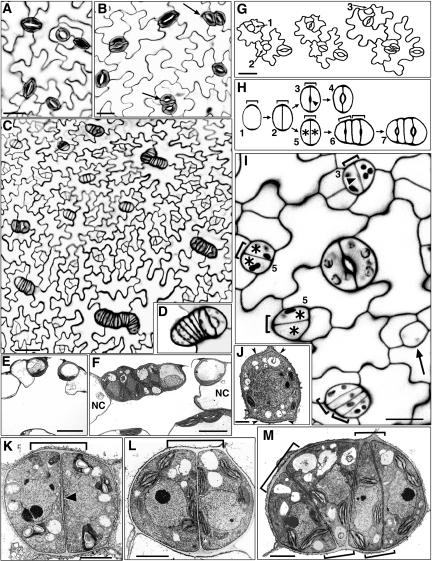
Figure 1.
Mutations in FLP Extend Precursor Cell Fate and Induce Extra Symmetric Divisions.
(A) Normal stomatal spacing in wild-type leaf. Confocal image of cell wall fluorescence (from propidium iodide).
(B) flp-1 stomatal clusters with four (arrows) or three (top left) guard cells. Confocal image.
(C) flp-7 developing epidermis showing elongated clusters. Confocal image.
(D) Cluster in flp-7 includes one mature stoma (right).
(E) Cross section of wild-type stoma over a substomatal cavity. The neighbor cells have more depth than the guard cells. Reprinted from Zhao and Sack (1999). Transmission electron micrograph (TEM).
(F) TEM cross section of flp-7 stomatal cluster that includes one mature stoma (right) and four other cells of the same depth as the guard cells. Cells outside the cluster (NC) are deeper than those inside.
(G) Drawing of the same flp-1 field that developed over several days. GMCs 1 and 2 divided symmetrically. GMC 2 daughter cells divided again. An asymmetric division correctly positioned the meristemoid (data not shown) that gave rise to GMC 3.
(H) Illustration of developmental stages in the wild type (1 to 4) and in flp-1 (1, 2, and 5 to 7). End cell wall thickening in GMC (stage 1, bracket) marks the future site of division. The two daughter cells from the GMC division normally form a stoma (note pore cell wall thickening at arrowhead in stage 3). However, in flp-1, these two daughter cells (asterisks, stage 5) divide again before developing pore thickenings (stage 6). Two paired brackets are shown in stage 6 to indicate that the first wall thickening present from the first symmetric division (stages 1 and 2) is later amplified laterally prior to and during the second symmetric division.
(I) Confocal image of developing flp-1 leaf epidermis. The GMC shown by the arrow (bottom right) does not yet have an end wall thickening. GMC division produces daughter cells with (arrowhead, top, stage 3) or without (asterisks, stage 5) pore wall thickenings. Note that all developing cell pairs shown are roughly comparable in size despite the presence or absence of the pore thickening. All cells that divided symmetrically have end wall thickenings (brackets) that presumably developed previously in GMC-like cells. The developing cluster at the bottom has not yet reached stage 6 because no pore thickenings are present.
(J) A wild-type GMC showing end wall thickenings (between pairs of arrowheads; stage 1). TEM from Zhao and Sack (1999).
(K) TEM of a developing wild-type stoma (stage 3) in paradermal view. The symmetric division of the GMC was correctly placed as shown by the new wall meeting the end wall thickenings (bracket). Arrowhead indicates pore wall thickening.
(L) TEM of a developing flp-1 cell pair (stage 5) in paradermal view. The symmetric division was correctly placed, but a pore wall thickening is absent, consistent with a disruption of guard cell differentiation.
(M) TEM of a developing flp-1 cluster at stage just prior to stage 6 (pore wall thickenings not yet formed). Note that the newly produced, oblique cell wall joins a wall thickening (bracket, top left) that is separated from that present before the extra round of divisions. The two brackets at the bottom indicate increased wall thickenings that probably developed before each of the cells at stage 5 divided.
Bars in (A), (B), (G), and (I) = 15 μm; bar in (C) = 25 μm; bars in (E) and (F) = 10 μm; bar in (J) = 1 μm; bars in (K) to (M) = 2 μm.
Whereas mutations in the above genes produce loose stomatal clusters with stomata arranged at different angles, clusters in the four lips (flp) mutant are mostly linear with stomata packed side by side (Yang and Sack, 1995; Nadeau and Sack, 2002b). This difference suggests that FLP has a novel function compared with earlier acting genes in the pathway.
To determine how FLP controls stomatal patterning, we analyzed stomatal development in flp mutants and established gene identity. We report that FLP is an atypical two-repeat (R2R3) MYB protein that is expressed late in the stomatal cell lineage and acts by restricting cell divisions before terminal differentiation. A closely related gene is partially redundant with FLP and both constitute a distinct and near basal clade of Arabidopsis R2R3 MYB proteins.
RESULTS
Phenotype of flp Alleles
All flp mutant alleles characterized exhibit stomata in direct contact. Alleles with weaker phenotypes, such as flp-1 and flp-2, typically contain two laterally aligned stomata that otherwise appear normal (Figure 1B). Clusters in severe alleles, flp-7 and flp-8, contain many more cells than in weaker alleles (Figures 1B to 1D), and these clusters have two cell types in addition to mature stomata. The first type includes developing or arrested guard cells recognized by pore wall thickenings that are not yet open. The second type includes cells whose identity could not be assigned using morphological criteria. In addition to larger clusters, severe alleles exhibit a higher frequency of clustered stomata (the number of clusters divided by the sum of normal stomata plus clusters). This parameter was measured to be 43% (±4.0% se), 25% (±1.5%), and 16% (±1.2%), for flp-7, flp-8, and flp-1, respectively. The presence of many normally spaced stomata presumably contributes to the viability and fecundity of flp plants.
flp Induces Ectopic Precursor Cell Division and Delays Stomatal Differentiation
The ontogeny of flp stomatal clusters was analyzed to evaluate how FLP controls stomatal patterning and development. The GMC is the last precursor cell in the pathway and develops from a meristemoid that is an earlier, self-renewing precursor. Several lines of evidence show that paired flp-1 stomata originate through extra symmetric divisions rather than through the production of new meristemoids and that these extra divisions probably take place in cells with an abnormally persistent GMC identity (Figure 1H).
First, the behavior of cells through time was followed using nondestructive impressions of the leaf surface. As in the wild type, normally spaced stomata in flp-1 developed after division of a GMC. By contrast, two adjacent stomata in flp arose when each progeny cell of a GMC divided again (Figures 1G and 1H). Thus, cells in flp-1 stomatal clusters are clonally related, meaning that each cluster can be traced back to a single original parent GMC (49 out of 50 cases).
A clonal origin is also consistent with cluster morphology. GMCs differ in size and shape from other epidermal cells and do not expand as much in their depth as other cells. Clusters derived from more than one parent cell would be more likely to have an irregular outline and heterogeneity in cell depth, but flp clusters usually have a smooth and continuous cell wall outline in both paradermal and cross-sectional views (Figures 1B to 1F). This is most readily explained by a clonal origin, by divisions that are mostly parallel to each other, and by more expansion along the long than the short axis of the cluster.
Next, we used cytological data to assess the identity of the daughter cells that divide abnormally. We hypothesized that these divisions might take place in persistent GMCs, in young guard cells, or in cells of mixed identity. As shown in Figures 1H and 1J, a specific marker for wild-type GMCs is the presence of end wall thickenings that predict the division plane (Zhao and Sack, 1999). These thickenings remain apparent in stomata (Figure 1K). A second novel wall thickening that appears during stomatal morphogenesis is a lens-shaped swelling that eventually forms the stomatal pore (arrowheads in Figures 1H, 1I, and 1K). In flp-1 plants, end wall thickenings were found in all first-generation GMCs (data not shown), suggesting that the first GMC division is normal regardless of subsequent daughter cell fate. Pore thickenings were also detected in many GMC daughter cells as would be expected, since most stomata in flp-1 plants are patterned and differentiate normally. However, other GMC daughter cells lacked pore thickenings even though the cells were of a size and shape normally associated with terminal differentiation (Figures 1H, 1I, and 1L). This suggests that guard cell differentiation is delayed in a fraction of GMC daughter cells in flp-1. In addition, the new cell walls present in developing clusters were observed to intersect end wall thickenings (Figures 1H, 1I, and 1M). Because end wall thickenings are normally only produced in GMCs, the presence of new thickenings in each daughter cell provides evidence for an abnormal reiteration of a GMC marker. These cytological data are consistent with a scenario in which flp-1 causes a persistence of GMC identity in daughter cells that later produce ectopic stomata and in which flp-1 delays the timing of the transition to guard cell fate. This scenario is further supported by observations that the divisions that produce the flp clusters are usually symmetric in size and in cell fate, characteristics normally only found in GMCs in the stomatal pathway.
A third set of data supporting this view comes from an analysis of the expression frequency of a guard cell–specific construct, the promoter of the Arabidopsis KAT1 gene driving the β-glucuronidase reporter (ProKAT1:GUS; Nakamura et al., 1995). As expected for a potassium channel involved in stomatal opening and closing, this promoter drove strong GUS staining in all mature guard cells in both the wild type and flp-1 (n = 590 and 507; Figures 2A and 2D). The onset of expression during development was then determined for wild-type plants using differential interference contrast optics needed for visualizing the thin new cell wall that forms after the symmetric division of the GMC. Staining was present in most young guard cells (78% of 99 cell pairs) but absent from GMCs, indicating that KAT1 expression starts soon after symmetric division.
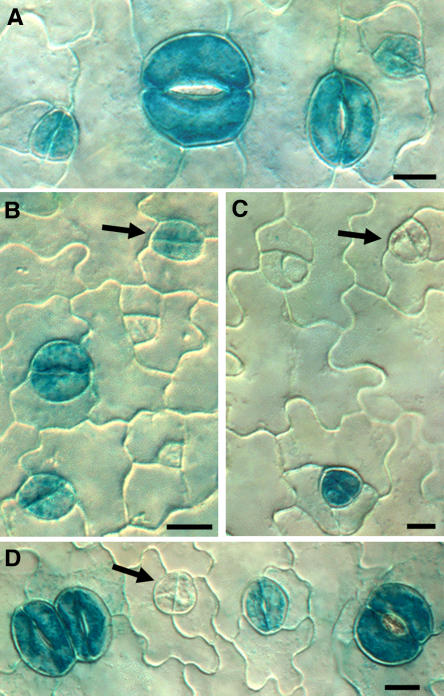
Figure 2.
Fewer GMC Daughter Cells Show Guard Cell–Specific ProKAT1:GUS Staining in flp-1.
(A) Wild type. GUS staining present in developing and mature stomata.
(B) Wild type. Arrow indicates staining in daughter cells produced by GMC division. Staining is absent from stomatal precursors (GMCs and meristemoids) in both the wild type and flp-1.
(C) and (D) flp-1. Arrows indicate daughter cells without staining that are likely to function as GMCs and divide one more time.
Bars in (A) and (B) = 7.5 μm; bars in (C) and (D) = 10 μm.
Because flp-1 plants have both normally patterned and clustered stomata, we scored ProKAT1:GUS staining separately in isolated cell pairs and in cells of developing clusters. The frequency of staining in young cell pairs in developing flp-1 clusters (four cells side by side) was roughly similar to that in young guard cells in wild-type plants (73 ± 1.5% se versus 78 ± 1.5%; n = 100 for each). By contrast, only 43 ± 1.1% (n = 248) of isolated cell pairs in flp-1 exhibited staining. It was not possible to predict at this stage whether these two cells were young guard cells that would have formed a stoma or whether each cell would have divided again symmetrically producing two stomata in contact. Side-by-side cell pairs, on the other hand, are very likely to be undergoing terminal differentiation and to develop into two adjacent stomata, which is the predominant cluster size in flp-1 (Yang and Sack, 1995). This probability is reflected in the same frequency of GUS staining as in young guard cells of wild-type plants. The reduced percentage of isolated cell pairs in flp-1 with GUS staining (Figure 2) suggests that some of these cells have not yet acquired a guard cell identity and might divide again. Together, these data are consistent with a model in which flp-1 clusters result from a reiteration of a GMC fate in cells that would normally differentiate directly into guard cells.
FLP Encodes an R2R3 MYB Protein
Using map-based cloning, FLP was found to be MYB124 (At1g14350), which encodes a two-repeat MYB putative transcription factor of previously unknown function (Figure 3). R2R3 MYB proteins possess two tandem imperfect MYB repeats of ∼50 amino acids each (Martin and Paz-Ares, 1997; Kranz et al., 2000; Stracke et al., 2001). Each repeat has three helices and three conserved Trp residues that help form the hydrophobic core of a helix-turn-helix structure used to bind target DNA (Ogata et al., 1992).
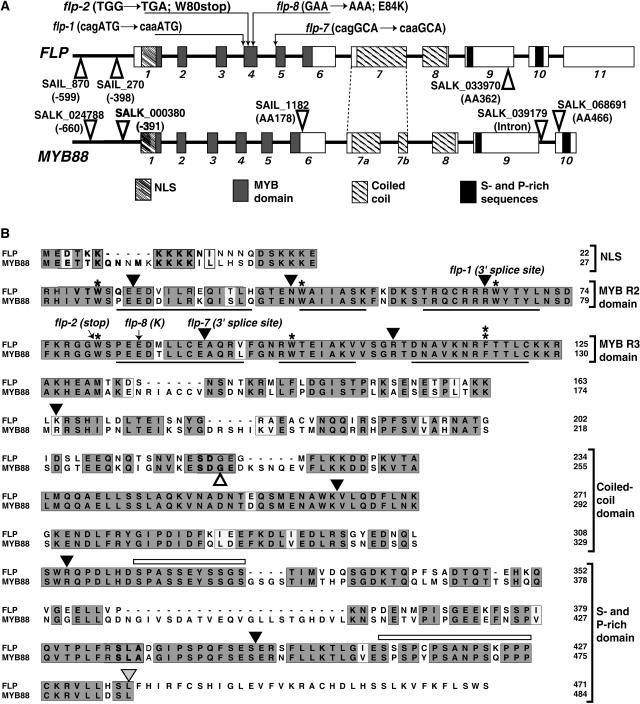
Figure 3.
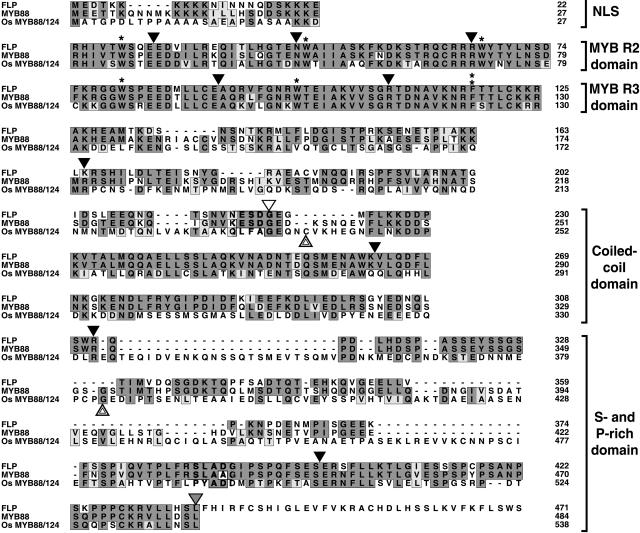
FLP and MYB88 Are Closely Related Arabidopsis R2R3 MYB Proteins.
(A) Gene structures. Exons are shown as boxes, introns as bold lines, domains by different fills, and T-DNA insertions by open triangles. NLS, predicted nuclear localization signal.
(B) Alignment of their amino acid sequences. Mutations in coding regions are indicated by arrows. The splice site lesions in flp-1and flp-7 are indicated above the positions of the respective introns. Arrowheads indicate the positions of introns. An arrowhead pointing directly to a residue signifies that the intron interrupts the codon. Black arrowheads, introns in both genes; gray arrowhead (residue 436 in FLP), intron specific to FLP; white arrowhead (residue 236 in MYB88), intron specific to MYB88. Single asterisks, conserved Trp (W) residues; double asterisk, Trp-to-Phe (W-to-F) substitution; black lines (in MYB domains), locations of three helices in each MYB repeat whose sites were predicted using PSIPRED (Jones, 1999). Sequences with dark gray shading, conserved residues; sequences with light gray shading, similar residues; open bars (in the S- and P-rich domain), regions rich in Ser and Pro residues.
The predicted full-length FLP protein is encoded by 11 exons and contains 471 amino acids. In addition to the R2R3 MYB domain, FLP also contains a predicted nuclear localization signal, a coiled-coil domain, and a C-terminal 163-residue region rich in Ser (16.6%) and Pro (9.8%) that might regulate transactivation (Martin and Paz-Ares, 1997; Jin and Martin, 1999).
Four flp alleles harbor G-to-A substitutions in different parts of the region coding for the MYB domain (Figure 3). The flp-8 allele contains a missense mutation in the first R3 helix that replaces a highly conserved, acidic Glu residue with a basic Lys residue. This Glu residue is present in 97% of all MYB proteins (Stracke et al., 2001). The flp-2 allele harbors a nonsense mutation that replaces the first R3 Trp codon with a termination codon leading to protein truncation.
The mutations in flp-1 and flp-7 are located in the 3′ splice site AG of introns 3 and 4, respectively. We confirmed the presence of splicing errors in these two alleles using RT-PCR. The analysis of flp-1 mRNA revealed only one mature transcript, which included intron 3. This transcript also showed a defect in the splicing of intron 5, in that another 5′ splice site four bases upstream of the normal site was used. The flp-7 mutation leads to various mis-splicing events in different transcripts, including the retention of either intron 4 or 5, the exclusion of exon 5, and the use of cryptic splice sites in exons 5 and 6. The predominant transcript resulted from the use of a newly produced 3′ AG splice site, with the A deriving from the G-to-A mutation and the G from exon 5, (i.e., cagGCA to caaGCA). All splicing errors in flp-1 and flp-7 plants are predicted to result in early translational termination in the third R2 helix and right after the first R3 helix, respectively, due to frameshift or intronic read-through. Because the flp-1, flp-2, and flp-7 alleles are all predicted to produce truncated proteins with incomplete MYB domains, these mutations are highly likely to disrupt protein activity.
Based on the identity of FLP, three more mutant alleles were identified in the Syngenta Arabidopsis Insertion Library (SAIL) and SALK collections of T-DNA insertions in Arabidopsis (Sessions et al., 2002; Alonso et al., 2003). We confirmed the locations of these T-DNA inserts in codon 362 within exon 9 (SALK_3397), at −599 where +1 is assigned to the A of ATG (SAIL_870) and at −398 (SAIL_270) (Figure 3A). All three alleles displayed flp-like paired stomata in contact. Their phenotypes resembled flp-1 and flp-2 much more than severe alleles (flp-7 and flp-8).
The five alleles with weaker phenotypes are all in a Columbia or C24 background. By contrast, both of the severe alleles, flp-7 and flp-8, are in a Landsberg erecta (Ler) background. This prompted us to determine whether the stronger stomatal phenotype depends upon the er mutation, a locus known to affect stomatal patterning (Shpak et al., 2005). Homozygous flp-7 Ler plants were crossed to homozygous FLP LER plants. F2 plants with clusters displayed a severe flp phenotype independent of the number of er copies, suggesting that phenotypic severity results either from the flp mutations themselves or from enhancers in the Ler background.
To further establish the identity of FLP, we transformed flp-1 plants with the 11.5-kb MYB124 genomic region, a construct that included 8.7 kb of 5′ and 0.4 kb of 3′ intergenic sequences. This transformation restored a wild-type stomatal phenotype in the primary leaves in 39 out of 40 independent transformants. The single remaining transformant displayed a highly reduced number of clusters. These findings confirm the identity of FLP and demonstrate that the included regulatory regions are sufficient to drive expression for functional complementation.
FLP Is Expressed at the Transition to Terminal Stomatal Differentiation
Using RT-PCR, FLP mRNA was detected in all shoot organs tested (Figure 4A). These data are consistent with the presence of stomata as well as flp stomatal clusters in vegetative and reproductive organs (Geisler et al., 1998). In addition to shoots, seedling roots also expressed FLP (Figure 4A), and green fluorescent protein (GFP) expression driven by the FLP promoter region (see below) was present in root tips (data not shown). However, no abnormal phenotype has yet been detected outside the stomatal pathway in flp plants.
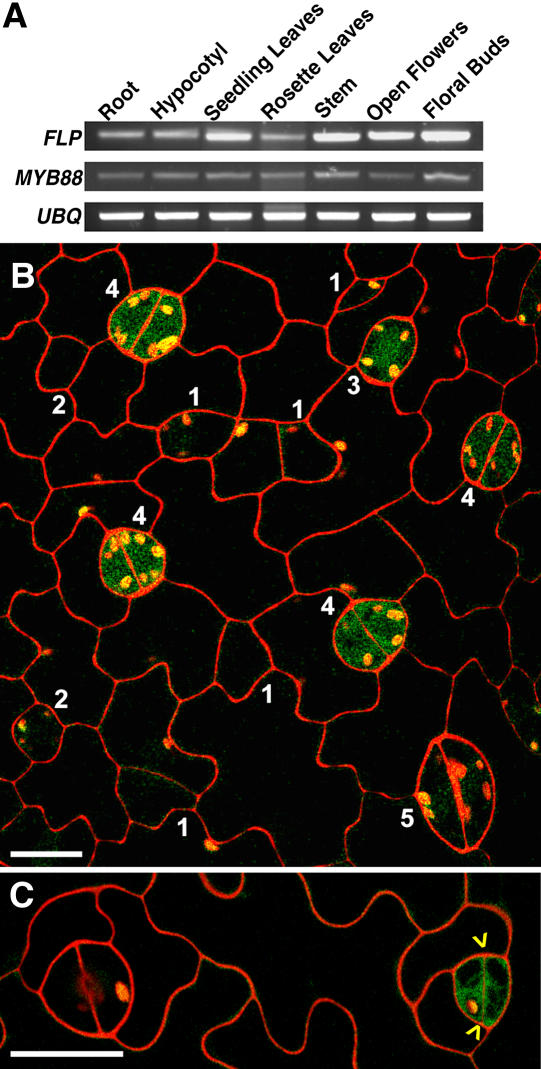
Figure 4.
FLP and MYB88 Expression throughout the Plant and in the Stomatal Pathway.
(A) Both FLP and MYB88 are expressed in all organs tested. Semiquantitative RT-PCR using ubiquitin for normalization.
(B) GFP driven by the FLP promoter in a wild-type plant. Fluorescence is present in a late GMC (stage 3) and in developing stomata (stage 4) but not in younger GMCs (stage 2), meristemoids (stage 1), or maturing stomata (stage 5). Note that the assigned stages here are different from those in Figures 1H and 1I.
(C) As in (B) except showing FLP expression in a newly divided GMC (yellow arrowheads point to new cell wall) but not in an older stoma with pore (left).
Bars in (B) and (C) = 15 μm.
To analyze the spatial and temporal expression of FLP during stomatal ontogeny, the 8.7-kb FLP 5′ region was used to drive a GUS-GFP fusion. In the developing leaf epidermis, GFP fluorescence was confined to late-stage GMCs and to young, still differentiating guard cells (Figures 4B and 4C). Fluorescence was absent in mature stomata, young GMCs, meristemoids, and other epidermal cells. We cannot rule out that native FLP expression differs from the above pattern since only the promoter was used, and some intron sequences dramatically affect MYB gene expression (Millar and Gubler, 2005). However, the GFP pattern that we found marks the same developmental window when abnormal stomatal flp phenotypes were detected.
An FLP Paralog Also Acts in Stomatal Patterning
Sequence similarity between members of the Arabidopsis MYB protein family is often restricted to the MYB domain (Stracke et al., 2001). Of all the currently known Arabidopsis R2R3 MYB genes, only MYB88 (At2g02820) has high sequence identity to FLP (Figure 3). The predicted proteins share 92% identity in their MYB domains and 71% identity overall. Their putative promoter sequences are 55% identical in the 0.9-kb regions proximal to the open reading frames. Also, the gene structures of MYB88 and FLP are highly similar except for two differences: exon 7 in FLP is represented by two exons in MYB88, and exon 11 in FLP is absent from MYB88 (Figure 3). Finally, global genome analysis (Blanc et al., 2000; Vision et al., 2000) as well as our more restricted, local analysis (data not shown) revealed a segmental duplication between a region of chromosome 1 that contains FLP and the part of chromosome 2 that contains MYB88. Thus, FLP and MYB88 are probably paralogs.
Like FLP, MYB88 is expressed in all plant organs based on RT-PCR analysis (Figure 4A). To determine whether MYB88 also functions in the stomatal pathway, we identified five different homozygous lines harboring T-DNA insertions in this locus. Two insertions are in the likely promoter region, two are in exons (6 and 10), and one is in the last intron (Figure 3A). All five lines showed a wild-type phenotype in stomatal distribution and in the gross appearance of the plants.
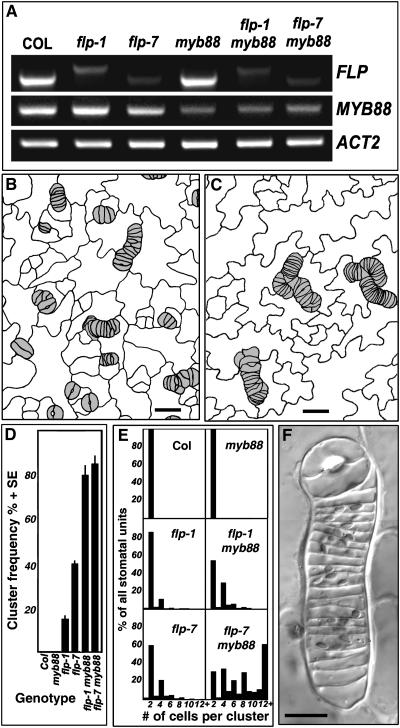
Two lines, SALK_039179 (insert in the last intron) and SALK_068691 (insert in exon 10), showed significantly reduced levels of MYB88 mRNA expression in RT-PCR (Figure 5A). Plants homozygous for the SALK_068691 insertion were crossed with flp-1 and with flp-7. Approximately one-sixteenth of segregating F2 plants from both crosses, the expected ratio for the double mutant class (Table 1), showed a stomatal phenotype that differed from flp-1 or flp-7 (Figures 5B to 5F). PCR analysis and the reduced levels of expression of both FLP and MYB88 confirmed that these plants were double mutants (Figure 5A).
Figure 5.
Exaggerated Stomatal Cluster Phenotype in flp myb88 Double Mutants.
(A) Semiquantitative RT-PCR showing reduced FLP and MYB88 expression in single mutants flp-1, flp-7, and myb88 (SALK_068691) and in double mutants. The shift in size of the flp-1 band is consistent with a splicing defect in this allele.
(B) and (C) Tracings of leaf surface showing cluster phenotypes in flp-1 myb88 and flp-7 myb88 double mutants, respectively.
(D) Mean frequencies of clusters per area as a function of genotype.
(E) The relative means of cells per cluster and of normal stomata in each genotype. The flp-7 myb88 double mutant has comparatively larger clusters.
(F) Differential interference contrast micrograph of a large cluster in flp-7 myb88.
Bars in (B) and (C) = 20 μm; bar in (F) = 10 μm.
Table 1.
Segregation Ratios of F2 Plants from flp×myb88 (SALK_068691) Crosses
| Phenotypes
|
||||
|---|---|---|---|---|
| flp Parent | Wild Type | flp | Exaggerated flp | χ2 Test |
| flp-1 | 769 | 167 | 60 | P ≥ 0.24 |
| flp-7 | 347 | 104 | 29 | P ≥ 0.26 |
Plants homozygous for the T-DNA insertion in MYB88 resemble wild-type plants, thus collapsing the 9:3:3:1 phenotypic ratio to 12:3:1.
The stomatal phenotype in leaves of the double mutants was more severe than in flp-1 or flp-7 as judged by the frequency and the size of the clusters (Figures 1 and 5). In addition, the phenotype of the flp-7 myb88 double mutant was more severe than the flp-1 myb88 double mutant, especially in cluster size (Figure 5). Cell clusters in both double mutants contained some developmentally arrested guard cells as judged by cell size and by chloroplast morphology (Figure 5F). Together, these phenotypes indicate that functional redundancy exists between these two genes in stomatal development.
To further probe a possible shared function, we transformed flp-1 plants with a 4.5-kb MYB88 genomic construct that included the 5′ and 3′ intergenic regions. In two separate experiments involving 44 and 55 independent transformants, 70 to 73% of all plants were complemented, ∼11% were partially complemented, and the remainder displayed a flp-1 phenotype. Because the MYB88 genomic region can drive expression sufficient to rescue flp-1, it is likely that the functions of MYB88 and FLP overlap. By contrast with the rescue of flp-1 by transgenic MYB88, the transformation of flp-1 with the FLP cDNA driven by the cauliflower mosaic virus 35S constitutive promoter failed to complement the mutant stomatal phenotype. This outcome could result from the presence of shared cis-acting elements in FLP and MYB88 that are absent from the 35S promoter.
FLP/MYB88-Like Genes Present in Many Flowering Plants
Our analysis of ESTs in databases indicates that many other dicots and monocots harbor genes closely related to MYB88 and FLP (Table 2). Sequence homology searches against the rice (Oryza sativa) genome revealed a gene we refer to as Os MYB88/124 (Figure 6). Nine out of the 11 introns in this gene are in positions identical to those in MYB88 and MYB124 (FLP). Also, the intron found in MYB88 but absent from MYB124 is present in Os MYB88/124 in an almost identical position. In addition, the rice protein shares slightly higher sequence identity with MYB88 (35.5%) than with MYB124 (33.3%).
Table 2.
Partial List of EST Sequences with Similarity to FLP and MYB88
| Monocotyledons | Dicotyledons |
|---|---|
| Acorus americanus: CK745173 | Brassica rapa: CO749412, CO750332, CO749907 |
| Hordeum vulgare: CK124529 | Glycine max: CA800624, BG507680, AW757235, AI900277, BG882813, CO981579, CO978628 |
| Oryza sativa: CA763233, CA763232 | Lactuca sativa: BQ987819, BQ993731, BU006944, BQ850844, BQ848657 |
| Saccharum officinarum: CA248397, CA248485, CA152057, CA077325, CA234814, CA207168, CA222270, CA230802, CA117471, CA167298, CA204462 | Solanum lycopersicum: BF096418, BF096904, AW622893, BF096910 |
| Malus domestica: CN910940, CN887651, CO901860, CO899320 | |
| Medicago sativa: CO515716 | |
| Sorghum bicolor: AW563279, BG947658, BM330092 | Medicago trunculata: AJ499212, BF518673, BQ124299, BQ123712, BF637579, BE204776, BG644656, BE204708 |
| Triticum aestivum: BQ743864 | Nicotiana benthamiana: CK290892 |
| Zea mays: BH784682, CC014152, BG837499 | Nicotiana sylvestris: BP751696 |
| Solanum tuberosum: CK274023 |
Figure 6.
Predicted Protein Sequence Alignments Comparing the Homology of a Rice Gene to FLP and MYB88.
The rice MYB88/124 gene (Os07g43420 and full-length cDNA, AK063426) shares with FLP/MYB88 the same presence or absence of W-to-F substitutions in the R3 domain (fourth asterisk from the top and double asterisk). Arrowheads indicate the positions of introns; an arrowhead pointing directly to a residue signifies that the intron interrupts the codon. Black arrowheads, intron locations found in all three genes; gray arrowhead, location of intron specific to FLP; white arrowhead, intron found only in MYB88; double arrowheads, introns specific to Os MYB88/124. Sequences with dark gray shading, conserved residues; sequences with light gray shading, similar residues. Protein domains (at right) are described in the legend to Figure 3.
Unique DNA Binding Domain
We then compared FLP and MYB88 to other Arabidopsis two-repeat MYB proteins, especially with respect to features of their MYB domains. MYB repeats are the defining motif of MYB proteins, and each repeat has three α-helices that form a helix-loop-helix structure involved in DNA binding (Martin and Paz-Ares, 1997). Each MYB repeat usually contains three regularly spaced and highly conserved Trp residues (Figure 7).
Figure 7.
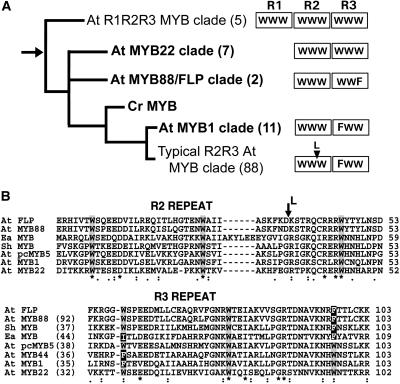
The MYB88/FLP Clade Is Phylogenetically Distinct from Most R2R3 Arabidopsis MYB Proteins.
(A) A phylogeny of selected clades of Arabidopsis R2R3 MYB proteins. The figure shows the relatively basal position of the MYB88/FLP clade and the hypothesized derivation of plant R2R3 MYB proteins through the loss of the R1 repeat from an ancestral R1R2R3 MYB protein. The root of the tree (arrow) is located between R1R2R3 and atypical R2R3 (latter in bold) MYB proteins and is close to the MYB88/FLP clade. This tree was adapted from Dias et al. (2003) and is supported by Bayesian (8000 trees sampled) and Bootstrap (500 replicates) analyses as described in their supplemental data. Number of clade members is in parentheses. Each box (right) represents a MYB repeat with either three regularly spaced Trp (W) residues or with a Phe (F) substitution. Typical, as well as a few atypical, R2R3 MYB proteins (MYB1 clade) have the F substitution in the first R3 W. Cr: Chlamydomonas reinhardtii. L at arrowhead: Leu insertion characteristic of typical R2R3 MYB proteins (Braun and Grotewold, 1999).
(B) Two ciliate proteins share a novel R3 substitution with FLP and MYB88. Sequence comparison of R2R3 MYB repeats in FLP and MYB88 with those in selected MYB proteins from Arabidopsis and ciliates. Ea MYB is an R2R3 MYB protein, and Sh MYB is an R1R2R3 MYB protein from the ciliates Euplotes aediculatus and Sterkiella histriomuscorum, respectively (Yang et al., 2003). Like FLP and MYB88, both ciliate proteins have the specific F substitution for the third R3 W. The Sh MYB lacks an F substitution found in the first R3 W, a substitution also present in MYB1 and MYB44, which are atypical R2R3 MYB proteins. Alignments were generated using ClustalW. Percentage of identity to FLP is specified in parentheses. Asterisks indicate identical residues, two dots conserved, and one dot semiconserved residues. The L and the arrow indicate where the Leu insertion (see above) would be present if these were typical R2R3 MYB proteins.
Two likely events during the evolution and proliferation of plant R2R3 MYB proteins were a substitution of a Phe for the first Trp in the R3 MYB repeat and then the insertion of a specific Leu (Dias et al., 2003). R2R3 MYB proteins that lack the Leu insertion are considered atypical. Atypical R2R3 MYB proteins that also lack the Trp-to-Phe substitution are considered to be relatively basal among plant MYB proteins that possess more than one repeat (Dias et al., 2003).
FLP and MYB88 are atypical MYB proteins that form a clade located close to the root of plant R1R2R3 and R2R3 MYB proteins (Figure 7A) (see supplemental data in Dias et al., 2003). Consistent with this basal position, FLP and MYB88 lack the Phe substitution in the first Trp of the R3 repeat. But unlike all other Arabidopsis R2R3 MYB proteins, the FLP/MYB88 clade has a Phe substitution in the third R3 Trp (Figure 3), a helix directly involved in DNA binding (Ogata et al., 1992).
DISCUSSION
FLP Limits Divisions Late in the Stomatal Cell Lineage
We have shown that FLP, a probable transcription factor, prevents more than one symmetric division from occurring at the end of the stomatal cell lineage. Normally, a single symmetric division takes place in the GMC and produces daughter cells that terminally differentiate into the two guard cells that make up the stoma. Guard cells probably arrest in G1 or exit the cell cycle (G0) because they do not divide and they retain a 2C level of DNA (Melaragno et al., 1993). In flp mutants, the GMC division appears normal, but the daughter cells divide symmetrically one or more times, which is abnormal. Because these extra divisions occur in cells that exhibit GMC traits and lack guard cell traits, loss-of-function mutations in FLP appear to prolong GMC identity. FLP expression apparently starts before GMC mitosis and ends after the initial specification of guard cells. It is therefore likely that FLP functions to limit division during this period directly by regulating cell cycle genes or indirectly by promoting a transition in cell fate.
A direct function in division control might be to influence the transcription of genes needed for cell cycle exit or for guard cell G1 arrest or G0 entry. This would resemble the role of the transcription factor Prospero in Drosophila neurogenesis, which functions to suppress the expression of positive regulators of cell proliferation (Li and Vaessin, 2000). As in FLP, loss-of-function mutations in Prospero induce an extra division at the end of a stem cell lineage, thereby causing two neurons to form abnormally in direct contact. Because FLP is expressed during the GMC-to-guard-cell transition, FLP's target might be a cell cycle regulator expressed just before the symmetric GMC division. One such candidate is the cyclin-dependent kinase B1;1, a kinase that regulates the G2/M transition. This gene is expressed in GMCs, and a dominant negative version introduced transgenically prevents GMC mitosis (Boudolf et al., 2004).
An indirect role in cell cycling could result from FLP acting in cell specification. Cell cycle exit might depend upon a progression in cell fate, such as if FLP were required to terminate a GMC fate or to promote a guard cell fate. A defect in either of these processes is consistent with the phenotypes of flp mutants where daughter cells initially display GMC rather than stomatal traits, suggesting that extra divisions result from prolonging a GMC fate.
Although most of the cells in clusters in weaker flp alleles become stomata or exhibit guard cell traits, cells of uncertain identity can be found in clusters in severe flp alleles and in flp myb88 double mutants, hinting at a possible role for FLP in cell specification. However, a role for FLP in cell specification might only be indirect since excess divisions generate cell packing constraints and might dilute out positive differentiation factors. Taken together, our data indicate that FLP is required primarily for the timeliness of the transition from cell cycling to terminal cell specification.
Functions in cell cycling and/or specification have been assigned to several plant R2R3 MYB proteins. GLABROUS1 (GL1) helps specify Arabidopsis trichomes and also regulates the transition from cell proliferation to trichome endocycling (Szymanski and Marks, 1998; Larkin et al., 2003). And DUO1 positively regulates mitosis in generative cells in pollen grains and is needed to form the two sperm cells (Rotman et al., 2005). Interestingly, both FLP and DUO1 regulate cell cycling in specific cell types and both affect mitosis related to a terminal symmetric division.
Comparison with Other Genes in the Stomatal Pathway
Mutations in FAMA, a basic helix-loop-helix (bHLH) protein, were recently shown to produce stomatal clusters (Bergmann et al., 2004) that resemble severe flp alleles. As some R2R3 MYB proteins interact with bHLH proteins in specifying epidermal cell fate (Bernhardt et al., 2005), it is attractive to hypothesize that FLP and MYB88 interact with FAMA in regulating stomatal patterning. If they do interact, however, it would be through domains yet to be identified, as neither FAMA nor FLP/MYB88 possess sequences previously demonstrated to be necessary for physical interactions between bHLH and MYB transcription factors (Heim et al., 2003; Zimmermann et al., 2004). Nevertheless, FLP/MYB88 and FAMA appear to be the only transcription factors with a demonstrated function in stomatal development.
FLP and MYB88 act later in the stomatal pathway after the GMC is specified by preventing the cellular amplification of a correctly positioned GMC. By contrast, genes that act earlier in the pathway, such as TMM, SDD1, and YODA, all negatively regulate the number of asymmetric divisions of stem cells and are required for meristemoid spacing (Figure 1G; GMC 3) (Berger and Altmann, 2000; Nadeau and Sack, 2003; Bergmann et al., 2004). Consistent with this analysis, both TMM and SDD1 are expressed in cells competent for asymmetric divisions (Nadeau and Sack, 2002a; von Groll et al., 2002), whereas FLP is expressed primarily in GMCs. The difference between early- and late-acting genes is emphasized by the finding that a single copy of a constitutively active gain-of-function YODA transgene is sufficient to suppress the phenotypes of tmm and sdd1 but not flp-1 (Bergmann et al., 2004). Thus, while all five genes are required for normal stomatal patterning, only the early-acting genes control the key asymmetric spacing division, whereas FLP and MYB88 only regulate spacing indirectly by restricting divisions at the end of the cell lineage.
Although division regulation is crucial for stomatal patterning, the terminal differentiation of stomata can be partially uncoupled from division (Jakoby and Schnittger, 2004). Mutations that block GMC mitosis or cytokinesis still permit the differentiation of stomatal traits (Nishihama et al., 2001; Falbel et al., 2003; Boudolf et al., 2004). Our results with flp show that the opposite defect, excess GMC divisions, also allows partial or complete stomatal differentiation.
Gene Duplication and Functional Redundancy
Gene duplication followed by divergence in sequence and function are thought to have contributed substantially to the existence of a large family of plant R2R3 MYB genes (Dias et al., 2003; Jia et al., 2003). Some pairs of similar MYB genes, such as GL1 and WEREWOLF, have nonoverlapping patterns of expression but are still capable of reciprocally complementing loss-of-function mutations in each locus when their promoters are swapped (Lee and Schiefelbein, 2001; Kirik et al., 2005). By contrast, extra copies of the MYB88 genomic region that were introduced transgenically rescued flp-1, indicating retention of key regulatory elements as well as of protein function.
A retained function of MYB88 in the stomatal pathway is also supported by the synergistic rather than additive phenotype found in flp myb88 double mutants. Although the disruption of MYB88 function on its own shows no abnormal stomatal phenotype, double mutants display more and larger clusters than flp alone. Thus, both genes probably overlap in function in the pathway by restricting divisions at the end of the stomatal cell lineage.
Although the cell-specific pattern of MYB88 expression is not yet known, an overlap in expression is likely, since there is significant sequence conservation in both promoter regions and since transformation with MYB88 driven by its native promoter rescues flp-1. One possible explanation for the failure of the native MYB88 gene to compensate for a loss of FLP function in the flp-1 single mutant is that this locus is regulated differently from the MYB88 transgene. For instance, expression at the native MYB88 locus might be constrained by the local chromatin structure, a constraint that might not operate at the site where the transgene integrated in rescued lines. These data also suggest the involvement of a gene dosage mechanism where a threshold level of FLP and/or MYB88 is required to correctly pattern all stomata. Dosage balance mechanisms are often found in plant transcriptional regulator pathways, and the modification of function and expression after gene duplication can be adaptive in providing enhanced regulation (Birchler et al., 2005; Moore and Purugganan, 2005).
Position of FLP/MYB88 Clade
The FLP/MYB88 clade is relatively basal and is close to the root of plant two- and three-repeat MYB proteins (Dias et al., 2003; Jiang et al., 2004). Similarly, a MYB88/124-like gene (Figure 6) was recently placed at the base of a phylogenetic tree derived from 53 rice R2R3 MYB genes, a tree based upon features of their DNA binding domains (Jia et al., 2004).
The MYB repeats of FLP and MYB88 differ from all other known plant two- and three-repeat MYB proteins in having a Trp-to-Phe substitution in the third helix of the R3 repeat, a region thought to be critical for DNA binding (Ogata et al., 1992). The only other nonplant eukaryotes known to possess MYB proteins with this substitution are ciliates, which are among the most phylogenetically ancient organisms for which MYB proteins have been documented (Yang et al., 2003) (Figure 7B). This raises the possibility that atypical R2R3 MYB proteins like FLP/MYB88 arose before the divergence of plants and ciliates. Together, these data indicate that the FLP/MYB88 clade evolved earlier than most Arabidopsis R2R3 MYB proteins but is an offshoot from the lineage that gave rise to the majority of plant two-repeat MYB proteins.
The widespread presence of genes like FLP and MYB88 in dicots and monocots suggests a conserved biochemical function. It remains to be seen whether this function is limited to stomatal development or whether these MYB proteins act more broadly, such as in coordinating cell specification and cycling in other pathways.
METHODS
flp Alleles and Plant Growth Conditions
Four flp alleles induced by ethyl methanesulfonate mutagenesis were isolated in a microscopy-based screen. The flp-1 allele is in an Arabidopsis thaliana Columbia (Col) gl1 background (Yang and Sack, 1995). The flp-2 allele is from Thomas Altmann (Max-Planck Institute of Molecular Plant Physiology, Golm, Germany) and is in a C24 background. The flp-7 and flp-8 alleles are in a Ler background. Splicing errors in flp-1 and in flp-7 mRNAs were detected by RT-PCR using primers FLP-2F (5′-CAAAGACAAAAGCACAAGACAATGC-3′) and FLP-3R (5′-GGTGTTTGAGTTGCTGTCTTTAGTC-3′) that were complementary to regions in exons 3 and 6.
In addition, three lines with T-DNA insertions in FLP were obtained from the SALK collection (Alonso et al., 2003) via the Arabidopsis Biological Resource Center at the Ohio State University and from the SAIL collection (Sessions et al., 2002). All three lines, SALK_33970, SAIL_270, and SAIL_870, are in a Col background. Gene-specific primers that were used for identifying plants that were homozygous for insertions were FLP XI-R (5′-AAACCTCGAGGGAGGAATGCAG-3′) and FLP-4F (5′-GAATTCAGCTATGACTAAAGACAG-3′) for SALK_33970 and F14L17-3 (5′-TGCATCCATGAATCGTGTATATT-3′) and FLP15-prom (5′-CTTCGGATCCTCCATTTTTCTTC-3′) for SAIL_270 and SAIL_870. The T-DNA–specific primers used were LBa1 for the SALK line (5′-TGGTTCACGTAGTGGGCCATCG-3′) and LB1 for the SAIL lines (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′).
Plants were grown on sterile agar medium or on soil (Promix, Premier Brands). Seeds destined for agar medium were surface-sterilized in 30% bleach for 20 min and rinsed with four changes of sterile distilled water before sowing. Plants were grown in a chamber or in a growth room at 20°C with a 16-h-light (∼100 μmol m−2 s−1)/8-h-dark cycle.
Phenotypic Analysis of flp
Phenotypes were evaluated from alleles that had been backcrossed three times (flp-1, flp-7, and flp-8) except for flp-2, SAIL_270, SAIL_870, and SALK_33970. Replicas of the epidermis were obtained from the abaxial epidermis of third and fourth leaves from 3- to 8-d-old flp-1 plants using dental impression material as described by Geisler et al. (2000). Electron microscopy was performed as detailed by Zhao and Sack (1999). Fluorescence from propidium iodide–stained tissue was viewed using a Nikon PCM 2000 confocal laser-scanning microscope (Nadeau and Sack, 2002a), and in many figures, the image was inverted. For scoring cluster frequency of different flp alleles, mature third leaves were cleared in 3:1 ethanol:glacial acetic acid, then cut along the midrib and stained with crystal violet. Each of 10 leaves per genotype was scored at six different leaf regions that were comparable for all genotypes. A total of 1000 stomatal units was scored per genotype. A stomatal unit consists of either a normally spaced stoma or an entire cluster of stomata in contact (Yang and Sack, 1995). An Olympus AX-70 light microscope and an Optronics Magnafire CCD camera were used to capture bright-field and differential interference contrast images.
Wild-type RLD1 plants stably transformed with a ProKAT1:GUS construct were obtained from M. Sussman and R. Hirsch (Nakamura et al., 1995). This transgene was introgressed into flp-1 and wild-type Col plants. Leaves and cotyledons from 8-d-old seedlings grown on agar were scored for GUS staining in different stages of the stomatal pathway. Whole seedlings were placed into 50 mM sodium phosphate buffer, pH 7.0, containing 0.1% (w/v) 5-bromo-4-chloro-3-indolyl-β-glucuronide, 1 mM K3Fe(CN)6, and 0.1% Triton X-100. After a 30-min vacuum infiltration, the tissue was incubated at 37°C for 2.75 h, then fixed and cleared in ethanol:acetic acid (3:1). The quantitative GUS staining analysis was performed twice with comparable results.
FLP Mapping, Complementation, and ESTs
F2 mapping populations were generated from two crosses, flp-1 (Col) × Ler and flp-7 (Ler) × Col. Using published polymorphic markers, FLP was localized to a region between g2358 and AthSRP54A. Based on BAC sequences, six new cleaved amplified polymorphic sequence markers were developed to localize FLP to a 30-kb region in BAC F14L17. Sequence analysis of the six annotated open reading frames in this region revealed a single point mutation in At1g14350 in each of the four ethyl methanesulfonate–generated flp alleles.
To confirm FLP identity, an 11.5-kb FLP genomic fragment that included the 8.7-kb upstream and 0.4-kb downstream intergenic sequences was amplified by PCR from BAC F14L17 using High-Fidelity Platinum Taq Polymerase (Invitrogen). Subsequent to cloning into the binary vector pCAMBIA 2200, this construct was introduced into flp-1 plants by Agrobacterium tumefaciens–mediated transformation.
The ESTs F19798 and N96570 were obtained from the Arabidopsis Biological Resource Center and were completely sequenced; N96570 was found to be a chimeric sequence of 3′ FLP cDNA and At3g45240.
MYB88
Five lines with T-DNA insertions in MYB88 were evaluated phenotypically: SALK_000380, _024788, _068691, and _039179 and SAIL_1182. Zygosity and localization of T-DNA inserts were determined using PCR. The gene-specific primers used for SALK_000380 and _024788 were MYB88-F (5′-TTCCCCACAAAACTCTAAACCGTAATA-3′) and MYB88-1R (5′-GGAATCATCGGAATGAAGAAGAATC-3′), for SALK_068691 and _039179 were MYB88-3F (5′-GAGGATAAAAGTAACCAAGAGGTGTTTC-3′) and MYB88-10R (5′-GATATGGCTGCAAACCTATGGAG-3′), and for SAIL_1182 were MYB88-21F (5′-GAACAGGATTGCTTGTTGTGTTAACTCAG-3′) and MYB88-22R (5′-GAAACACCTCTTGGTTACTTTTATCCTC-3′).
The comparative phenotypes of SALK_068691/flp double mutants and of single mutants were evaluated from plants grown on soil. The extent of clustering (Figures 5D and 5E) was scored from a total of 477 to 666 stomatal units from at least five third leaves for each genotype. The flp-7/SALK_068691 double mutants are viable and usually set seed.
To analyze whether MYB88 shares similar or overlapping function with FLP, a 4.5-kb genomic fragment that included the 0.9-kb upstream and 1.2-kb downstream intergenic sequences was amplified by PCR from Col genomic DNA. This fragment was cloned into pCAMBIA 2200 and introduced into flp-1 plants by Agrobacterium-mediated transformation. Phenotypic analysis was performed on the fifth leaf of transformants that were transferred to soil for 2 weeks after selection. Complementation of flp with the genomic MYB88 region was confirmed in two separate sets of transformants.
Semiquantitative RT-PCR
Using TRIzol reagent (Invitrogen), total RNA was isolated from roots, hypocotyls, and leaves of 10-d-old Col gl1 seedlings grown on agar. Total RNA was also isolated from rosette leaves, stems, open flowers, and floral buds of Col gl1 plants grown for 4 weeks on soil. First-strand cDNAs were generated using ThermoScript (Invitrogen) and oligo(dT)20 primer. Several rounds of PCR-based amplification of the controls (UBQ and ACT2) were performed to normalize the amounts of cDNAs needed in the subsequent amplification of FLP and MYB88. The primers for amplifying ubiquitin (Shin et al., 2002) were UBQ-F (5′-GATCTTTGCCGGAAAACAATTGGAG-3′) and UBQ-R (5′-CGACTTGTCATTAGAAAGAAAGAGATAACAGG-3′), for FLP were FLP-2F (5′-CAAAGACAAAAGCACAAGACAATGC-3′) and FLP-3R (5′-GGTGTTTGAGTTGCTGTCTTTAGTC-3′), and for MYB88 were MYB88-1F (5′-GATTCTTCTTCATTCCGATGATTCC-3′) and MYB88-25R (5′-CTGAGTTAACACAACAAGCAATCCTGTTC-3′).
For determining expression levels in flp, myb88, and double mutants (Figure 5A), the primers used for FLP were the same as above. However, those used for MYB88 were MYB88-3F (5′-GAGGATAAAAGTAACCAAGAGGTGTTTC-3′) and MYB88-4R (5′-GAGTTCTTTAACACATCATGACCAGTTG-3′), and for ACTIN2 were ACT2-F (5′-GGCTCCTCTTAACCCAAAGGC-3′) and ACT2-R (5′-CACACCAT6CACCAGAATCCAGC-3′). The numbers of cycles used were 25 for the controls and 30 for FLP and MYB88.
Microscopy-Based Expression Analysis
An FLP promoter-reporter construct was created by replacing the cauliflower mosaic virus 35S promoter in pCAMBIA 1303 with the 8.7-kb upstream region of FLP in frame with the GUS-GFP coding sequence. The construct was sequenced to confirm the fusion sites and then introduced into Col gl1 plants by Agrobacterium-mediated transformation. Transformed T1 seedling leaves were stained for GUS activity, and T2 seeds were obtained from 21 independent T1 transformants showing strong GUS staining. Approximately 20 T2 seedlings that showed GFP fluorescence from each of the T1 lines were used to analyze the pattern of FLP promoter-driven fluorescence. Confocal microscopy was performed as described by Nadeau and Sack (2002a) on the first and second leaves of 8- to 9-d-old plants that had been grown on agar plates. Although the intensity of GFP fluorescence varied among the transformants, all showed the same qualitative pattern of fluorescence. Control plants transformed with a 35S:GUS-GFP construct exhibited fluorescence in all leaf epidermal cells as shown previously (Nadeau and Sack, 2002a).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g14350 (FLP), At2g02820 (MYB88), and Os07g43420 (rice).
Acknowledgments
We thank the Arabidopsis Biological Resource Center for T-DNA insertional mutants, M. Yang for initial mapping data, D. Kwon and S. Jones for technical assistance, and H. Vaessin, E. Grotewold, E. Braun, and A. Schnittger for critical discussions and comments on the manuscript. This work was funded by grants from the USDA (99353048098) and the National Science Foundation (IBN9505687).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Fred D. Sack (sack.1@osu.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.034116.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Berger, D., and Altmann, T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14, 1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Bergmann, D.C. (2004). Integrating signals in stomatal development. Curr. Opin. Plant Biol. 7, 26–32. [DOI] [PubMed] [Google Scholar]
- Bergmann, D.C., Lukowitz, W., and Somerville, C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497. [DOI] [PubMed] [Google Scholar]
- Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132, 291–298. [DOI] [PubMed] [Google Scholar]
- Birchler, J.A., Riddle, N.C., Auger, D.L., and Veitia, R.A. (2005). Dosage balance in gene regulation: Biological implications. Trends Genet. 21, 219–226. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Barakat, A., Guyot, R., Cooke, R., and Delseny, M. (2000). Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12, 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf, V., Barroco, R., Engler Jde, A., Verkest, A., Beeckman, T., Naudts, M., Inze, D., and De Veylder, L. (2004). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16, 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, E.L., and Grotewold, E. (1999). Newly discovered plant c-myb-like genes rewrite the evolution of the plant myb gene family. Plant Physiol. 121, 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias, A.P., Braun, E.L., McMullen, M.D., and Grotewold, E. (2003). Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol. 131, 610–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbel, T.G., Koch, L.M., Nadeau, J.A., Segui-Simarro, J.M., Sack, F.D., and Bednarek, S.Y. (2003). SCD1 is required for cell cytokinesis and polarized cell expansion in Arabidopsis thaliana. Development 130, 4011–4024. [DOI] [PubMed] [Google Scholar]
- Geisler, M., Nadeau, J., and Sack, F.D. (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler, M., Yang, M., and Sack, F.D. (1998). Divergent regulation of stomatal initiation and patterning in organ and suborgan regions of the Arabidopsis mutants too many mouths and four lips. Planta 205, 522–530. [DOI] [PubMed] [Google Scholar]
- Heim, M.A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Jakoby, M., and Schnittger, A. (2004). Cell cycle and differentiation. Curr. Opin. Plant Biol. 7, 661–669. [DOI] [PubMed] [Google Scholar]
- Jia, L., Clegg, M.T., and Jiang, T. (2003). Excess non-synonymous substitutions suggest that positive selection episodes occurred during the evolution of DNA-binding domains in the Arabidopsis R2R3-MYB gene family. Plant Mol. Biol. 52, 627–642. [DOI] [PubMed] [Google Scholar]
- Jia, L., Clegg, M.T., and Jiang, T. (2004). Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 134, 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., Gu, X., and Peterson, T. (2004). Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 5, R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Kirik, V., Lee, M.M., Wester, K., Herrmann, U., Zheng, Z., Oppenheimer, D., Schiefelbein, J., and Hulskamp, M. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132, 1477–1485. [DOI] [PubMed] [Google Scholar]
- Kranz, H., Scholz, K., and Weisshaar, B. (2000). c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J. 21, 231–235. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Brown, M., and Schiefelbein, J.W. (2003). How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 54, 403–430. [DOI] [PubMed] [Google Scholar]
- Lee, M., and Schiefelbein, J. (2001). Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Li, L., and Vaessin, H. (2000). Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 14, 147–151. [PMC free article] [PubMed] [Google Scholar]
- Martin, C., and Paz-Ares, J. (1997). MYB transcription factors in plants. Trends Genet. 13, 67–73. [DOI] [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B.M., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., and Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R.C., and Purugganan, M.D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8, 122–128. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2002. a). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2002. b). Stomatal development in Arabidopsis. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0066, http://www.aspb.org/publications/arabidopsis/.
- Nadeau, J.A., and Sack, F.D. (2003). Stomatal development: Cross talk puts mouths in place. Trends Plant Sci. 8, 294–299. [DOI] [PubMed] [Google Scholar]
- Nakamura, R., McKendree, W.J., Hirsch, R., Sedbrook, J., Gaber, R., and Sussman, M. (1995). Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol. 109, 371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama, R., Ishikawa, M., Araki, S., Soyano, T., Asada, T., and Machida, Y. (2001). The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev. 15, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, K., Hojo, H., Aimoto, S., Nakai, T., Nakamura, H., Sarai, A., Ishii, S., and Nishimura, Y. (1992). Solution structure of a DNA-binding unit of Myb: A helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl. Acad. Sci. USA 89, 6428–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman, N., Durbarry, A., Wardle, A., Yang, W.C., Chaboud, A., Faure, J.E., Berger, F., and Twell, D. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15, 244–248. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, B., Choi, G., Yi, H., Yang, S., Cho, I., Kim, J., Lee, S., Paek, N.C., Kim, J.H., and Song, P.S. (2002). AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J. 30, 23–32. [DOI] [PubMed] [Google Scholar]
- Shpak, E.D., McAbee, J.M., Pillitteri, L.J., and Torii, K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., and Marks, M.D. (1998). GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10, 2047–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumoto, Y.M., Apperly, J.A., Gao, F.B., and Raff, M.C. (2002). Posttranscriptional regulation of p18 and p27 Cdk inhibitor proteins and the timing of oligodendrocyte differentiation. Dev. Biol. 245, 224–234. [DOI] [PubMed] [Google Scholar]
- Vision, T.J., Brown, D.G., and Tanksley, S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- von Groll, U., Berger, D., and Altmann, T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14, 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M., and Sack, F.D. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7, 2227–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T., Perasso, R., and Baroin-Tourancheau, A. (2003). Myb genes in ciliates: A common origin with the myb protooncogene? Protist 154, 229–238. [DOI] [PubMed] [Google Scholar]
- Zhao, L., and Sack, F.D. (1999). Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am. J. Bot. 86, 929. [PubMed] [Google Scholar]
- Zimmermann, I.M., Heim, M.A., Weisshaar, B., and Uhrig, J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40, 22–34. [DOI] [PubMed] [Google Scholar]