Abstract
FtsH protease is important in chloroplast biogenesis and thylakoid maintenance. Although bacteria contain only one essential FTSH gene, multiple genes exist in cyanobacteria and higher plants. However, the functional significance of FTSH multiplication in plants is unclear. We hypothesized that some FTSH genes may be redundant. To test this hypothesis, we generated double mutant combinations among the different FTSH genes in Arabidopsis thaliana. A double mutant of ftsh1 and ftsh8 showed no obvious phenotypic alterations, and disruption of either FTSH1 or FTSH5 enhanced the phenotype of the ftsh2 mutant. Unexpectedly, new phenotypes were recovered from crosses between ftsh2 and ftsh8 and between ftsh5 and ftsh1, including albinism, heterotrophy, disruption of flowering, and severely reduced male fertility. These results suggest that the duplicated genes, FTSH1 and FTSH5 (subunit type A) and FTSH2 and FTSH8 (subunit type B), are redundant. Furthermore, they reveal that the presence of two types of subunits is essential for complex formation, photosystem II repair, and chloroplast biogenesis.
INTRODUCTION
FtsH in Escherichia coli is a membrane-bound ATP-dependent metalloprotease complex that is essential for survival (Saikawa et al., 2004, and references therein; Okuno et al., 2004). Unlike Clp protease, the FtsH proteolytic and ATPase domains are found on the same 71-kD polypeptide. The N terminus contains two transmembrane α-helices, which anchor the protein to the plasma membrane. The hydrophobic domains are followed by the ATPase domain, which relates this protein to the AAA+ superfamily of proteins (Neuwald et al., 1999). The proteolytic domain is found in the C terminus of the protein, and it contains the zinc binding motif H-E-X-X-H, which serves as the active site of the protease. The crystal structure of the ATPase domain suggests that FtsH forms ringlike hexamers, with the ATP binding motifs facing the center of the ring (Krzywda et al., 2002; Niwa et al., 2002). However, the relative arrangement of the proteolytic domain with respect to that of ATPase is still unknown. Nevertheless, it is assumed that like other ATP-dependent proteases, the active sites of FtsH are self-compartmentalized and thus excluded from the cytoplasmic environment. Translocation of the substrate proteins to the proteolytically active site requires ATP hydrolysis and is achieved by unfolding of the substrate proteins through the ring structure. Although the precise mechanism of translocation and unfolding remains unclear, these processes occur bidirectionally, from either end of the substrate protein.
FtsH-like complexes are also found in eukaryotic cells. Yeast mitochondria contain three FtsH homologs, known as AAA proteases, which are integral to the inner membrane (reviewed in Langer, 2000). One of these homologs, Yme11, forms a homohexamer (iAAA) whose catalytic domains face the intermembrane space; the two other homologs, Yta10 and Yta12, form a heterohexamer (mAAA) that is oppositely oriented toward the matrix. Although Yta10 and Yta12 are highly similar to each other, they are both essential for the formation of active mAAA, and one cannot compensate for the loss of the other (Arlt et al., 1998).
Whereas all bacterial genomes contain a single FTSH gene, cyanobacteria have four such genes (Mann et al., 2000). Thus, it appears that multiplication of FTSH genes correlates with the evolution of oxygenic photosynthesis. This trend is maintained in higher plants. The Arabidopsis thaliana nuclear genome contains 12 such genes (Sokolenko et al., 2002), and the rice (Oryza sativa) genome contains at least nine of them (Yu et al., 2005). This multiplication of higher plant FTSH genes occurred after chloroplasts had diverged from their prokaryotic progenitors, by duplication of the different cyanobacterial genes (see phylogenetic trees in Sakamoto et al., 2003; Yu et al., 2004). Moreover, the Arabidopsis and rice genomes contain genes that are more related to the yeast homologs than to the cyanobacterial homologs, suggesting that these derived from α-proteobacteria, the ancestors of today's mitochondria. Of the 12 FTSH genes in Arabidopsis, the products of three (FTSH3, FTSH4, and FTSH10) can be targeted to the mitochondria; the other nine (FTSH1, FTSH2, FTSH5 to FTSH9, FTSH11, and FTSH12) are capable of entering the chloroplast, as revealed by transient expression assay of green fluorescent protein fusions (Sakamoto et al., 2003). Nevertheless, separation of thylakoid membrane proteins by two different two-dimensional PAGE procedures, followed by mass spectrometry analysis, revealed that only four isozymes, FtsH1, FtsH2, FtsH5, and FtsH8, had accumulated in Arabidopsis leaves grown under optimal conditions (Sinvany-Villalobo et al., 2004; Yu et al., 2004). Furthermore, these isomers were the only FtsH proteins identified in a comprehensive proteome analysis of thylakoids (see The Plastid Proteome Database, http://cbsusrv01.tc.cornell.edu/users/ppdb/) (Friso et al., 2004). Of these, FtsH2 is by far the most abundant species, followed by FtsH5, FtsH8, and FtsH1 (Sinvany-Villalobo et al., 2004). In terms of phylogenetic relations, FTSH1 and FTSH5 are duplicated genes (type A), and so are FTSH2 and FTSH8 (type B). It is possible that the other chloroplast-predicted isozymes are expressed only at very low levels, at specific developmental stages, or under specific environmental conditions. To date, chloroplast FtsH proteases have been linked to the degradation of unassembled thylakoid proteins (Ostersetzer and Adam, 1997) and to the degradation of the D1 protein of the photosystem II (PSII) reaction center during its repair from photoinhibition (Lindahl et al., 2000; Bailey et al., 2002; Sakamoto et al., 2002).
Analysis of different FTSH knockout mutants revealed that mutations in FTSH2 result in leaf variegation, in which green sectors containing normal chloroplasts are found adjacent to white sectors containing plastids that have not developed into chloroplasts (Chen et al., 2000; Takechi et al., 2000), and increased sensitivity to photoinhibition (Bailey et al., 2002; Sakamoto et al., 2002). The ftsh5 mutant shows a similar, although much weaker, phenotype (Sakamoto et al., 2002). ftsh1 and ftsh8 mutants show no phenotypic alterations (Sakamoto et al., 2003). Apparently, this differential response to the inactivation of different FTSH genes correlates with the level of accumulation of their products, with FtsH2 being the most abundant, and FtsH5, FtsH8, and FtsH1 accumulating to ∼60, 50, and 10%, respectively, of the level of FtsH2 (Sinvany-Villalobo et al., 2004). The mutant phenotypes suggest FtsH's involvement in chloroplast biogenesis and the development of the internal membrane network of thylakoids.
In fact, the variegated phenotype represents two phenotypes: a wild-type phenotype in the green sectors and a mutant phenotype in the white sectors. A threshold model has been proposed by the Rodermel group to explain this dual phenotype within a leaf having a single mutant genotype (Yu et al., 2004). According to this model, slight variations in gene expression in different cells push the level of the remaining FtsH proteins either above or below the threshold needed for the development of normal chloroplasts. The variegated phenotype, as well as the lack of any phenotypic alteration in the ftsh1 and ftsh8 mutants, suggests redundancy between the different FTSH genes. This is further supported by the recent observations that overexpression of FTSH8 in an ftsh2 mutant background restores the wild-type phenotype (Yu et al., 2004) and that ftsh2 accumulates higher mRNA levels of FTSH1, FTSH5, and FTSH8 than does the wild type (Zaltsman et al., 2005).
To thoroughly challenge the hypothesis that the different FTSH genes are redundant, we generated a series of double mutants in these genes. Characterization of the double mutants demonstrated, unexpectedly, that two types of FtsH proteins must be present to allow the accumulation of FtsH protease, chloroplast development, and the repair of PSII during photoinhibition, and that the duplicated genes within each type are partially redundant.
RESULTS
Mutants Lacking Both FtsH1 and FtsH8 Develop and Function Normally
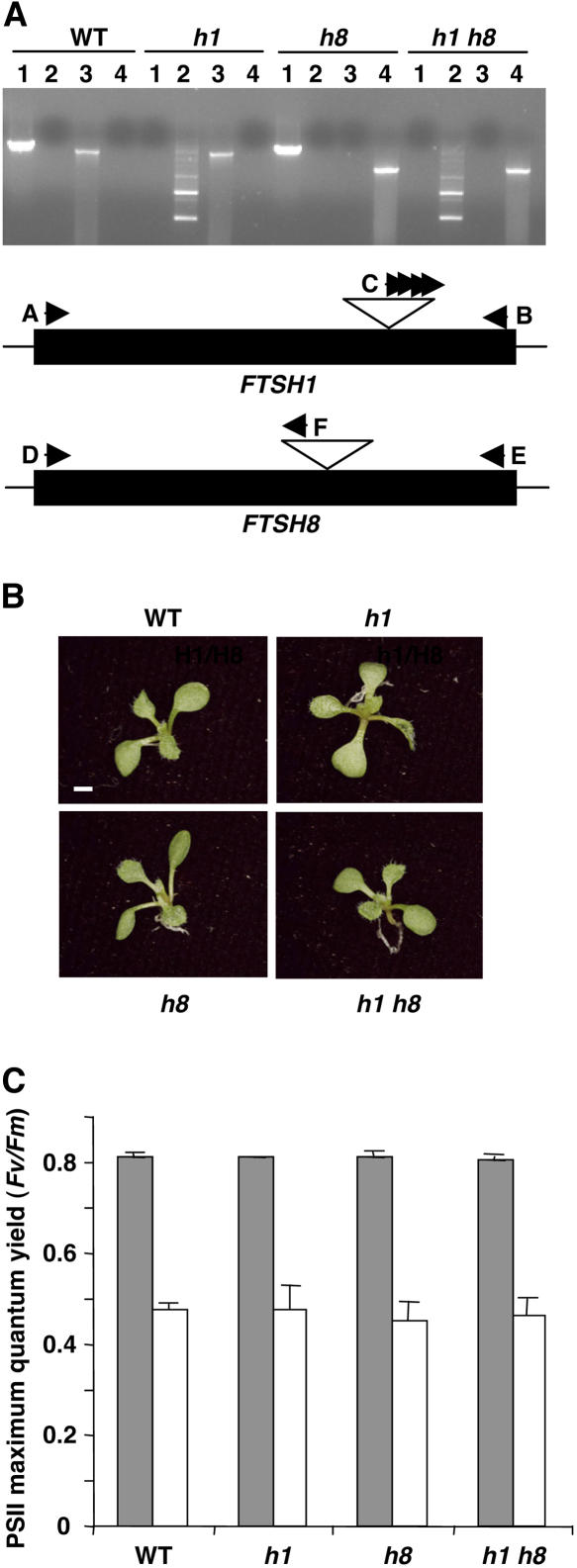
Disruption of the genes encoding either FtsH1 or FtsH8 by T-DNA insertions had no effect on the development of chloroplasts in Arabidopsis leaves or their sensitivity to high light intensity (Sakamoto et al., 2003). To test whether these mutations had a synergistic effect, we crossed the two null mutants. PCR analysis of the progeny was used to identify double mutants homozygous for both mutations (Figure 1A). In wild-type plants, PCR products could be obtained only with primers corresponding to the 5′ and 3′ ends of the respective genes. By contrast, in homozygous double mutant lines, these bands were absent, and only products of amplification with primers corresponding to the T-DNA and one of the ends of the relevant genes were observed. Segregation and PCR analysis of F3 seedlings confirmed that the double mutants were indeed homozygous (data not shown). Nevertheless, they did not differ from wild-type plants in appearance (Figure 1B) or in their sensitivity to photoinhibition (Figure 1C) throughout their life cycle when grown on either plates or soil.
Figure 1.
Plants Lacking Both FtsH1 and FtsH8 Look and Function Normally.
(A) PCR analysis of the F2 progeny of a cross between ftsh1 and ftsh8 homozygous mutants. The black boxes are schematic representations of the two genes. White triangles represent the T-DNA insertions and their relative positions in the genes. Black arrowheads represent the hybridization locations of the different primers used in the analysis. Lanes 1 on the gel represent amplification with primers A and B; lanes 2, amplification with primers B and C; lanes 3, amplification with primers D and E; lanes 4, amplification with primers D and F. The different homozygous genotypes are indicated above the gel (e.g., h1 is a homozygous mutant of FTSH1).
(B) Visual phenotypes of 2-week-old seedlings with different genotypes. Seedlings were grown on agar plates supplemented with sucrose. Bar = 1 mm.
(C) PSII maximum quantum yield, measured by chlorophyll fluorescence analysis of different mutants, before (gray columns) and after (white columns) a 90-min exposure to high light intensity. Values are means of six plants. Error bars indicate sd.
The Phenotype of ftsh2 Is Enhanced in the Absence of Either FTSH5 or FTSH1
Because disruption of either FTSH2 or FTSH5 resulted in some degree of leaf variegation, it was expected that a double mutant of these would demonstrate a more severe phenotype, and this was indeed the case (Sakamoto et al., 2002). Although the ftsh1 single mutant did not demonstrate any effect on phenotype, we tested whether disruption of this gene in an ftsh2 mutant background would have an effect similar to that observed with the ftsh2 ftsh5 double mutant. Thus, a homozygous mutant for the disruption of FTSH2 was crossed with homozygous mutants for the disruption of either FTSH5 or FTSH1. PCR analysis of F2 plants identified several lines homozygous for the mutations in FTSH1 and FTSH2 (Figure 2A). As shown in Figure 2B, disruption of FTSH1 enhanced the phenotype of the ftsh2 mutant. Plants grown on agar plates supplemented with sucrose were approximately half the size of wild-type or ftsh2 plants. Although the cotyledons of the double mutants were green, similar to those of the single ftsh2 mutant, their leaves demonstrated a decreased proportion of green sectors relative to the single mutant. Thus, the phenotype of the ftsh1 ftsh2 double mutant is similar to that of ftsh2 ftsh5, but slightly less severe (Figure 2B). The enhancement of the ftsh2 mutant phenotype by disruption of the FTSH1 gene suggested that FtsH1 does contribute to the pool of FtsH isomers that are important for chloroplast biogenesis.
Figure 2.
Loss of Either FTSH1 or FTSH5 Enhances the ftsh2 Mutant Phenotype.
(A) PCR analysis of the F2 progeny of a cross between homozygous ftsh1 and ftsh2 mutants. Other symbols are as in Figure 1.
(B) Visual phenotypes of 3-week-old seedlings with different genotypes. Seedlings were grown on agar plates supplemented with sucrose. Bar = 1 mm.
(C) Chlorophyll fluorescence analysis of different mutants before (gray columns) and after (white columns) a 90-min exposure to high light intensity. Values are means of six plants. Error bars indicate sd.
To test the effect of the different mutations on another aspect of FtsH function, involvement in the repair from photoinhibition, plants were subjected to chlorophyll fluorescence analysis. As shown in Figure 2C, and in accordance with previous studies, the ftsh2 mutant was more susceptible to photoinhibition than the wild type. Moreover, similar to the visible phenotypes, both the ftsh1 and ftsh5 mutations enhanced the ftsh2 mutant's susceptibility to photoinhibition.
ftsh2 ftsh8 Double Mutants Are Unable to Develop Normal Chloroplasts
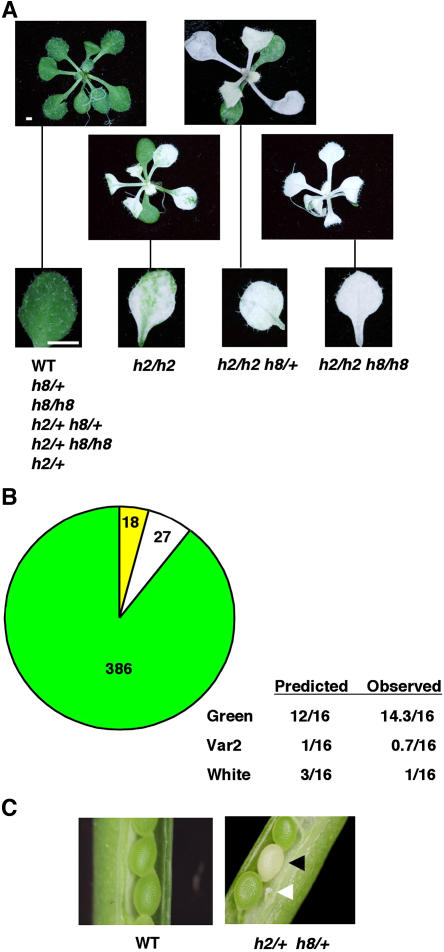
As the ftsh8 mutant itself demonstrated no detectable phenotypic alterations (Sakamoto et al., 2003), we sought to analyze whether it would have an effect in the background of the ftsh2 mutant. Single homozygous mutants, disrupted at the type B duplicated genes, either FTSH2 or FTSH8, were crossed and the progeny analyzed. F2 seeds were sown on agar plates supplemented with sucrose, and four phenotypes were observed: wild type, variegated like the ftsh2 mutant, and completely white seedlings having either green or white cotyledons (Figure 3A). Genotyping these plants revealed that a single wild-type allele of FTSH2 was sufficient to confer a wild-type phenotype, regardless of the genotype at the FTSH8 locus. The presence of one or two mutant alleles of FTSH8 converted the variegated homozygous ftsh2 mutant to a completely white seedling, but the presence of a single wild-type FTSH8 allele resulted in green cotyledons. Scoring the proportions of the different phenotypes demonstrated that the white seedlings were underrepresented compared with the expected 3:16 ratio (h2/h2 h8/+ and h2/h2 h8/h8) (Figure 3B). To further test this, we examined siliques of a double heterozygous line (h2/+ h8/+). As shown in Figure 3C and Table 1, all seeds of wild-type plants looked green and normal. By contrast, siliques of h2/+ h8/+ and h2/+ h8/h8 plants segregated some white seeds as well as aborted ones.
Figure 3.
A Cross between Homozygous Mutants in FTSH2 and FTSH8.
(A) Phenotypes of 3-week-old F2 seedlings and their leaves. The genotypes associated with these phenotypes, as revealed by PCR analyses similar to those shown in Figures 1 and 2, are indicated below the images. Mutant and wild-type alleles are indicated by lowercase letters and +, respectively. Bars = 1 mm.
(B) Segregation of phenotypes in the progeny of an h2/+ h8/+ double mutant. Seedlings looking like the wild type are represented by green, variegated ones (Var2) are represented by yellow, and white ones are represented by white. The numbers of seedlings corresponding to each phenotype are indicated within the chart. Predicted and observed proportions are also indicated.
(C) Open siliques from selfing a heterozygous line for disruption of both FTSH2 and FTSH8. Normal seeds are green, a white seed is indicated by a black arrowhead, and an aborted seed is indicated by a white arrowhead.
Table 1.
Segregation of Seed Phenotypes
| Seed Phenotype
|
|||
|---|---|---|---|
| Genotype | Green | White | Aborted |
| Wild type | 100 | 0 | 7 |
| h2/+ h8/+ | 84 | 4 | 20 |
| h2/+ h8/h8 | 271 | 50 | 85 |
Different genotypes were allowed to self-pollinate, their siliques were opened, and their seeds were analyzed.
Homozygous ftsh2 ftsh8 Double Mutants Are Infertile
A few h2/h2 h8/h8 plants could be maintained on plates containing sucrose and eventually flowered (Figures 4A and 4B). Their flowers appeared normal, with the exception of their color (Figure 4C). Examination of their reproductive organs revealed that their anthers were smaller than those of the wild type (Figure 4D). The stigma of the mutant was similar in size to the wild-type one (Figures 4E and 4F), but no pollen was observed on it. Apparently, no fertilization occurred in the mutant, and as a result, the mutant siliques were much smaller than those of wild-type plants (Figures 4G and 4H) and contained no seeds (Figures 4I and 4J).
Figure 4.
Phenotype of ftsh2 ftsh8 Homozygous Double Mutants.
(A) and (B) Six- and 9-week-old plants, respectively.
(C) Flowers of the wild type (left) and a double mutant (right).
(D) Anthers of the wild type (left) and a double mutant (right).
(E) and (F) Stigmas of the wild type and a double mutant, respectively.
(G) and (H) Wild-type (top) and mutant (bottom) siliques.
(I) and (J) Wild-type and mutant open siliques, respectively.
White arrowheads indicate aborted seeds. Bars = 0.5 mm.
ftsh1 ftsh5 Double Mutants Resemble the ftsh2 ftsh8 Double Mutants
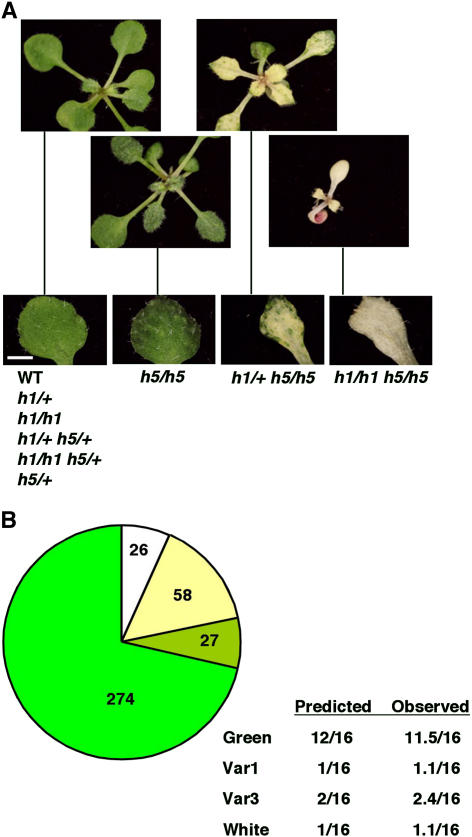
As the mutant phenotype of ftsh5 is weaker than that of ftsh2 (Sakamoto et al., 2002), we were interested in determining whether its phenotype could be enhanced by the disruption of another FTSH gene. Here again, we crossed heterozygous mutants of the type A duplicated genes FTSH5 and FTSH1 and analyzed the F2 progeny. When sown on plates supplemented with sucrose, four phenotypes were observed (Figure 5A). A wild-type phenotype was observed in seedlings carrying at least one wild-type allele of FTSH5, and slightly variegated first true leaves were found in seedlings homozygous for the loss of FTSH5. Much more severe variegation was observed in h1/+ h5/h5, but these seedlings still had green cotyledons. Seedlings homozygous for both mutations were very small, had both white cotyledons and leaves, and could hardly be maintained, even on sucrose. As shown in Figure 5B, the different phenotypes segregated close to the expected ratios.
Figure 5.
A Cross between Homozygous Mutants in FTSH1 and FTSH5.
(A) Phenotypes of 3-week-old F2 seedlings and their corresponding single leaves are shown. The genotypes corresponding to these phenotypes, as revealed by PCR analyses similar to those shown in Figures 1 and 2, are indicated below the images. Bar = 1 mm.
(B) Segregation of phenotypes in the progeny of an h1/+ h5/+ double mutant. Seedlings looking like the wild type are represented by green, slightly variegated ones (Var1) are represented by light green, severely variegated ones (Var3) are represented by light yellow, and white ones are represented by white. The numbers of seedlings corresponding to each phenotype are indicated within the chart. Predicted and observed proportions are also indicated.
Comparison between the Different Genotypes
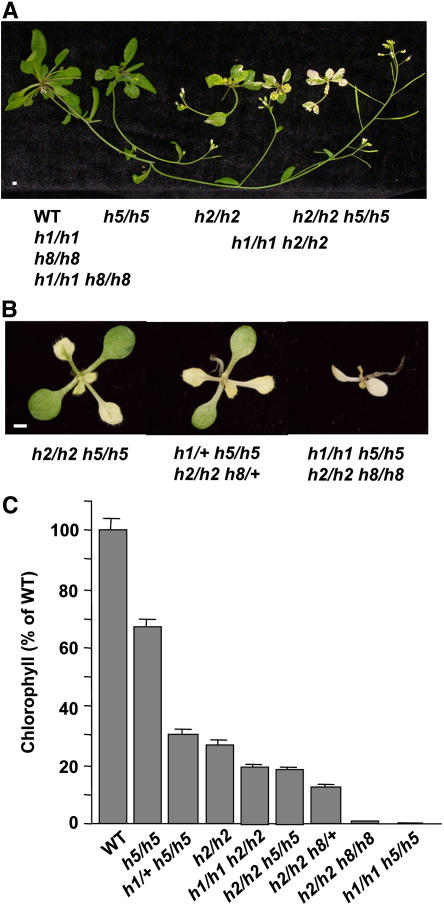
To compare the phenotypes observed in the different crosses, we germinated all of the genotypes on plates. The less severely affected phenotypes were then transferred to soil (Figure 6A), whereas the more severely affected ones were left on the plates (Figure 6B). A more quantitative record of the differences in variegation between the different genotypes was obtained by measuring the chlorophyll content in 6-week-old seedlings (Figure 6C). Among the homozygous mutants, loss of FTSH1 and FTSH8 had no effect, ftsh1 ftsh2 was more variegated than the ftsh2 mutant, and ftsh2 ftsh5 looked quite similar. ftsh1 ftsh5 and ftsh2 ftsh8 mutants were completely white and grew only on sucrose. The presence of wild-type alleles of either FTSH8 or FTSH5 allowed chloroplast development in the cotyledons, and two wild-type alleles allowed the beginning of chloroplast development in sectors of the leaves.
Figure 6.
Comparison of the Different Mutants.
All genotypes were sown on agar plates supplemented with sucrose.
(A) The stronger phenotypes were transferred to soil. Five-week-old seedlings are shown. Bar = 1 mm.
(B) Two-week-old seedlings grown on plates. Bar = 1 mm.
(C) Chlorophyll content in 6-week-old seedlings of the different genotypes. Wild-type seedlings and all genotypes having a similar phenotype had 1.3 μg chlorophyll/g fresh weight. Values are means of three replicates. Error bars indicate sd.
Disruption of Duplicated Genes Prevents the Accumulation of Other FTSH Gene Products
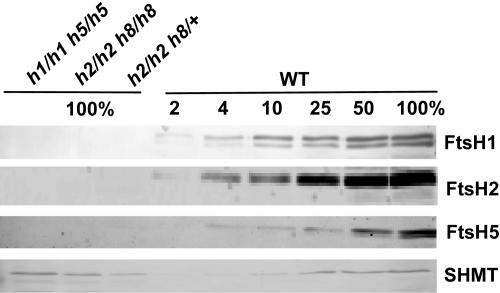
It has been shown previously that disruption of either the FTSH2 or FTSH5 gene results in a coordinated reduction in the level of the other FtsH proteins (Sakamoto et al., 2003). Thus, we were interested to determine whether the more severe phenotypes observed here were accompanied by a further decrease in the level of products of the remaining FTSH genes. To test this, immunoblot analysis was performed on completely white seedlings. As shown in Figure 7, inactivation of either FTSH1 and FTSH5 or FTSH2 and FTSH8 prevented the accumulation of the remaining gene products. If there is any FtsH protein left, its level should be <2% of wild-type levels. Thus, we conclude that in the absence of one type of subunit, subunits of the other type cannot form an active complex and are probably degraded.
Figure 7.
FtsH Levels in Double Mutants.
Proteins were extracted from the wild type, homozygous double mutants of ftsh1 and ftsh5, and homozygous and heterozygous mutants of ftsh2 and ftsh8. They were then separated by SDS-PAGE, blotted, and reacted with antibodies against FtsH protease or the mitochondrial protein SHMT. Lanes with wild-type samples, containing 2 to 100% of the mutant samples, are on the right side of the blot. The 100% samples are equivalent to 3.3 mg of tissue fresh weight.
DISCUSSION
The multiplication of FTSH genes in photosynthetic organisms in general, and in higher plants in particular, raises a question regarding its functional significance. A partial answer to this question lies in the fact that the products of some of these genes are targeted to mitochondria and some are targeted to chloroplasts (Sakamoto et al., 2003). Of the nine gene products that can be targeted to chloroplasts, only four have been detected there (Sinvany-Villalobo et al., 2004; Yu et al., 2004). Nevertheless, it is still not clear why multiple copies of highly related genes are needed. The simplest hypothesis is that different genes are expressed in different organs. However, expression studies have shown similar expression of the different FTSH genes in different plant organs (Yu et al., 2004, 2005). The other hypothesis, that different genes are expressed in response to different environmental stimuli, has also not been corroborated experimentally. All FTSH genes are upregulated in response to exposure to high light, and some also respond to changes in temperature, either high or low. However, no differential response of FTSH genes to temperature and high light has been detected (Sinvany-Villalobo et al., 2004), excluding the possibility of gene specialization in response to different environmental conditions, at least to those tested.
Another peculiar behavior is the plant's response to the inactivation of different FTSH genes. Of the four aforementioned genes, inactivation of FTSH1 and FTSH8 has no phenotypic consequence (Sakamoto et al., 2003), whereas loss of FTSH2 and FTSH5 results in higher and lower degrees, respectively, of leaf variegation and sensitivity to photoinhibition (Chen et al., 2000; Takechi et al., 2000; Sakamoto et al., 2002). This differential behavior correlates fairly well with the absolute expression level of these genes, as revealed by single-channel hybridization experiments (Sinvany-Villalobo et al., 2004). These results suggest that, under normal physiological conditions, FTSH1 and FTSH8 have no function in the chloroplast. This suggestion was further corroborated here when a double mutant of FTSH1 and FTSH8 showed a wild-type phenotype (Figure 1). Nevertheless, inactivation of FTSH1 in the background of an ftsh2 mutant further aggravated the latter's phenotype (Figure 2), suggesting that FtsH1 does contribute to the pool of FtsH isozymes.
The functionality of FtsH1 was further supported by the observation that the mild phenotype of the ftsh5 mutant became practically lethal when the FTSH1 gene was inactivated (Figure 5). A similar phenomenon was observed for the gene pair FTSH2 and FTSH8 (Figure 3), suggesting that FtsH8 is also functional. Even a single mutant allele of either FTSH1 or FTSH8 severely enhanced the phenotype of the ftsh5 or ftsh2 mutant, respectively (Figures 3 and 5). A comparison between the different 6-week-old mutants allowed the establishment of a hierarchy in the severity of phenotypes attributable to the inactivation of FTSH genes, from mild to severe: ftsh5, ftsh2, ftsh1 ftsh2, ftsh2 ftsh5, and the two lethal ones, ftsh1 ftsh5 and ftsh2 ftsh8 (Figure 6). As only ftsh2 and ftsh5 single mutants showed phenotypic alterations, in a scenario of mere additive genetic interaction, the ftsh2 ftsh5 double mutant would be expected to display the most severe phenotype. Strikingly, this was not the case. Rather, the most severe combinations were of the duplicated pairs of genes FTSH1 and FTSH5, and FTSH2 and FTSH8. These results suggest that each pair constitutes a separate type of subunit and that a functional FtsH complex is composed of subunits of type A (FtsH1 and FtsH5) and type B (FtsH2 and FtsH8). In the absence of either type, no active complex accumulates, as was confirmed by the immunoblot in Figure 7. However, within each type, the subunits are interchangeable. This suggestion is supported by the recent observation that overexpression of FTSH8 in the background of an ftsh2 mutant restores the wild-type phenotype (Yu et al., 2004), as does overexpression of FTSH1 in the background of an ftsh5 mutant (Yu et al., 2005). By contrast, subunits from the different types cannot compensate for each other's loss (S. Rodermel, personal communication).
An estimation of the relative quantities of the different FtsH subunits suggests that type B subunits (FtsH2 + FtsH8) are twice as abundant as type A subunits (FtsH1 + FtsH5) (Sinvany-Villalobo et al., 2004). As FtsH subunits form hexamers, this stoichiometry prompted us to propose a model in which each hexamer is composed of two type A and four type B subunits. The relative levels of the different subunits within a type can vary and are determined by their relative levels of expression and their epistatic relations. In this respect, the chloroplast FtsH protease complex is reminiscent of the yeast mitochondrial mAAA protease, in which the two closely related proteins Yta10 and Yta12 are both required for proper function (Arlt et al., 1998).
The variegated phenotype of the ftsh mutants suggested that FtsH protease is essential for chloroplast biogenesis and the formation of thylakoids. This suggestion is further supported by the results from the double mutants presented in this work (Figure 6). Mechanisms of thylakoid formation are very poorly understood (for a recent review, see Waters and Pyke, 2005), and how FtsH protease might be involved in this process is not at all clear. In this respect, it would be interesting to test whether it is the proteolytic function of FtsH that is needed or whether ATPase activity alone is sufficient.
A striking feature of the variegated ftsh2 mutant is that the degree of variegation is dependent on growth conditions and that it decreases with development (Zaltsman et al., 2005). Consistent with this observation, ftsh1/+ ftsh5/ftsh5 or ftsh2/ftsh2 ftsh8/+ seedlings developed a few pale green sectors in the youngest leaves when grown for long enough periods (data not shown). However, the appearance of green sectors was restricted to plants having one wild-type allele of either FTSH1 or FTSH8, respectively. It is also worth noting that the inactivation of one allele of FTSH1 enhanced the phenotype of the ftsh5 mutant far beyond that expected from the wild-type and mild phenotypes of the single mutants ftsh1 and ftsh5, respectively. This implies that the relative levels of the different FtsH isozymes observed by Sinvany-Villalobo et al. (2004) represent only the wild type. In mutant plants, the remaining subunits are likely to accumulate to different levels according to two factors: degradation of noncomplexed subunits, as suggested previously (Sakamoto et al., 2003), and increased expression of the remaining subunits as a compensation mechanism, as shown recently in the ftsh2 mutant (Zaltsman et al., 2005).
As already mentioned, double mutants comprising either type A or type B subunits were lethal and grew only heterotrophically. Interestingly, although they failed to develop chloroplasts, when these mutants were grown on sucrose, they developed normal roots and leaves and even reached flowering. Thus, it appears that chloroplast FtsH proteins and normal chloroplasts are not essential for morphogenesis or organogenesis. Although the color of the mutant flowers differed from that of the wild type (Figure 4C), their stigmas looked normal. However, no pollen was observed on the stigma (Figure 4F), and the anthers were smaller than wild-type anthers and never opened (Figure 4D). Arabidopsis plants can be grown to seed under sterile conditions; however, yield is usually poor (McCourt and Keith, 1998). Lower seed yields were indeed observed in our control wild-type plants grown on sucrose. Nevertheless, our albino double mutants were completely seedless. This observation is consistent with the absence of seeds in other Arabidopsis albino mutants (Hsieh and Goodman, 2005; Shimada et al., 2005) and suggests that the lack of seeds should be attributed to the lack of chloroplasts and not to the lack of FtsH protease.
In summary, the results of this work demonstrate that two types of subunits are necessary for the chloroplast FtsH protease to function. The subunits comprising the FtsH complex are encoded by two pairs of duplicated genes. The complex itself is expected to contain two type A subunits and four type B subunits. With respect to function, two levels can be observed: (1) repair of PSII from photoinhibition and probably other proteolytic degradation processes related to protein quality control; and (2) biogenesis of chloroplasts, probably through participation in the formation of thylakoids. The first function has been characterized but is not yet fully understood in mechanistic terms. The second function has been proposed but never totally understood. This will be the focus of future research.
METHODS
Plant Material
Arabidopsis thaliana plants were grown in either Kekkila peat (Finland) or on 0.7% (w/v) agar plates containing Murashige and Skoog medium, supplemented with 3% (w/v) sucrose. Temperature was held constant at 22°C, RH was 70%, and light intensity was 100 μE·m−2·s−1 for 16 h/d. To allow flowering, plants grown on plates were transferred to sterile flasks containing the medium described above. Wild-type lines used in this study were Columbia and Wassilewskija ecotypes. T-DNA insertion lines for FTSH1, FTSH2, FTSH5, and FTSH8 were described previously (Sakamoto et al., 2003). Single mutants were backcrossed to the wild type three times before generation of double mutants to segregate away additional mutations. Double mutants were obtained by crossing these lines and screening the F2 population. Cosegregation between the insertions and the respective phenotypes was verified.
DNA Isolation and PCR
DNA was isolated from a single leaf by its homogenization with a single 3-mm glass bead in a 1.5-mL Eppendorf tube for 10 s in an Ultramat 2 instrument (SDI). A 500-μL aliquot of a cold solution containing 0.2 M Tris-HCl, pH 9.0, 0.4 M LiCl, 25 mM EDTA, and 1% SDS was added to the tube, and the tube was vortexed for 15 s and then kept on ice for 5 min. A 500-μL aliquot of phenol:chloroform:isoamyl alcohol (25:24:1) was then added, and the tube was vortexed and then centrifuged at full speed in a microfuge for 5 min. The upper aqueous phase was transferred to a new tube containing 500 μL of isopropanol, kept at −20°C for 30 min, and centrifuged at full speed in a microfuge for 10 min. The pellet was air-dried and dissolved in 50 μL of water. DNA (150 ng) was subjected to 40 cycles of PCR with gene-specific primers and Ex Taq polymerase (TaKaRa). Products of the reactions were then separated on a 1% agarose gel and stained with ethidium bromide. The following primers, designed to a Tm of 65°C, were used: for FTSH1, 5′-TCTCGTTTGTAGCTTCTTCTCTCTCAGAA-3′ and 5′-CTCAGCTTAACCGTCAATAAAGAGACTCA-3′; for FTSH2, 5′-TGAACACAACAACCAAACAGAGGTTAAGT-3′ and 5′-ATCAGACATTCCAAATGTTGTTACCATCT-3′; for FTSH5, 5′-CAATTCCAATTCAAAGCTCCTTCC-3′ and 5′-GGCTTGTCCGTCAATGAAAAGACT-3′; for FTSH8, 5′-CTGCTTCATCGGCTTGTCTTCTC-3′ and 5′-TCGGAATTCATCGCCTGACATAGT-3′; for T-DNA in the ftsh1 mutant, 5′-CACGTCTTCAAAGCAAGTGGATTGATGTG-3′; and for T-DNA in all other mutants, 5′-CCAAGCCTCGCTAGTCAAAAGTGTA-3′.
Chlorophyll Fluorescence Measurements
For chlorophyll fluorescence measurements, third or fourth true leaves, which were not shaded by other leaves, were excised from seedlings. The detached leaves were irradiated with 1300 μmol·m−2·s−1, using 500-W halogen lamps, through a 25-mm water filter to avoid heating. Chlorophyll fluorescence was measured at room temperature using a pulse amplitude–modulated fluorometer (mini-PAM; Heinz Walz). Minimal chlorophyll fluorescence (F0) was measured after 10 min of dark adaptation. The intensity of the modulated measuring light was 0.1 μmol·m−2·s−1. Maximal chlorophyll fluorescence (FM) was measured during a 1-s pulse of nonmodulated light from a halogen lamp (8000 μmol·m−2·s−1). The maximum quantum yield of PSII electron transport was calculated using the following equation: FV/FM = (FM − F0)/FM, where FV is variable chlorophyll fluorescence.
Phenotypic Analyses
To determine chlorophyll concentrations, a set of three plants was used for each line. Fresh weight was determined, and the plants were gently shaken in 80% acetone for 12 h at 4°C. Chlorophyll concentration was determined spectroscopically using a standard procedure and normalized to the fresh weight. DNA concentration was determined using the ND-1000 spectrophotometer (NanoDrop Technologies). Images of seedlings were taken using a DXM1200 Nikon digital camera through an optical binocular. SDS-PAGE was performed on 15% gels, and immunoblot analysis was performed using a polyclonal antibody against FtsH1, which recognizes the different isomers equally well, as described previously (Zaltsman et al., 2005).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT1G50250 (FTSH1), AT2G30950 (FTSH2), AT5G42270 (FTSH5), and AT1G06430 (FTSH8).
Acknowledgments
We thank Rick Vierstra for access to the T-DNA insertion line collection, which allowed us to isolate the ftsh1 mutant, E. Glaser for the SHMT antibody, and W. Sakamoto for the FtsH2 and FtsH5 antibodies. This work was supported by grants from the Israel Science Foundation, the U.S.–Israel Binational Agricultural Research and Development Fund, and the U.S.–Israel Binational Science Foundation to Z.A. A.Z. was supported by a fellowship from the Horowitz Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zach Adam (zach@agri.huji.ac.il).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035071.
References
- Arlt, H., Steglich, G., Perryman, R., Guiard, B., Neupert, W., and Langer, T. (1998). The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 17, 4837–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, S., Thompson, E., Nixon, P.J., Horton, P., Mullineaux, C.W., Robinson, C., and Mann, N.H. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277, 2006–2011. [DOI] [PubMed] [Google Scholar]
- Chen, M., Choi, Y., Voytas, D.F., and Rodermel, S. (2000). Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 22, 303–313. [DOI] [PubMed] [Google Scholar]
- Friso, G., Giacomelli, L., Ytterberg, A.J., Peltier, J.B., Rudella, A., Sun, Q., and van Wijk, K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, M.-H., and Goodman, H.M. (2005). The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol. 138, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywda, S., Brzozowski, A.M., Verma, C., Karata, K., Ogura, T., and Wilkinson, A.J. (2002). The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 A resolution. Structure 10, 1073–1083. [DOI] [PubMed] [Google Scholar]
- Langer, T. (2000). AAA proteases: Cellular machines for degrading membrane proteins. Trends Biochem. Sci. 25, 247–251. [DOI] [PubMed] [Google Scholar]
- Lindahl, M., Spetea, C., Hundal, T., Oppenheim, A.B., Adam, Z., and Andersson, B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12, 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, N.H., Novac, N., Mullineaux, C.W., Newman, J., Bailey, S., and Robinson, C. (2000). Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 479, 72–77. [DOI] [PubMed] [Google Scholar]
- McCourt, P., and Keith, K. (1998). Sterile techniques in Arabidopsis. In Arabidopsis Protocols, J. Martinez-Zapater and J. Salinas, eds (Totowa, NJ: Humana Press), pp. 13–17. [DOI] [PubMed]
- Neuwald, A.F., Aravind, L., Spouge, J.L., and Koonin, E.V. (1999). AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43. [PubMed] [Google Scholar]
- Niwa, H., Tsuchiya, D., Makyio, H., Yoshida, M., and Morikawa, K. (2002). Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8. Structure 10, 1415–1423. [DOI] [PubMed] [Google Scholar]
- Okuno, T., Yamada-Inagawa, T., Karata, K., Yamanaka, K., and Ogura, T. (2004). Spectrometric analysis of degradation of a physiological substrate sigma32 by Escherichia coli AAA protease FtsH. J. Struct. Biol. 146, 148–154. [DOI] [PubMed] [Google Scholar]
- Ostersetzer, O., and Adam, Z. (1997). Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: The possible role of the FtsH protease. Plant Cell 9, 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikawa, N., Akiyama, Y., and Ito, K. (2004). FtsH exists as an exceptionally large complex containing HflKC in the plasma membrane of Escherichia coli. J. Struct. Biol. 146, 123–129. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Tamura, T., Hanba-Tomita, Y., Murata, M., and Sodmergen (2002). The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7, 769–780. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Zaltsman, A., Adam, Z., and Takahashi, Y. (2003). Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15, 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, H., Ohno, R., Shibata, M., Ikegami, I., Onai, K., Ohto, M., and Takamiya, K. (2005). Inactivation and deficiency of core proteins of photosystems I and II caused by genetical phylloquinone and plastoquinone deficiency but retained lamellar structure in a T-DNA mutant of Arabidopsis. Plant J. 41, 627–637. [DOI] [PubMed] [Google Scholar]
- Sinvany-Villalobo, G., Davydov, O., Ben-Ari, G., Zaltsman, A., Raskind, A., and Adam, Z. (2004). Expression in multigene families. Analysis of chloroplast and mitochondrial proteases. Plant Physiol. 135, 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko, A., Pojidaeva, E., Zinchenko, V., Panichkin, V., Glaser, V.M., Herrmann, R.G., and Shestakov, S.V. (2002). The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41, 291–310. [DOI] [PubMed] [Google Scholar]
- Takechi, K., Sodmergen, Murata, M., Motoyoshi, F., and Sakamoto, W. (2000). The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol. 41, 1334–1346. [DOI] [PubMed] [Google Scholar]
- Waters, M., and Pyke, K. (2005). Plastid development and differentiation. In Plastids, S.G. Moller, ed (Oxford, UK, Blackwell Publishing), pp. 30–59.
- Yu, F., Park, S., and Rodermel, S.R. (2004). The Arabidopsis FtsH metalloprotease gene family: Interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37, 864–876. [DOI] [PubMed] [Google Scholar]
- Yu, F., Park, S., and Rodermel, S.R. (2005). Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol. 138, 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman, A., Feder, A., and Adam, Z. (2005). Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts—Implications for thylakoid formation and photosystem II maintenance. Plant J. 42, 609–617. [DOI] [PubMed] [Google Scholar]