Abstract
Plant nuclear genomes encode hundreds of predicted organellar RNA binding proteins, few of which have been connected with their physiological RNA substrates and functions. In fact, among the largest family of putative RNA binding proteins in plants, the pentatricopeptide repeat (PPR) family, no physiologically relevant RNA ligands have been firmly established. We used the chloroplast-splicing factor CAF1 to demonstrate the fidelity of a microarray-based method for identifying RNAs associated with specific proteins in chloroplast extract. We then used the same method to identify RNAs associated with the maize (Zea mays) PPR protein CRP1. Two mRNAs whose translation is CRP1-dependent were strongly and specifically enriched in CRP1 coimmunoprecipitations. These interactions establish CRP1 as a translational regulator by showing that the translation defects in crp1 mutants are a direct consequence of the absence of CRP1. Additional experiments localized these interactions to the 5′ untranslated regions and suggested a possible CRP1 interaction motif. These results enhance understanding of the PPR protein family by showing that a PPR protein influences gene expression through association with specific mRNAs in vivo, suggesting an unusual mode of RNA binding for PPR proteins, and highlighting the possibility that translational regulation may be a particularly common function of PPR proteins. Analogous methods should have broad application for the study of native RNA–protein interactions in both mitochondria and chloroplasts.
INTRODUCTION
Modulation of plant organellar gene expression involves a diversity of posttranscriptional mechanisms, including group I and group II intron splicing, RNA editing, endonucleolytic and exonucleolytic RNA processing, and regulated translation and RNA stability (Barkan and Goldschmidt-Clermont, 2000; Monde et al., 2000; Zerges, 2000; Choquet and Wollman, 2002; Binder and Brennicke, 2003; Barkan, 2004). This complexity is reflected by the existence of hundreds of nuclear genes encoding predicted organellar RNA binding proteins, many of which are specific to plants (Aubourg et al., 1999; Fedoroff, 2002; Landsberger et al., 2002; Lorkovic and Barta, 2002; Vermel et al., 2002; Belostotsky, 2003; Nickelsen, 2003; Ostheimer et al., 2003; Lurin et al., 2004; Nakamura et al., 2004). Some organellar RNA binding proteins have been detected through their ability to bind RNA in vitro and others have surfaced through genetic screens, but most are unstudied. Furthermore, in only a few cases have the direct, physiological RNA substrates of specific RNA binding proteins been firmly established.
The pentatricopeptide repeat (PPR) family is a particularly interesting family of putative RNA binding proteins (Small and Peeters, 2000; Lurin et al., 2004). There are ∼450 predicted PPR proteins in Arabidopsis thaliana, most of which are predicted to be targeted to mitochondria or chloroplasts. PPR-encoding genes are also found in animals and fungi, albeit in smaller numbers. Although genetic data implicate several PPR proteins in organellar RNA metabolism (Barkan et al., 1994; Fisk et al., 1999; Auchincloss et al., 2002; Bentolila et al., 2002; Desloire et al., 2003; Hashimoto et al., 2003; Kazama and Toriyama, 2003; Koizuka et al., 2003; Meierhoff et al., 2003; Williams and Barkan, 2003; Xu et al., 2004; Yamazaki et al., 2004; Kotera et al., 2005) and several PPR proteins have been shown to bind nucleic acids in vitro (Ikeda and Gray, 1999; Lahmy et al., 2000; Mancebo et al., 2001; Nakamura et al., 2003; Lurin et al., 2004), evidence for the in vivo association between PPR proteins and specific RNA ligands has been lacking.
Previously, we used a coimmunoprecipitation assay to show that the maize (Zea mays) chloroplast-splicing factors CRS1, CAF1, and CAF2 are associated with their genetically defined intron targets in chloroplast extract (Ostheimer et al., 2003): RNA purified from immunoprecipitation reactions was applied to slot blots and probed to detect introns whose splicing was disrupted in the corresponding mutant background. Here, we modify this approach by using the coimmunoprecipitated RNA to probe a full genome chloroplast microarray. In this way, RNA ligands of specific proteins can be identified even in the absence of prior clues. This approach is referred to here as RIP-chip (for RNA immunoprecipitation and chip hybridization) in analogy to the term ChIP-chip, which is used for microarray analysis of chromatin immunoprecipitates.
We first demonstrate the fidelity of the RIP-chip assay with an experiment involving CAF1, whose RNA substrates are known. We then use the assay to acquire new information about CRP1, a genetically characterized member of the PPR family in maize. CRP1 is required for the translation of the chloroplast petA mRNA and for the accumulation of processed chloroplast petB and petD mRNAs; these mRNAs encode subunits of the cytochrome b6f complex, which fails to accumulate in crp1 mutants (Barkan et al., 1994; Fisk et al., 1999). In addition, a role for CRP1 in the translation of psaC mRNA (encoding a photosystem I subunit) was suggested by the loss of psaC mRNA from polysomes and the decreased abundance of the photosystem I complex in crp1 mutants; however, we were unable to measure psaC translation rates with pulse-labeling experiments and so did not reach a firm conclusion about the role of CRP1 in psaC expression (Barkan et al., 1994).
These effects on RNA metabolism suggested that CRP1 might interact with RNA, but CRP1 did not detectably cofractionate with RNA during size-fractionation of stromal extract (Fisk et al., 1999; Fisk, 2000). Furthermore, the complexity of the mutant phenotype made it unclear which molecular defects were direct effects of the absence of CRP1 and which might be indirect effects. In this study, application of the RIP-chip assay clarified these issues by showing CRP1 to be associated with the petA and psaC mRNAs in native chloroplast extract. These results, considered in conjunction with the molecular defects in crp1 mutants, provide strong evidence that CRP1 indeed activates the translation of both the petA and psaC mRNAs and that the translation defects in crp1 mutants are a direct, rather than an indirect, consequence of the loss of CRP1.
These findings show that a representative PPR protein influences the expression of specific RNAs through close proximity with those mRNAs; this has been hypothesized to be a general feature of PPR proteins (Small and Peeters, 2000) but has not previously been demonstrated. The results further suggest that translational regulation may be a particularly common function of PPR proteins. Finally, the organization of a motif that is shared by the two RNAs bound by CRP1 suggests an unusual mode of RNA recognition for CRP1 and, by extension, for other members of the PPR family.
RESULTS
Experimental Design
A microarray was designed with 248 overlapping PCR fragments representing the complete maize chloroplast genome. The PCR fragments were assigned identifying numbers according to the position of their left terminus (i.e., lower residue number) in the maize chloroplast genome sequence (Maier et al., 1995) (see Supplemental Table 1 online); thus, fragment numbers reflect approximate positions along the genome and provide a convenient way to display data according to chromosomal position. Each fragment was spotted in five copies.
cDNA-based labeling methods were used in prior studies involving arrays to identify RNAs that copurify with cytosolic RNA binding proteins (Tenenbaum et al., 2000; Hieronymus and Silver, 2003; Shepard et al., 2003; Gerber et al., 2004; Inada and Guthrie, 2004). However, we found that the direct coupling of dyes to the RNA (Gupta et al., 2003) resulted in higher signal-to-noise ratios than cDNA-based methods (data not shown). The improved results with the direct labeling method are to be expected, given its lack of bias against small RNA fragments, 3′ regions, and structured RNAs, all of which are poor substrates for reverse transcription.
Microarray analyses typically involve the simultaneous hybridization of differentially labeled experimental and reference samples to the same array. In the assays used here, the RNA purified from the immunoprecipitation pellet served as the experimental sample. Results obtained with three types of reference sample were compared: (1) RNA purified from the supernatant of the same immunoprecipitation; (2) RNA purified from total stromal extract; and (3) RNA purified from the pellet of an immunoprecipitation involving an antibody that does not bind a chloroplast RNA binding protein. The first of these methods revealed the enrichment of specific RNAs most distinctly and was chosen as our standard procedure; indeed, the depletion of an RNA from the supernatant and its simultaneous enrichment in the pellet would be expected to result in the largest change in ratio. Comparison with a stromal RNA reference gave similar results, but the magnitude of the enrichment peaks was somewhat lower (see Supplemental Figure 1 online). Comparison with a mock immunoprecipitate was least useful because of the uniformly low signals in the reference channel; this resulted in difficulty detecting many spots and also in spurious high experimental:reference ratios as a result of the low reference values.
RNAs from immunoprecipitation pellets and supernatants were labeled with the red fluorescing dye Cyanine 5 (Cy5) and the green fluorescing dye Cyanine 3 (Cy3), respectively. After hybridization and array scanning, a small number of red spots, representing RNAs that were enriched by immunoprecipitation, were observed on a background of green spots, representing all detectable chloroplast RNAs (Figure 1).
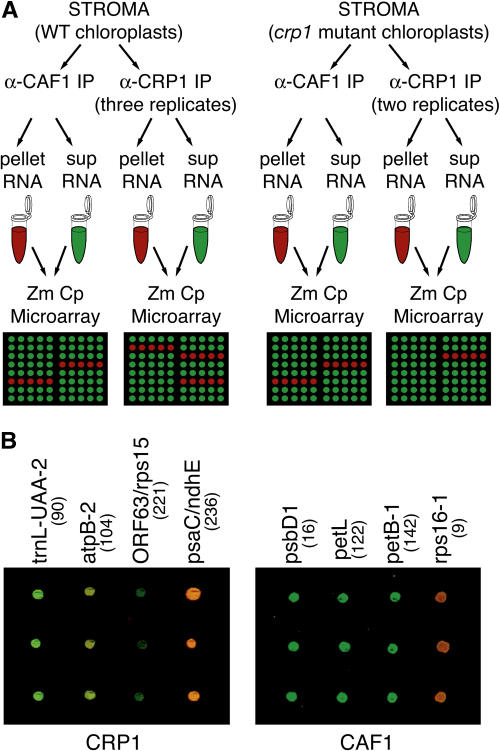
Figure 1.
Overview of the RIP-Chip Experiments.
(A) Experimental design. Chloroplast stroma from either wild-type or crp1 mutants was used for immunoprecipitation (IP) with the indicated antisera. RNA purified from immunoprecipitation pellets or supernatants (sup) was labeled with Cy5 or Cy3, respectively, combined, and used to probe the maize chloroplast (Zm Cp) microarray. Replicate data sets for the wild-type extract derive from two independent immunoprecipitations, one of which was used for two labeling/hybridization reactions. Replicate data sets for the crp1 mutant extract derive from two independent immunoprecipitations.
(B) Excerpts of merged F635:F532 images from a CRP1 and a CAF1 experiment involving wild-type stroma. Fragment names are indicated, and fragment numbers are in parentheses. Each DNA fragment is represented five times on the array, in clusters of two and three spots. These excerpts show three-spot clusters.
Genome-Wide Analysis of RNA Ligands of the Chloroplast-Splicing Factor CAF1
CAF1 is a nucleus-encoded protein that is required for the splicing of several group II introns in maize chloroplasts (Ostheimer et al., 2003). Slot-blot hybridizations of RNAs that coimmunoprecipitate with CAF1 showed it to be associated with introns whose splicing it facilitates (Ostheimer et al., 2003). However, the probe set used for the slot blots was not comprehensive, so additional targets could not be excluded. The RIP-chip assay assesses all possible RNA partners and should allow the detection of other RNA ligands, should they exist. Additionally, the associations between CAF1 and several introns were known to survive immunoprecipitation, so CAF1 was a suitable choice for optimizing the RIP-chip method.
Antibody to CAF1 was used for immunoprecipitation with chloroplast stroma from wild-type maize seedlings or from crp1 mutant plants. The crp1 mutant material provided a replicate in the CAF1 experiment as well as a control for the CRP1 experiment described below. Mutations in crp1 and caf1 influence the metabolism of different RNAs, so it was anticipated that CAF1–RNA interactions would be unaltered in crp1 mutants; indeed, this is what was observed.
RNAs purified from the pellet and supernatant of each immunoprecipitation were labeled with Cy5 and Cy3, respectively, combined, and used to probe the chloroplast microarray. The ratio of Cy5 to Cy3 fluorescence (F635:F532) for each spot reflects the degree to which corresponding RNAs were enriched by immunoprecipitation relative to other spots on the same array. It is not, however, an absolute enrichment ratio because of inevitable differences in the labeling efficiencies of the pellet and supernatant RNAs and differences in the fluorescence characteristics of the two dyes. Spots whose Cy3 signal (supernatant RNA) was not significantly above background were excluded from subsequent data analyses (see Methods). At least three replicate spots per array yielded data for most DNA fragments (see Supplemental Table 2 online). Figure 1 provides an overview of this experimental design.
Figure 2A (gray bars) shows the distribution of enrichment ratios for all array fragments in the two CAF1 experiments. Almost all of the most highly enriched sequences derive from the petD, trnG, ycf3, and rps16 introns (black bars in Figure 2A and maps in Figure 2B), which were shown previously to require CAF1 for their splicing and to coimmunoprecipitate with CAF1 in a slot-blot hybridization assay (Ostheimer et al., 2003). Furthermore, spots corresponding to these regions are reproducibly found in this highly enriched fraction. These positives stand out upon casual inspection of the fluorescence images (Figure 1B; data not shown). Their enrichment is dependent on CAF1 antibody, because they were not among the most highly enriched sequences in CRP1 immunoprecipitates (see below).
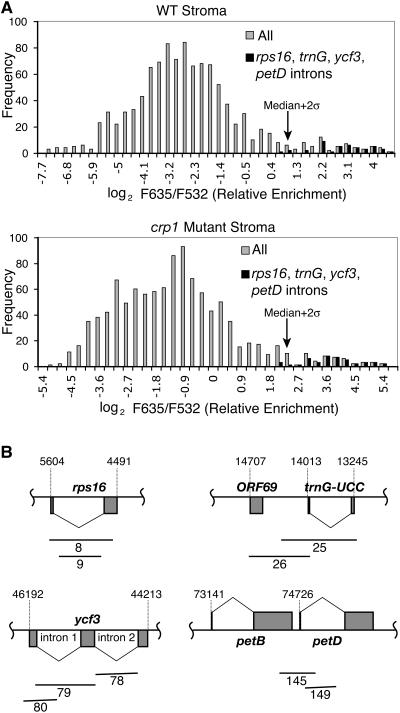
Figure 2.
Distributions of Enrichment Ratios in CAF1 RIP-Chip Experiments.
(A) Histograms showing the distribution of enrichment ratios in CAF1 RIP-chip assays involving wild-type or crp1 stroma. F635/F532 is the ratio of fluorescence deriving from the pellet and supernatant RNAs, respectively. Gray bars show the distribution of ratios for all array elements; black bars show the distribution for the subset of elements diagrammed in (B).
(B) Map positions of array fragments corresponding to the most highly enriched sequences in CAF1 RIP-chip assays. Fragment numbers are shown below each fragment. Residue numbers within the chloroplast genome (Maier et al., 1995) are indicated.
Relative enrichment values are mapped onto the chloroplast genome in Figure 3 by plotting the median enrichment ratio for replicate spots as a function of fragment number. The four most prominent peaks correspond to the rps16, trnG, ycf3, and petD introns. In addition to these known CAF1 substrates, this plot reveals the strong enrichment of one segment of the trans-spliced rps12 group II intron (fragment 130). Less prominent peaks correspond to the intronless gene trnV-GAC and to three genes with group II introns: rpl2, ndhB, and rpl16. The rpl16 intron requires CAF1 for its splicing, so enrichment here likely reflects an authentic interaction. The enrichment of the rpl2, rps12, and ndhB introns is intriguing in that the splicing of these introns is reduced (albeit not eliminated) in caf1 mutants (Ostheimer et al., 2003). In light of these new data, it seems likely that CAF1 plays a nonessential role in their splicing, perhaps by acting redundantly with the closely related splicing factor CAF2 (Ostheimer et al., 2003).
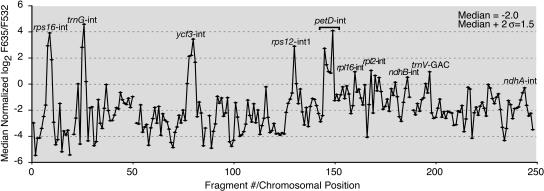
Figure 3.
CAF1 RIP-Chip Data Plotted According to Genomic Position.
Log2-transformed F635/F532 ratios were normalized between replicate CAF1 RIP-chip data sets (see Methods). The median normalized values for replicate spots across the two replicate experiments are plotted according to fragment number. Fragments for which fewer than two spots per array yielded an F532 signal above background were excluded and appear as gaps in the curve. The normalized data used to generate this curve are provided in Supplemental Table 2 online. Supplemental Figure 2 online shows analogous plots for the two individual experiments without normalization and illustrates the high degree of reproducibility. The enrichment values in the petD intron region show a bimodal peak in which F635/F532 correlates with the proportion of the fragment containing intron sequence (see Supplemental Table 1 online). Enrichment of the petD, rps16, trnG, and ycf3 introns was previously shown by slot-blot analysis of RNAs that coimmunoprecipitate with CAF1 (Ostheimer et al., 2003). The slight enrichment of rpl2 and ndhB intron sequences was confirmed by slot-blot hybridizations of pellet and supernatant RNAs derived from an independent CAF1 immunoprecipitation (data not shown).
The ndhA intron fails to splice in caf1 mutants (Ostheimer et al., 2003) but was not represented among the most highly enriched sequences (Figure 3, fragments 242 to 244). Therefore, either CAF1 influences the splicing of the ndhA intron indirectly or the affinity of CAF1 for this intron is too low to detect with this assay. Nonetheless, the unambiguous results with the petD, trnG, ycf3, and rps16 introns show RIP-chip to be effective for identifying RNAs associated with a specific chloroplast protein.
CRP1 Is Associated with Two mRNAs Whose Translation Is CRP1-Dependent
CRP1 was chosen as a particularly interesting candidate for RIP-chip analysis for the following reasons. (1) CRP1 is among only a handful of genetically characterized PPR proteins and therefore provides a useful model for elucidating the general properties of proteins in this large and poorly understood family. (2) Although genetic and in vitro evidence suggests that PPR proteins typically interact with RNA in vivo, only for the human PPR protein LRP130 has an in vivo association with RNA been demonstrated; even then, the identity of the RNA is unknown (Mili and Pinol-Roma, 2003). Identification of CRP1's RNA ligands would thus fill an important gap in our understanding of PPR proteins. (3) In contrast with CAF1, which binds group II introns, the genetically defined targets of CRP1 are mRNAs (Barkan et al., 1994). Thus, analysis of CRP1 should indicate whether RIP-chip will be useful for analyzing protein–mRNA interactions in chloroplasts.
Three replicate data sets were collected for CRP1. Controls for this experiment used CRP1 antibody with stroma from crp1mutant chloroplasts (two replicates) and CAF1 antibody with the same wild-type and mutant stromal preparations (Figure 1A). Use of the crp1 mutant extract was necessary because the CRP1 antibody cross-reacts with several proteins (perhaps closely related PPR proteins) even after affinity purification; the crp1 mutant extract lacks CRP1 but retains the cross-reacting proteins (Figure 4A) and so can be used to distinguish CRP1-associated RNAs from those associated with the cross-reacting proteins. The integrity of the crp1 mutant extract is illustrated by the fact that its use for a CAF1 RIP-chip assay yielded results similar to those with wild-type extract (Figure 2; see Supplemental Figure 2 online).
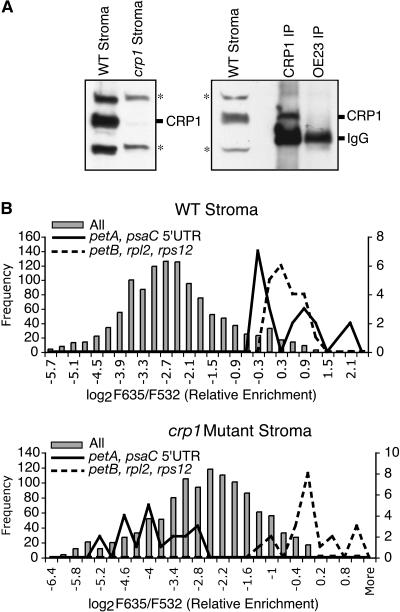
Figure 4.
Representative CRP1 RIP-Chip Data.
(A) Immunoblots probed with the affinity-purified CRP1 antibody used in the RIP-chip assays. The left panel shows an immunoblot of chloroplast stroma from wild-type and crp1 mutant seedlings. Cross-reacting proteins are indicated with asterisks. The right panel shows an immunoblot of an α-CRP1 immunoprecipitation involving wild-type stroma; an immunoprecipitation with a different antibody (α-OE23) is shown as a negative control. IgGs in the immunoprecipitates are detected by the secondary antibody used to probe the immunoblot.
(B) Histograms showing the distribution of enrichment ratios in representative experimental and control CRP1 RIP-chip assays. Gray bars show the distribution of ratios for all array elements and are drawn to the frequency scale at left. Solid lines show the distribution for replicate spots overlapping the petA 5′ UTR (fragments 113 and 114) and the psaC 5′ UTR (fragments 233 and 235); the positions of these fragments are diagrammed in Figure 6C. Dashed lines show the distribution for spots corresponding to fragments 139, 142, 169, and 194, which derive from the petB, rpl2, and rps12 genes. The line plots are drawn to the frequency scale at right.
Histograms showing the distribution of enrichment ratios for one CRP1 RIP-chip experiment involving wild-type extract and one involving crp1 mutant extract are shown in Figure 4B. The wild-type data show a bimodal distribution, with the upper peak including ∼7% of the spots on the array. Notably, the fragments at the very highest end of the distribution were derived from the 5′ regions of the psaC and petA genes, the two genes for which translation defects had been detected in crp1 mutants (Barkan et al., 1994). These stand out as red spots upon casual inspection of the fluorescence image (Figure 1B; data not shown). The distribution of enrichment ratios for replicate spots derived from two fragments overlapping the psaC 5′ untranslated region (UTR) and two overlapping the petA 5′ UTR is shown by the solid lines in Figure 4B. These were consistently found at the top of the distribution in the wild-type data set but not in the crp1 mutant data set. These results suggested that CRP1 is associated with RNAs from the psaC and petA regions.
The upper peak of the wild-type data set also included sequences from several other regions, primarily the rps12, rpl2, and petB genes. These latter sequences, however, were found at the high end of the distribution of both the wild-type and mutant data sets (dashed lines in Figure 4B and additional data shown below), indicating that their enrichment is independent of CRP1. The petB and rpl2 exon sequences were not highly enriched in the CAF1 RIP-chip experiments, so their enrichment here suggests that they are associated with the cross-reacting proteins detected by the CRP1 antibody.
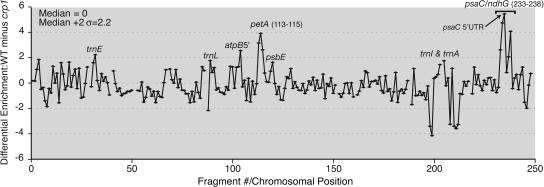
Figure 5 displays the combined data from all of the CRP1 assays as the difference between the normalized median enrichment ratios in the experimental (wild-type stroma) versus control (crp1 mutant stroma) assays, plotted according to fragment number. This representation maps onto the chloroplast genome the degree to which each sequence was enriched in a CRP1-dependent manner. The two highest peaks derive from the petA and psaC regions: both peaks are >2 sd above the median, both include several fragments, and both peak values derive from 5′ UTR sequences. These results provide strong evidence that CRP1 is associated with the petA and psaC mRNAs in vivo and that these are its highest affinity ligands. These results also suggest that CRP1 binding is localized to the 5′ regions of both mRNAs.
Figure 5.
CRP1 RIP-Chip Data Plotted According to Genomic Position.
The log2-transformed enrichment ratios (F635:F532) were normalized between the three CRP1 RIP-chip assays involving wild-type stroma and the two control assays with crp1 mutant stroma. The difference between the median normalized values for replicate spots in the experimental and control data sets is plotted as a function of fragment number. Fragments for which fewer than two spots per array yielded an F532 signal above background were excluded and appear as gaps in the curve. The data used to generate this curve are provided in Supplemental Table 3 online. Supplemental Figure 3 online shows analogous plots for the five individual experiments without data normalization. The valley within the psaC peak region is artifactual: the F532 signal/base pair for fragment 236 (the low point) is substantially lower than that for the fragments that overlap it, indicating that the DNA in this spot is defective.
We subjected the data to additional analyses to more rigorously evaluate these conclusions and to assess the significance of the less prominent peaks. First, a t test was used to evaluate the significance of the difference between the normalized enrichment values obtained with wild-type versus crp1 mutant stroma. Second, the data were analyzed with a ranking procedure that avoids the need to normalize data across replicate assays and that has been suggested for analysis of ChIP-chip data (Buck and Lieb, 2004): (1) the percentile rank of each spot was expressed as the fraction of spots on the same array with a lower enrichment ratio; (2) the median percentile rank among replicate spots across replicate arrays was calculated; and (3) a t test was performed to evaluate the significance of the difference between the ranks observed with wild-type versus crp1 mutant stroma. Median percentile ranks and P values are provided in Supplemental Table 4 online.
Table 1 summarizes these analyses for those DNA fragments that ranked in the top 15% of the wild-type data sets according to both types of calculation. Fragments in Table 1 are ordered according to the magnitude of their differential enrichment from wild-type versus crp1 mutant stroma. P values were calculated to evaluate the null hypothesis that sequences corresponding to each fragment were not differentially enriched from wild-type versus crp1 mutant extract. A P value of 3 × 10−4 or less provides confidence that enrichment is significant, as this cutoff would be anticipated to yield fewer than 0.1 false positives among the 248 fragments analyzed. The P values derived from both methods of analysis (i.e., normalized enrichment ratio and percentile rank) indicate highly significant CRP1-dependent enrichment of fragments from the psaC and petA regions (psaC fragments 233 to 235, 237, and 238; petA fragments 113 to 115). The fact that three or more adjacent fragments were coenriched in each region is not reflected in the already low P values for individual fragments and further increases confidence that the enrichment of petA and psaC sequences is authentic. The genomic context of these fragments is shown in Figure 6C.
Table 1.
Top-Ranking Fragments in CRP1 RIP-Chip Assays
| Wild-Type Stroma
|
crp1 Mutant Stroma
|
Comparison: Wild-Type versus crp1 Stroma
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fragment Name | Fragment Numbera | Median Log2 Ratio (E)b | Median Percentile Rankc | nbc | Median Log2 Ratio (E)b | Median Percentile Rankc | nbc | Differential Enrichment (EWT – Ecrp1) | P (Enrichment Ratio)d | P (% Rank)d |
| psaC/ndhE | 235 | 1.82 | 1.00 | 15 | −3.61 | 0.16 | 9 | 5.43 | 5.3E-09 | 4.3E-10 |
| psaC-2 | 234 | 0.66 | 0.96 | 10 | −4.06 | 0.13 | 7 | 4.72 | 6.4E-05 | 4.2E-07 |
| ndhG-1 | 238 | 0.89 | 0.98 | 15 | −3.17 | 0.35 | 7 | 4.06 | 2.3E-04 | 1.6E-04 |
| cemA/petA | 114 | 0.68 | 0.99 | 15 | −3.22 | 0.38 | 8 | 3.9 | 2.4E-05 | 1.1E-06 |
| cemA | 113 | −0.25 | 0.94 | 15 | −3.39 | 0.15 | 10 | 3.14 | 4.6E-05 | 1.5E-09 |
| psaC-1 | 233 | −0.32 | 0.94 | 15 | −2.92 | 0.34 | 9 | 2.6 | 1.2E-04 | 1.3E-04 |
| petA-1 | 115 | −0.36 | 0.93 | 12 | −2.96 | 0.28 | 8 | 2.6 | 1.5E-05 | 1.0E-08 |
| atpB5′ | 104 | −1 | 0.86 | 15 | −3.54 | 0.24 | 9 | 2.54 | 1.8E-03 | 5.7E-07 |
| ndhE/ndhG | 237 | 0.2 | 0.95 | 15 | −1.86 | 0.74 | 8 | 2.06 | 5.9E-05 | 3.0E-04 |
| trnI/trnA | 205 | −0.16 | 0.95 | 15 | −1.9 | 0.54 | 10 | 1.74 | 1.4E-03 | 1.4E-04 |
| ndhA-3 | 244 | 0.07 | 0.94 | 15 | −1.17 | 0.88 | 9 | 1.24 | 1.4E-01 | 6.7E-02 |
| rps12-3 | 190 | 0.38 | 0.94 | 15 | −0.74 | 0.88 | 10 | 1.12 | 2.0E-01 | 3.1E-02 |
| rps18/rpl20 | 129 | −0.87 | 0.87 | 15 | −1.99 | 0.75 | 8 | 1.12 | 9.7E-02 | 9.3E-03 |
| Rpl2-1 | 167 | −0.23 | 0.93 | 15 | −1.03 | 0.85 | 10 | 0.8 | 1.5E-01 | 4.2E-03 |
| petA3′ | 116 | −0.39 | 0.92 | 11 | −1.17 | 0.80 | 5 | 0.78 | 2.8E-02 | 1.0E-01 |
| ndhA intron | 243 | 0.25 | 0.96 | 15 | −0.39 | 0.91 | 10 | 0.64 | 5.5E-02 | 5.1E-06 |
| Rrn16/trnI | 201 | −0.95 | 0.88 | 9 | −1.42 | 0.70 | 6 | 0.47 | 7.9E-02 | 1.3E-04 |
| psbH-2 | 140 | −0.18 | 0.91 | 15 | −0.62 | 0.93 | 10 | 0.44 | 8.6E-01 | 4.9E-01 |
| trnA intron | 207 | −0.59 | 0.90 | 15 | −0.97 | 0.85 | 10 | 0.38 | 3.0E-01 | 8.2E-02 |
| rps18 | 128 | −0.12 | 0.94 | 14 | −0.47 | 0.92 | 9 | 0.35 | 2.3E-01 | 1.6E-01 |
| petB-1 | 142 | 0.38 | 0.95 | 15 | 0.17 | 0.99 | 10 | 0.21 | 9.2E-01 | 7.5E-01 |
| Rpl2-2 | 169 | −0.01 | 0.95 | 15 | −0.12 | 0.98 | 9 | 0.11 | 3.7E-01 | 7.1E-04 |
| rps12 int1-1 | 131 | −0.02 | 0.94 | 15 | −0.11 | 0.92 | 10 | 0.09 | 5.1E-01 | 3.4E-01 |
| petB-2 | 139 | 0.36 | 0.97 | 15 | 0.51 | 0.98 | 10 | −0.15 | 6.8E-01 | 2.7E-01 |
| trnK intron | 6 | −0.41 | 0.92 | 15 | −0.05 | 0.96 | 10 | −0.36 | 5.0E-01 | 3.4E-01 |
| petB intron | 144 | −0.22 | 0.91 | 15 | 0.17 | 0.95 | 10 | −0.39 | 2.6E-01 | 8.4E-03 |
| ycf3-4 | 80 | −0.65 | 0.90 | 15 | 0.02 | 0.96 | 9 | −0.67 | 4.1E-02 | 1.3E-05 |
| rps125′ | 194 | 0.63 | 0.98 | 15 | 1.62 | 1.00 | 10 | −0.99 | 1.2E-02 | 5.1E-04 |
Fragments ranking in the top 15% for both median normalized enrichment ratio (E) and median percentile rank are listed and ordered according to the magnitude of their differential enrichment from wild-type versus crp1 stroma (EWT – Ecrp1). Fragments whose differential enrichment is highly significant (P < 3E-04) according to both methods are highlighted in boldface; this P-value cutoff would be anticipated to yield fewer than 0.1 false positives among the 248 array fragments.
Fragment numbers reflect chromosomal position (see Supplemental Table 1 online).
E = median (log2 F635:F532) normalized across three replicate experiments with wild-type stroma and two with crp1 mutant stroma. Replicate experiments constitute a total of n replicate spots with F532 above background.
Median percentile ranks for three (wild type) or two (crp1) replicate experiments compared with other spots on the same array. Elements were ranked within a data set according to the nonnormalized F635:F532 value; median rank for n replicate spots in replicate experiments is shown.
P values were calculated with a t test (two-tailed, unequal variance).
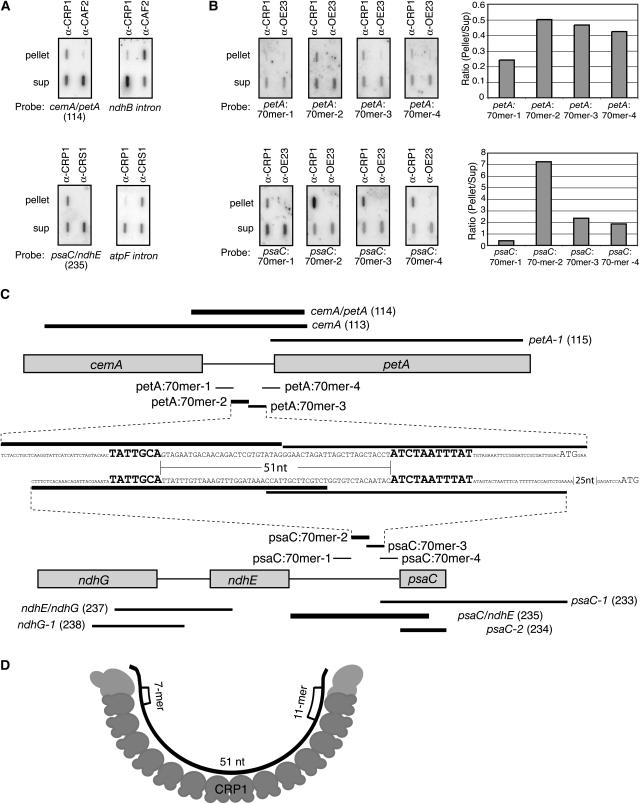
Figure 6.
Fine-Mapping of RNAs That Coimmunoprecipitate with CRP1.
(A) Verification that petA and psaC RNAs are enriched in CRP1 coimmunoprecipitates. Coimmunoprecipitations and RNA extractions were performed as for the RIP-chip assays; the RNAs were then analyzed by slot-blot hybridization with the indicated probes. The top panels, showing CRP1-dependent enrichment of petA 5′ sequences, are duplicate blots of RNAs from parallel CRP1 and CAF2 immunoprecipitations. The CAF2 immunoprecipitation was included as a negative control; the ndhB intron is a known CAF2 substrate (Ostheimer et al., 2003) and was analyzed to demonstrate the integrity of the CAF2 pellet RNA. The bottom panels, showing CRP1-dependent enrichment of psaC 5′ sequences, are duplicate blots of RNAs from parallel CRP1 and CRS1 immunoprecipitations. The CRS1 immunoprecipitation was included as a negative control; the atpF intron is a known CRS1 substrate (Ostheimer et al., 2003) and was analyzed to demonstrate the integrity of the CRS1 pellet RNA. The psaC/ndhE (fragment 235) and cemA/petA (fragment 114) probes correspond to the two highest ranking fragments in the CRP1 RIP-chip experiments and are diagrammed in (C). Sup, supernatant.
(B) Fine-mapping of CRP1-associated RNAs from the psaC and petA 5′ UTRs. Immunoprecipitation with a different antibody (α-OE23) served as a negative control. Duplicate slot blots were prepared as in (A) except that RNase inhibitor was omitted from the immunoprecipitation reactions. Blots were probed with 5′ end-labeled 70-mer oligonucleotides as diagrammed in (C). Results were quantified with a phosphorimager and plotted at right.
(C) Summary of CRP1 RIP-chip and slot-blot data for the psaC and petA regions. The PCR fragments on the microarray (identified by name and number) and the 70-mer oligonucleotides used for slot-blot probing are indicated with lines whose thickness reflects the degree to which the sequences were enriched in CRP1 immunoprecipitates. The nucleotide (nt) sequences shown span the pair of 70-mers from each region that detected the most highly enriched sequences; they are aligned to highlight the 7-mer and 11-mer motifs (large boldface letters), with start codons shown in large lightface letters.
(D) Model for RNA recognition by CRP1. The cartoon illustrates how a CRP1 monomer might bind the RNA encompassed by the shared 7-mer and 11-mer motifs in the psaC and petA 5′ UTRs. The 14 repeating units in CRP1 represent its 14 tandem PPR motifs. The RNA is shown bound to the concave face of the predicted PPR superhelix; this predicted surface of CRP1 appears well-suited for RNA binding (Williams and Barkan, 2003).
The data suggested that RNAs from several other regions might also be enriched in a CRP1-dependent manner (e.g., atpB [fragment 104] and trnI/trnA [fragment 205]). However, the P values for the enrichment ratio data (∼10−3) provide less confidence that these peaks are CRP1-dependent compared with those for the petA and psaC regions (∼10−5 and 10−8, respectively). Additional experimentation will be required to evaluate the physiological relevance of these and the other smaller peaks revealed by the plot in Figure 5.
CRP1 is required for the accumulation of processed mRNAs with termini mapping in the petB/petD intergenic region (Barkan et al., 1994). The RIP-chip data did not, however, provide evidence for an interaction between CRP1 and those sequences. The failure to detect those interactions cannot be attributed to low-quality array spots, as fragments spanning this region and the flanking petD intron detected enriched RNAs in the CAF1 assays (see above). Therefore, either CRP1's interactions in this region are of too low an affinity to detect, or the defects in petB/petD RNA metabolism in crp1 mutants are an indirect effect of the absence of CRP1.
Together, the RIP-chip data provide strong evidence that CRP1 is associated with the petA and psaC RNAs. Those findings were confirmed and extended in the slot-blot hybridization assays described below.
Higher Resolution Mapping of Coimmunoprecipitated RNAs
Overlapping fragments encompassing the 5′ UTRs of psaC or petA showed the greatest CRP1-dependent enrichment in the RIP-chip experiments (Table 1, Figures 5 and 6C), suggesting that CRP1 is bound to these 5′ UTRs and that enrichment of adjacent sequences is attributable to tethering. Slot blots of RNA purified from immunoprecipitation pellets and supernatants were probed with the PCR fragments that detected the most highly enriched sequences in the RIP-chip assays (Figure 6A); the results verified that RNAs from these regions are highly enriched in CRP1 immunoprecipitates but not in immunoprecipitates generated with other antisera.
Slot-blot hybridization assays were then performed to more precisely map the coimmunoprecipitated RNAs. To increase the resolution of these assays, we aimed to decrease the size of coimmunoprecipitated RNA fragments by omitting the RNase inhibitor that was otherwise included in the immunoprecipitation protocol. Duplicate slot blots were hybridized with a series of 70-mer oligonucleotides from within each 5′ UTR (Figure 6B). In the petA 5′ UTR, peak enrichment was detected by a pair of adjacent 70-mers (petA:70mer-2 and petA:70mer-3), which together span 140 nucleotides (Figure 6C). In the psaC 5′ UTR, peak enrichment was detected by a pair of overlapping 70-mers (psaC:70mer-2 and psaC:70mer-3), which together span 125 nucleotides (Figure 6C). Binding sites for CRP1 in these 5′ UTRs make good biological sense, because CRP1 is required to activate the translation of the downstream open reading frames (Barkan et al., 1994).
Identification of a Possible CRP1 Recognition Motif
The 70-mer hybridization data localized the likely CRP1 interaction sites to ∼140 nucleotides in each of the two 5′ UTRs. Strikingly, two shared motifs, of 7 and 11 nucleotides, are found in these small regions (Figure 6C). Furthermore, the relative positions of the two motifs are identical in both regions: the 7-mer is upstream of the 11-mer, and the two motifs are separated by exactly 51 nucleotides. The 11-mer motif is found at only these two sites in the chloroplast genome; the 7-mer motif is found at numerous other sites but is clustered in the ndhE–psaC intergenic region, where it is represented four times. Outside of the psaC and petA 5′ UTRs, there are no other transcribed regions in which any 6 contiguous nucleotides of the 7-mer are found within 100 nucleotides of any 8 consecutive nucleotides of the 11-mer. Therefore, it seems unlikely to be coincidental that the two ∼140-nucleotide sequences that are most strongly enriched in CRP1 coimmunoprecipitations share 7-mer and 11-mer sequence elements with identical spacing. These findings suggest that CRP1 might interact with the 69-nucleotide regions spanned by the shared motifs in the psaC and petA 5′ UTRs.
DISCUSSION
Plant nuclear genomes are predicted to encode many hundreds of organellar RNA binding proteins, the majority of which lack orthologs in nonplant species: notably, >400 genes in Arabidopsis are predicted to encode organelle-targeted PPR proteins (Lurin et al., 2004). Among the few such proteins that have been studied, the understanding of physiological function and mechanism is generally quite limited. In this study, we document the utility of a microarray-based strategy for identifying the in vivo RNA ligands of organellar RNA binding proteins, and we use the method to identify RNAs associated with the chloroplast PPR protein CRP1. The data show that CRP1 is associated with the 5′ UTRs of two mRNAs: the petA mRNA, whose translation was known to be CRP1-dependent, and the psaC mRNA, whose translation was suspected to be CRP1-dependent. These findings add an important element to the previous body of data concerning the PPR family by showing that a member of the family exhibits strong and specific interactions in vivo with RNAs whose expression it regulates. These results add weight to the proposal that PPR proteins can serve as sequence-specific RNA binding proteins (Small and Peeters, 2000), and they suggest that the RIP-chip approach will be a useful tool for identifying RNA ligands of other members of this large and interesting protein family.
The RIP-Chip Assay
The RIP-chip method provides a powerful adjunct to biochemical and genetic approaches for identifying the physiological substrates of organellar RNA binding proteins. Methods that seek binding partners through in vitro assays are subject to artifact when molecules are taken out of their physiological context, and mutant phenotypes can be difficult to interpret because of pleiotropic effects or the presence of redundant genes. The RIP-chip method can be used as a starting point in the absence of other clues, or it can be used to test hypotheses based on mutant phenotypes or in vitro assays.
Hybridization of coimmunoprecipitated RNA to arrayed genes has been used in several recent studies involving nonplant species. The method described here incorporates two technical differences that we believe to be advantageous. First, we found the direct coupling of fluorescent dyes to the immunoprecipitated RNA to be superior to the cDNA-based labeling methods used previously (Tenenbaum et al., 2000; Hieronymus and Silver, 2003; Shepard et al., 2003; Gerber et al., 2004; Inada and Guthrie, 2004). Second, we found that the use of RNA from the immunoprecipitation supernatant as a reference sample revealed the enrichment of specific RNAs more unambiguously than the use of total input RNA (see Supplemental Figure 1 online) or RNA from a mock immunoprecipitation (data not shown).
Although statistical tests of RIP-chip data can provide strong evidence for particular RNA–protein associations, confidence is greatest when the mutational loss of a protein affects the metabolism of the same RNAs that are identified in RIP-chip assays. This condition was met for both CRP1 and CAF1: five of CAF1's six genetically defined intron targets rose to the top of the RIP-chip data, as did both mRNAs whose translation is dependent on CRP1. However, even in the absence of informative mutants, the RIP-chip assay can be used to direct attention to likely RNA substrates, whose validity can then be tested in other ways. This assay provides an entrée into the in vivo function of a protein when genetic analysis is problematic as a result of, for example, an embryo-lethal mutant phenotype or the presence of functionally redundant genes. We have used the RIP-chip assay with several other PPR proteins and with members of several other protein families. Each experiment has identified a distinct set of highly enriched sequences that correspond well with other information about the immunoprecipitated protein (our unpublished data). Thus, we anticipate that the method has the potential to yield useful information about the RNA ligands of a wide variety of organellar RNA binding proteins.
This study used polyclonal antisera generated to recombinant proteins, as have our prior published (Ostheimer et al., 2003) and unpublished RNA coimmunoprecipitation experiments. Each of the eight antisera we have used to date have coimmunoprecipitated RNAs whose metabolism is disrupted in the corresponding mutant background; this suggests that polyclonal antisera will not generally disrupt native RNA–protein interactions. Nonetheless, a negative result in an RNA coimmunoprecipitation assay should not be taken as strong evidence for the absence of an interaction. An alternative to custom antisera for organisms that are amenable to genetic transformation is the expression of tagged proteins. Indeed, tagged proteins were used for RIP-chip studies involving cytosolic mRNAs in yeast (Hieronymus and Silver, 2003; Shepard et al., 2003; Gerber et al., 2004; Inada and Guthrie, 2004). Because aberrantly high protein concentrations could result in nonnative RNA–protein interactions, the native promoter should be used to drive the expression of tagged proteins for RIP-chip assays.
RNA Recognition by PPR Proteins
Results of genetic and in vitro analyses have suggested that PPR tracts might represent a new class of RNA binding motif (Small and Peeters, 2000; Lurin et al., 2004; Nakamura et al., 2004). Structural modeling further suggested that RNA binding might take place along the length of a surface formed by stacked helical repeating units (Small and Peeters, 2000; Williams and Barkan, 2003). The recent finding that a different helical hairpin repeat motif, the PUM-HD, binds RNA in this general manner adds plausibility to these ideas (Edwards et al., 2001; Wang et al., 2002). Results presented here document an association between a PPR protein and specific RNA sequences in vivo. These interactions must be of high affinity, as they survive extract preparation and immunoprecipitation.
The assays used here do not indicate whether CRP1 binds directly to the petA and psaC RNAs or binds indirectly through interaction with another protein. However, in light of the biochemical, genetic, and structural data summarized above, it seems likely that the interactions are direct. Comparison of the ∼140-nucleotide segments in the petA and psaC mRNAs that are most strongly enriched in CRP1 immunoprecipitations revealed shared features that might constitute a CRP1 recognition element: identical 7-mer and 11-mer sequences separated by 51 nucleotides. Characterized single-stranded RNA binding proteins recognize short (∼5 to 10 nucleotides) motifs that are contiguous, or nearly so (Antson, 2000; Perez-Canadillas and Varani, 2001; Messias and Sattler, 2004). The long and precise spacing between the two conserved sequence elements in the CRP1-associated RNAs suggests that they are not recognized by typical globular RNA binding domains but rather by an elongated protein surface. Indeed, the 14 tandem PPR motifs in CRP1 are predicted to form a single, elongated domain, based on the structure of the related TPR motif (Fisk et al., 1999; Small and Peeters, 2000; D'Andrea and Regan, 2003). Consecutive helical hairpin repeats in a TPR domain stack upon one another to form a concave substrate binding surface. Helical packing geometry in TPR tracts is determined by several conserved hydrophobic amino acids (D'Andrea and Regan, 2003). PPRs have hydrophobic residues at analogous positions (Small and Peeters, 2000), but their helical packing is likely to be less compact as a result of the substitution of amino acids with large side chains for those with small side chains at two conserved positions of close helical contact (Fisk, 2000). Based on this, on the established dimensions of a TPR protein (Das et al., 1998), and on the structure of RNA in complex with TRAP, a different modular repeat protein (Hopcroft et al., 2004), it seems likely that each PPR motif could accommodate 3 or 4 nucleotides of single-stranded RNA and that the 14 repeats in CRP1 could bind ∼50 nucleotides along the length of their predicted substrate binding face. RNA associated with the structurally related PUM-HD is more extended than this as a result of the intercalation of aromatic amino acids (Wang et al., 2002), but aromatic residues are lacking from CRP1's predicted substrate binding surface (Williams and Barkan, 2003).
Thus, a CRP1 homodimer or heterodimer seems likely to be able to accommodate a single-stranded RNA encompassing the conserved 7-mer and 11-mer motifs and the intervening 51 nucleotides. Given the uncertainties about the protein and RNA geometries, it is even plausible that these 69 nucleotides bind along the surface of a CRP1 monomer (see model in Figure 6D). This binding might be aided by amino acids that are C terminal to the PPR tract in CRP1, which appear well-suited for RNA binding (data not shown).
In summary, the presence of identical 7-mer and 11-mer motifs separated by 51 nucleotides in both the psaC and petA 5′ UTRs, and the predicted dimensions of CRP1's PPR tract, lead us to favor a model in which the RNA between the conserved motifs is bound in single-stranded form along an elongated monomeric or dimeric protein surface. Structural data and structure–function studies of PPR–RNA interactions will be required to evaluate these possibilities. Such studies with CRP1 have been hampered by the fact that recombinant CRP1 is exceedingly difficult to generate as a soluble protein in Escherichia coli; exploration of a wide variety of expression vectors, host strains, and induction conditions have yielded, at best, microaggregates that elute in the void volume when subjected to size-exclusion chromatography (data not shown).
PPR Proteins as Translational Regulators
crp1 mutants have reduced levels of the cytochrome b6f and the photosystem I core complexes (Barkan et al., 1994). Three molecular lesions were detected that could account for these protein deficiencies: (1) a defect in the translation of the petA mRNA, encoding cytochrome f, was shown through pulse-labeling and polysome assays; (2) a defect in the translation of the psaC mRNA, encoding a photosystem I subunit, was suspected based on its reduced association with polysomes; and (3) the absence of monocistronic petD mRNA (which encodes a cytochrome b6f subunit) was correlated with reduced petD translation rates (Barkan et al., 1994).
The complexity of this phenotype, and the precedent from work in Chlamydomonas reinhardtii for secondary effects on the synthesis of photosynthetic complex subunits when the synthesis of a key subunit is disrupted (Choquet et al., 2001), made it uncertain which defects reflect CRP1's direct function. The findings described here clarify this situation: CRP1 is tightly associated with the 5′ UTRs of both the petA and psaC mRNAs, providing strong evidence that CRP1 directly affects the translation of these two mRNAs. No association was detected between CRP1 and the petB/petD intergenic region, so it remains uncertain whether CRP1's role there is direct or indirect. The mechanism through which CRP1 activates translation will be interesting to explore; the proximity of the CRP1 binding sites to start codons on both regulated mRNAs suggests that it might act by influencing local RNA structure and/or by recruiting a component of the basal translation machinery.
These results highlight the possibility that translational regulation may be a particularly common function of PPR proteins. Roles in the translation of specific organellar mRNAs are now established for all three genetically characterized PPR-encoding genes for which translational effects were assayed: maize crp1, yeast pet309 (Manthey and McEwen, 1995), and Neurospora cya-5 (Coffin et al., 1997). TBC2, a PPR-like protein in Chlamydomonas chloroplasts, is also a translational activator (Auchincloss et al., 2002). Given this trend, it seems possible that some of the PPR proteins that have been shown to influence only RNA processing (Pruitt and Hanson, 1991; Iwabuchi et al., 1993; Bentolila et al., 2002; Hashimoto et al., 2003; Kazama and Toriyama, 2003; Meierhoff et al., 2003; Akagi et al., 2004; Komori et al., 2004) may also regulate translation.
PPR domains are anticipated to be structurally similar to the PUM-HD, which forms a sequence-specific RNA binding surface through the stacking of repeating α-helical units (Edwards et al., 2001; Wang et al., 2001, 2002). Functionally characterized proteins with the PUM-HD bind to the 3′ UTRs of specific mRNAs and repress their translation (Spassov and Jurecic, 2003). It is intriguing that PPR proteins are likewise implicated in translational regulation, albeit to date as translational activators. It will be interesting to discover whether this structure/function parallel between the PUM-HD and the PPR domain is merely coincidental or has mechanistic significance.
METHODS
Microarray Construction
A total of 248 overlapping PCR products were generated that cover the entire maize (Zea mays) plastid chromosome (inverted repeat represented once), with three small gaps attributable to failed PCR procedures. The name, fragment number, and coordinates of each fragment (Maier et al., 1995) (GenBank accession number X86563) are shown in Supplemental Table 1 online. DNA samples were dried, resuspended in 3× SSC (1× SSC is 150 mM sodium chloride, 15 mM sodium citrate, pH 7.0) and 0.1 M betaine and arrayed from a 384-well microtiter plate onto poly-l-Lys-ES–coated microscope slides (Erie Scientific) by a custom-built robot (Shalon et al., 1996; DeRisi et al., 1997) at the University of Oregon Genomics and Proteomics Facility. Each sample was spotted five times using different needle sets for the first three and last two print runs. Arrays were postprocessed within a few weeks before use, as follows. They were rehydrated by suspension over a 40°C water bath for ∼10 s until the spots glistened, snap-dried on a heat block (80°C) for 30 s, and UV cross-linked to polylysine in a Stratalinker (Stratagene) at 250 mJ. The array surface was blocked by submersion in a solution of 4.2 g of succinic anhydride dissolved in 235 mL of 1-methyl-2-pyrrolidinone and 10.5 mL of 1 M sodium borate, pH 8.0, with gentle shaking (50 rpm) for 15 min at 25°C. The arrays were then submerged in water for 2 min at 95°C to denature the DNA, rinsed briefly in 95% ethanol, dried by centrifugation, and stored in the dark.
Immunoprecipitation and RNA Labeling
The inbred line B73 (Pioneer HiBred) was used as the source of wild-type chloroplasts; the crp1-1 mutant line was described previously (Barkan et al., 1994; Fisk et al., 1999). Preparation of stromal extract, immunoprecipitations, and RNA extraction were performed as described previously (Ostheimer et al., 2003). The CRP1 and CAF1 antisera were described by Fisk et al. (1999) and Ostheimer et al. (2003), respectively. RNase inhibitor was omitted from the immunoprecipitation reactions used for the slot-blot hybridization assays in Figure 6B to decrease the size of coprecipitated RNAs and thereby increase the resolution of the assay.
RNA from each pellet or supernatant was ethanol-precipitated and resuspend in 30 μL of water. Fifteen microliters of pellet RNA and 5 μL of supernatant RNA were labeled with Cy5 and Cy3, respectively, using the Micromax ASAP RNA labeling kit (Perkin-Elmer Life Sciences) as follows. Volumes were brought to 18 μL by the addition of ASAP labeling buffer. Two microliters of Cy3 or Cy5 chemical labeling reagent was added, and the mixture was incubated at 85°C for 15 min. The reactions were then cooled to 4°C for 5 min and stopped by the addition of 5 μL of ASAP stop solution. The labeled RNA was purified using a Qiaquick PCR purification spin column (Qiagen). Elution from the column was performed with 100 μL of water, and the eluate was dried by vacuum centrifugation. Although gene expression profiling experiments routinely include controls in which the fluorophore on the experimental and reference samples are swapped (dye swaps), such controls have not been used in landmark studies involving analogous RNA immunoprecipitation assays (Gerber et al., 2004; Inada and Guthrie, 2004) or the related ChIP-chip assay (Lieb et al., 2001). In the CRP1 assays, positives were defined as sequences that were significantly more enriched from wild-type extract than from crp1 mutant extract; dye bias and other artifacts of the procedure cannot account for differential enrichment from the two extracts. Furthermore, we assessed potential dye bias by probing an array with a mixture of Cy3- and Cy5-labeled stromal RNA; F635:F532 showed only small fluctuations across all array fragments (<2-fold) (see Supplemental Figure 4 online), in contrast with the 8- to 64-fold enrichment seen for positives noted here.
For the slot-blot hybridization experiments, one-sixth of the RNA purified from each immunoprecipitation pellet and one-twelfth of the RNA purified from the corresponding supernatant was applied to a nylon membrane with a slot-blot manifold and hybridized to specific radiolabeled probes, as described previously (Ostheimer et al., 2003). Seventy-mer oligonucleotide probes were radiolabeled at their 5′ ends with [γ-32P]ATP and T4 polynucleotide kinase. PCR fragments were body-labeled with [32P]dCTP by the random priming method. Slot blots were hybridized in 7% SDS, 0.5 M Na-phosphate, pH 7, at 60 and 55°C for PCR probes and 70-mer probes, respectively. Blots were washed in 1× SSC, 0.1% SDS at the temperature used for hybridization.
Microarray Hybridization and Washing
The dried, labeled pellet and supernatant RNAs were each resuspended in 30 μL of hybridization buffer III supplied with the Micromax ASAP kit (Perkin-Elmer), warmed to 50°C, pooled, and heated to 50°C for 3 min. After a brief centrifugation to spin down the drops, the mixture was pipetted onto a microarray that had been prewarmed to 55°C. A cover slip was applied, and the slide was placed in a microarray hybridization chamber (Corning) and incubated at 58°C overnight. After hybridization, the slide was submerged in 0.01% SDS, 0.5× SSC until the cover slip slipped off the surface. The slide was then agitated at 50 rpm on a horizontal shaker for 15 min at room temperature in the same buffer. The array was transferred to a solution of 0.01% SDS, 0.06× SSC and shaken at room temperature for 15 min, then washed once more by shaking in 0.06× SSC at room temperature. Slides were dried by centrifugation for 3 min at 550 rpm in a Beckmann Coulter Allegra 6 tabletop centrifuge. Slides were scanned with a GenePix 4000B microarray scanner (Axon Instruments) with laser intensities chosen to maximize signals while avoiding pixel saturation.
Microarray Data Analysis
Data were imported into GenePix Pro 6.0 (Axon Instruments) and filtered to remove elements that did not meet the following criteria: (1) the F532 (supernatant RNA) signal-to-noise ratio (calculated by GenePix Pro 6.0) must be >2; (2) >50% of pixels in the F532 channel must have a fluorescence >2 sd above background. Fragments for which at least two of the five replicate spots on each array did not pass both tests were not included in subsequent analyses. Local background was calculated according to the default method in GenePix Pro 6.0, and the background-subtracted data were used to calculate the median of ratios (F635:F532). The log2-transformed value for the median of ratios is referred to in the figures as relative enrichment ratio. In Figures 3 and 5, data sets from replicate experiments were normalized according to the median log2 (median of ratios) value for all spots on each array.
Accession Numbers
Array design and data from this article have been deposited at ArrayExpress under accession numbers A-MEXP-164, E-MEXP-318, and E-MEXP-282.
Supplementary Material
Acknowledgments
We are indebted to Eric Johnson for his guidance during array preparation and his suggestions concerning data analysis and to Peter O'Day and Nicholas Stiffler for help implementing data analysis schemes. We also thank Eric Johnson for suggesting the term “RIP-chip,” Dan Graham and Doug Turnbull for help with array printing, Nigel Walker and Rodger Voelker for help with bioinformatics, Susan Belcher for preparing stromal extract, Kenny Watkins for help with the CAF1 immunoprecipitations, and Ian Small for useful comments on the manuscript. This work was supported by grants to A.B. from the National Science Foundation (MCB-0314597 and DBI-0421799) and by a postdoctoral fellowship to C.S.-L. from the Deutsche Forschungsgemeinschaft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alice Barkan (abarkan@molbio.uoregon.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.034454.
References
- Akagi, H., Nakamura, A., Yokozeki-Misono, Y., Inagaki, A., Takahashi, H., Mori, K., and Fujimura, T. (2004). Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 108, 1449–1457. [DOI] [PubMed] [Google Scholar]
- Antson, A.A. (2000). Single-stranded-RNA binding proteins. Curr. Opin. Struct. Biol. 10, 87–94. [DOI] [PubMed] [Google Scholar]
- Aubourg, S., Kreis, M., and Lecharny, A. (1999). The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 27, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchincloss, A., Zerges, W., Perron, K., Girard-Bascou, J., and Rochaix, J.-D. (2002). Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell Biol. 157, 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (2004). Intron splicing in plant organelles. In Molecular Biology and Biotechnology of Plant Organelles, H. Daniell and C. Chase, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 281–308.
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Barkan, A., Walker, M., Nolasco, M., and Johnson, D. (1994). A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13, 3170–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky, D. (2003). Unexpected complexity of poly(A)-binding protein gene families in flowering plants: Three conserved lineages that are at least 200 million years old and possible auto- and cross-regulation. Genetics 163, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila, S., Alfonso, A., and Hanson, M. (2002). A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 99, 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder, S., and Brennicke, A. (2003). Gene expression in plant mitochondria: Transcriptional and post-transcriptional control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, M.J., and Lieb, J.D. (2004). ChIP-chip: Considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83, 349–360. [DOI] [PubMed] [Google Scholar]
- Choquet, Y., and Wollman, F.A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529, 39–42. [DOI] [PubMed] [Google Scholar]
- Choquet, Y., Wostrikoff, K., Rimbault, B., Zito, F., Girard-Bascou, J., Drapier, D., and Wollman, F.A. (2001). Assembly-controlled regulation of chloroplast gene translation. Biochem. Soc. Trans. 29, 421–426. [DOI] [PubMed] [Google Scholar]
- Coffin, J.W., Dhillon, R., Ritzel, R.G., and Nargang, F.E. (1997). The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr. Genet. 32, 273–280. [DOI] [PubMed] [Google Scholar]
- D'Andrea, L.D., and Regan, L. (2003). TPR proteins: The versatile helix. Trends Biochem. Sci. 28, 655–662. [DOI] [PubMed] [Google Scholar]
- Das, A.K., Cohen, P.T.W., and Barford, D. (1998). The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Desloire, S., et al. (2003). Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4, 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, T.A., Pyle, S.E., Wharton, R.P., and Aggarwal, A.K. (2001). Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105, 281–289. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N. (2002). RNA-binding proteins in plants: The tip of an iceberg? Plant J. 5, 452–459. [DOI] [PubMed] [Google Scholar]
- Fisk, D.G. (2000). CRP1: Founding Member of a Novel Protein Family That Functions in Organellar Gene Expression. PhD dissertation (Eugene, OR: University of Oregon).
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18, 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, A.P., Herschlag, D., and Brown, P.O. (2004). Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2, E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, V., Cherkassky, A., Chatis, P., Joseph, R., Johnson, A., Broadbent, J., Erickson, T., and DiMeo, J. (2003). Directly labeled mRNA produces highly precise and unbiased differential gene expression data. Nucleic Acids Res. 31, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M., Endo, T., Peltier, G., Tasaka, M., and Shikanai, T. (2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36, 541–549. [DOI] [PubMed] [Google Scholar]
- Hieronymus, H., and Silver, P.A. (2003). Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 33, 155–161. [DOI] [PubMed] [Google Scholar]
- Hopcroft, N.H., Manfredo, A., Wendt, A.L., Brzozowski, A.M., Gollnick, P., and Antson, A.A. (2004). The interaction of RNA with TRAP: The role of triplet repeats and separating spacer nucleotides. J. Mol. Biol. 338, 43–53. [DOI] [PubMed] [Google Scholar]
- Ikeda, T., and Gray, M. (1999). Characterization of a DNA-binding protein implicated in transcription in wheat mitochondria. Mol. Cell. Biol. 19, 8113–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada, M., and Guthrie, C. (2004). Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc. Natl. Acad. Sci. USA 101, 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi, M., Kyozuka, J., and Shimamoto, K. (1993). Processing followed by complete editing of an altered mitochondrial atp6 RNA restores fertility of cytoplasmic male sterile rice. EMBO J. 12, 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama, T., and Toriyama, K. (2003). A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 544, 99–102. [DOI] [PubMed] [Google Scholar]
- Koizuka, N., Imai, R., Fujimoto, H., Hayakawa, T., Kimura, Y., Kohno-Murase, J., Sakai, T., Kawasaki, S., and Imamura, J. (2003). Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 34, 407–415. [DOI] [PubMed] [Google Scholar]
- Komori, T., Ohta, S., Murai, N., Takakura, Y., Kuraya, Y., Suzuki, S., Hiei, Y., Imaseki, H., and Nitta, N. (2004). Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). Plant J. 37, 315–325. [DOI] [PubMed] [Google Scholar]
- Kotera, E., Tasaka, M., and Shikanai, T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330. [DOI] [PubMed] [Google Scholar]
- Lahmy, S., Barneche, F., Derancourt, J., Filipowicz, W., Delseny, M., and Echeverria, M. (2000). A chloroplastic RNA-binding protein is a new member of the PPR family. FEBS Lett. 480, 255–260. [DOI] [PubMed] [Google Scholar]
- Landsberger, M., Lorkovic, Z.J., and Oelmuller, R. (2002). Molecular characterization of nucleus-localized RNA-binding proteins from higher plants. Plant Mol. Biol. 48, 413–421. [DOI] [PubMed] [Google Scholar]
- Lieb, J., Liu, X., Botstein, D., and Brown, P. (2001). Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28, 327–334. [DOI] [PubMed] [Google Scholar]
- Lorkovic, Z., and Barta, A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, R.M., Neckermann, K., Igloi, G.L., and Koessel, H. (1995). Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 251, 614–628. [DOI] [PubMed] [Google Scholar]
- Mancebo, R., Zhou, X., Shillinglaw, W., Henzel, W., and Macdonald, P. (2001). BSF binds specifically to the bicoid mRNA 3′ untranslated region and contributes to stabilization of bicoid mRNA. Mol. Cell. Biol. 21, 3462–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey, G.M., and McEwen, J.E. (1995). The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14, 4031–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff, K., Felder, S., Nakamura, T., Bechtold, N., and Schuster, G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15, 1480–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messias, A.C., and Sattler, M. (2004). Structural basis of single-stranded RNA recognition. Acc. Chem. Res. 37, 279–287. [DOI] [PubMed] [Google Scholar]
- Mili, S., and Pinol-Roma, S. (2003). LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell. Biol. 23, 4972–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monde, R., Schuster, G., and Stern, D. (2000). Processing and degradation of chloroplast mRNA. Biochimie 82, 573–582. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Meierhoff, K., Westhoff, P., and Schuster, G. (2003). RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 270, 4070–4081. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Schuster, G., Sugiura, M., and Sugita, M. (2004). Chloroplast RNA-binding and pentatricopeptide repeat proteins. Biochem. Soc. Trans. 32, 571–574. [DOI] [PubMed] [Google Scholar]
- Nickelsen, J. (2003). Chloroplast RNA-binding proteins. Curr. Genet. 43, 392–399. [DOI] [PubMed] [Google Scholar]
- Ostheimer, G., Williams-Carrier, R., Belcher, S., Osborne, E., Gierke, J., and Barkan, A. (2003). Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J. 22, 3919–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Canadillas, J.-M., and Varani, G. (2001). Recent advances in RNA-protein recognition. Curr. Opin. Struct. Biol. 11, 53–58. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D., and Hanson, M.R. (1991). Transcription of the Petunia mitochondrial CMS-associated Pcf locus in male sterile and fertility-restored lines. Mol. Gen. Genet. 227, 348–355. [DOI] [PubMed] [Google Scholar]
- Shalon, D., Smith, S.J., and Brown, P.O. (1996). A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 6, 639–645. [DOI] [PubMed] [Google Scholar]
- Shepard, K.A., Gerber, A.P., Jambhekar, A., Takizawa, P.A., Brown, P.O., Herschlag, D., DeRisi, J.L., and Vale, R.D. (2003). Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA 100, 11429–11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I., and Peeters, N. (2000). The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47. [DOI] [PubMed] [Google Scholar]
- Spassov, D.S., and Jurecic, R. (2003). The PUF family of RNA-binding proteins: Does evolutionarily conserved structure equal conserved function? IUBMB Life 55, 359–366. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, S., Carson, C., Lager, P., and Keene, J. (2000). Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. USA 97, 14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermel, M., Guermann, B., Delage, L., Grienenberger, J., Marechal-Drouard, L., and Gualberto, J. (2002). A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc. Natl. Acad. Sci. USA 99, 5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., McLachlan, J., Zamore, P.D., and Hall, T.M. (2002). Modular recognition of RNA by a human pumilio-homology domain. Cell 110, 501–512. [DOI] [PubMed] [Google Scholar]
- Wang, X., Zamore, P.D., and Hall, T.M. (2001). Crystal structure of a Pumilio homology domain. Mol. Cell 7, 855–865. [DOI] [PubMed] [Google Scholar]
- Williams, P., and Barkan, A. (2003). A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36, 675–686. [DOI] [PubMed] [Google Scholar]
- Xu, F., Morin, C., Mitchell, G., Ackerley, C., and Robinson, B.H. (2004). The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: Mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 382, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, H., Tasaka, M., and Shikanai, T. (2004). PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 38, 152–163. [DOI] [PubMed] [Google Scholar]
- Zerges, W. (2000). Translation in chloroplasts. Biochimie 82, 583–601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.