Abstract
The processes accompanying endosymbiosis have led to a complex network of interorganellar protein traffic that originates from nuclear genes encoding mitochondrial and plastid proteins. A significant proportion of nucleus-encoded organellar proteins are dual targeted, and the process by which a protein acquires the capacity for both mitochondrial and plastid targeting may involve intergenic DNA exchange coupled with the incorporation of sequences residing upstream of the gene. We evaluated targeting and sequence alignment features of two organellar DNA polymerase genes from Arabidopsis thaliana. Within one of these two loci, protein targeting appeared to be plastidic when the 5′ untranslated leader region (UTR) was deleted and translation could only initiate at the annotated ATG start codon but dual targeted when the 5′ UTR was included. Introduction of stop codons at various sites within the putative UTR demonstrated that this region is translated and influences protein targeting capacity. However, no ATG start codon was found within this upstream, translated region, suggesting that translation initiates at a non-ATG start. We identified a CTG codon that likely accounts for much of this initiation. Investigation of the 5′ region of other nucleus-encoded organellar genes suggests that several genes may incorporate upstream sequences to influence targeting capacity. We postulate that a combination of intergenic recombination and some relaxation of constraints on translation initiation has acted in the evolution of protein targeting specificity for those proteins capable of functioning in both plastids and mitochondria.
INTRODUCTION
The earliest information regarding mitochondrial protein targeting was obtained predominantly from nonplant species, and studies of plastid targeting were conducted independently. Although extensive detail is available on the process of plastid protein targeting and import, our understanding of the degree of overlap that exists between mitochondrial and plastid protein traffic has expanded in detail only recently. The numerous investigations of mitochondrial presequence features, compared with plastid targeting peptides, have provided some general properties that distinguish proteins destined for the two cellular compartments (reviewed in Peeters and Small, 2001; Zhang and Glaser, 2002). Plant mitochondrial targeting is conditioned by a fairly long (∼40 to 60 amino acids) N-terminal peptide generally rich in Arg and Ser, with a number of aliphatic residues such as Leu and Ala, to form an amphiphilic α helix. Similarly, the chloroplast targeting peptide is generally long (averaging 58 amino acids), rich in Ser and Ala, and low in acidic amino acids. Other than these general features, no defining amino acid sequence or pattern is evident to distinguish mitochondrial and plastid targeting.
During the endosymbiotic process, transfer of mitochondrial and plastid genes to the nucleus was followed by acquisition of necessary regulatory and protein targeting information upstream of the newly integrated organellar sequence. Attainment of regulatory information to allow proper expression within the nucleus is likely the more complicated process in this interorganellar DNA exchange, whereas the development of targeting capability may be somewhat straightforward (Martin and Herrmann, 1998). To some extent, the process involves intergenic recombination, resulting in shared presequences among distinct organellar proteins (reviewed in Brennicke et al., 1993; Kadowaki et al., 1996). Evidence of common presequences can still be found in present-day plant genomes (Elo et al., 2003). However, the acquisition of targeting information may also occur by a more fortuitous, random process of local sequence incorporation (Baker and Schatz, 1987; Lucattini et al., 2004).
The plant cell produces numerous proteins that are targeted and apparently functional in both mitochondria and plastids. Apparently, the endosymbiotic transfer of genetic information from progenitor mitochondrial and plastid forms to the nucleus has permitted the substitution and redirection of proteins originally synthesized in one organelle to the other, likely involving a cytosolic intermediate stage (Martin and Herrmann, 1998). Evidence includes a nucleus-encoded ribosomal protein of plastid origin now targeted to mitochondria (Adams et al., 2002) as well as a ribosomal protein of mitochondrial origin now targeted to the plastid (Gallois et al., 2001). In fact, several nucleus-encoded genes involved in organellar DNA and RNA metabolism (Elo et al., 2003) and gene expression (Peeters and Small, 2001; Lurin et al., 2004) appear to be shared by the mitochondrion and plastid (Elo et al., 2003). In the case of organellar DNA polymerase, biochemical evidence also supports its dual localization to mitochondria and plastids (Kimura et al., 2002; Sakai et al., 2004).
The dual targeting of nucleus-encoded proteins to both mitochondria and plastids appears to be accomplished in at least two ways. The first involves the incorporation of dual transcription or translation start sites conferring distinct targeting specificity to the encoded protein (reviewed in Small et al., 1998). Alternatively, one translation start site may exist, with the targeting presequence conferring both mitochondrial and plastid targeting capacity. Although these various dual-targeting strategies have generally been viewed as distinct, we postulate that they have emerged from a similar evolutionary process involving the acquisition of what are now discrete, functionally separable plastid and mitochondrial targeting domains.
Here, we present evidence that the recombinational acquisition of protein targeting information may be accompanied by a relaxation in translation initiation controls to permit influence by the upstream 5′ transcript leader sequence in some cases. Amino acids that facilitate dual targeting appear to be those most likely to appear in randomly derived sequences. Our findings suggest that the evolution of dual targeting in these particular cases may, in part, rely on the ability to initiate translation at a non-ATG start codon.
RESULTS
Sequence Conservation Exists within the Predicted Untranslated Leader Region of Duplicate Organellar Protein Genes
Study and alignment of two duplicate Arabidopsis thaliana genes, DNA Polymerase γ1 (POLγ1) and POLγ2, predicted to encode organellar γ-type DNA polymerase, revealed not only a high degree of sequence conservation (Elo et al., 2003) but unusual features located within the 5′ region of the genes. Translation is annotated to initiate at a Met located at corresponding locations within both genes, and the amino acid sequences immediately following the predicted initiator codon in both genes showed sequence divergence, consistent with their predicted function as targeting presequences. However, within the predicted untranslated leader region (UTR) sequence immediately adjacent to the initiator Met of each gene, a surprising level of nucleotide, amino acid, and reading frame conservation was apparent (Figure 1). The observed sequence conservation within a region not predicted to be translationally active was unexpected.
Figure 1.
UTR Sequence Alignment Data from Arabidopsis and Rice Organellar DNA Polymerase Genes.
(A) Alignment of putative UTR DNA and amino acid sequences from two Arabidopsis organellar DNA polymerase genes, POLγ1 and POLγ2. Identity is indicated by a vertical line between the sequences. Codons and their corresponding amino acids in the UTR of POLγ2 that were altered in this work are indicated above the sequence with numbers. Codons preceding the annotated +1 codon are indicated with negative numbers.
(B) Amino acid alignment of putative UTR sequences from the rice organellar genes OsPolγ1 and OsPolγ2. Annotated start codons are shaded in gray.
We reasoned two possible explanations for sequence alignment upstream of the translation start codon of two homologous, unlinked genes. One explanation is that the second gene copy arose by relatively recent gene duplication, so that sequence divergence within the UTR is not yet extensive. Alternatively, the predicted UTR sequence might be a functional part of the gene. We were able to identify within the rice (Oryza sativa) genome sequence database two genes that appeared to represent homologs of Arabidopsis POLγ1 and POLγ2. Both rice genes were also predicted to encode organellar DNA polymerase proteins. As in the case of Arabidopsis, the two rice genes shared a high degree of amino acid sequence similarity and likely arose by gene duplication. More importantly, the annotated UTR sequence of both rice genes, when compared as translated sequences, showed even more extensive predicted amino acid sequence alignment than was observed in Arabidopsis (Figure 1). Gaps in the alignment seemed to occur in nucleotide triplets, again supporting our assumption that this region is translated. These observations suggest that the sequences annotated as UTRs in both rice and Arabidopsis genes likely function as part of the genes, perhaps as amino acid coding sequence.
The Putative UTR Sequence Influences Protein Targeting
To investigate the function of the identified putative UTR sequences of POLγ1 and POLγ2 in Arabidopsis, we conducted organellar protein targeting experiments that used the green fluorescent protein (GFP) reporter gene in leaf particle bombardment assays analyzed by confocal laser scanning microscopy.
Gene sequences that encode only the predicted initiator Met and targeting presequence of POLγ1 (clone TP1) and POLγ2 (clone TP2) proteins, 100 and 113 amino acids in length, respectively, were fused to the GFP reporter and expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The results suggested that POLγ1 was dual targeted to mitochondria and plastids and POLγ2 was targeted to plastids only. These results were previously shown with particle bombardment (Elo et al., 2003), and similar results obtained by stable transformation are shown for POLγ2 in Figure 2A (POLγ1 data not shown). Chloroplast targeting conferred by the POLγ2 targeting presequence was most pronounced within the smaller plastids found in epidermal cells but was also evident in a green/yellow coloration of mature plastids within subepidermal cells.
Figure 2.
Organellar Protein Targeting Experiments Using a GFP Reporter Gene in Transformation Experiments and Analyzed by Confocal Laser Scanning Microscopy.
(A) Mature leaf cells from a stable transformant containing construct TP2, comprising the CaMV 35S promoter and a translational fusion of the POLγ2 targeting presequence with GFP. Magnification ×600.
(B) Fusions of the POLγ2 native promoter/UTR sequence with the POLγ2 targeting presequence and GFP are shown expressed in stably transformed mature (m) and young (y; 1-mm) leaves of Arabidopsis. Leaves shown are from a single plant. Magnification ×200.
(C) Enhanced magnification of the young leaf cells in (B). The arrow indicates mitochondria, seen as small punctuate green structures. Magnification ×600.
(D) Enhanced magnification of mature leaf cells in (B). The arrow indicates mitochondria. Magnification ×600.
When the CaMV 35S promoter was replaced by the native promoter and UTR sequences from the corresponding genes, the POLγ1 protein chimera retained its dual-targeting features (data not shown), but the POLγ2 protein, previously shown to be plastid targeting, now showed mitochondrial or dual targeting (Figures 2B to 2D). In young, meristematic tissue of 1-mm leaves, we observed clear evidence of dual targeting (Figures 2B and 2C), whereas in mature leaves of the same line, plastid targeting was not evident (Figures 2B and 2D). This observation suggests a lower level of gene expression in mature leaves, complicating the visualization of plastid targeting over background autofluorescence. Together, our observations suggested a functional influence of the sequences upstream of the predicted initiator codon in protein targeting, at least in the case of POLγ2.
Protein Targeting of POLγ2 Relies on Sequences Upstream of the Annotated ATG Start
To investigate the influence of sequences upstream of POLγ2 on protein targeting, we developed several constructs. The first construct, designated CP2, contained 294 nucleotides of the sequence immediately upstream of the annotated initiator ATG. Specifically, this construct was designed to contain the POLγ2 targeting presequence together with the putative native promoter/UTR sequence, fused to GFP and placed under the control of the CaMV 35S promoter. The 294-nucleotide stretch is predicted to contain no alternative upstream ATG codons (data not shown). This construct was in keeping with the truncated constructs described below with which it was compared.
To more carefully test the requirements for translation initiation of the POLγ2 sequence, directed mutations were introduced to the 294-nucleotide CP2 clone. The first three mutations, producing clones CP2*-27, CP2*-16, and CP2*-5, introduced stop codons at predicted in-frame amino acids 27, 16, and 5, respectively, upstream of the annotated initiator Met codon. Each stop codon change involved the substitution of a single nucleotide (Figure 3).
Figure 3.
Depiction of Gene Constructs Developed for the Analysis of POLγ2 Protein Targeting.
Gene constructs were developed with a GFP reporter gene (green), the POLγ2 targeting presequence nucleotides (blue) including one intron (bent line), and upstream leader sequence (red). Introduced stop codons are indicated by asterisks, the annotated translation initiation site and the next two codons are indicated by (MAM), and artificially introduced ATG codons are indicated by (M). The changes of the CTG codon at −7 to CTC and ATG are also indicated. Clones shown were developed in the pCAMBIA1302 vector. Cp designates chloroplast localization, and Mt designates mitochondrial localization.
These four constructs were used in Arabidopsis leaf particle bombardment experiments with protein localization assessed by confocal laser scanning microscopy. The results of these experiments are shown in Figure 4. We found that the 35S promoter driving the expression of the 294-nucleotide UTR, together with the predicted targeting presequence, directed dual targeting of the protein (Figure 4A, clone CP2). This result was consistent with that obtained with the native promoter (Figure 2) and implied that translation initiation occurs upstream of the annotated Met. Bombardment analysis of clones CP2*-27 and CP2*-16 also resulted in dual targeting of GFP (Figure 4B; CP2*-27 data not shown). However, clone CP2*-5 resulted in plastid localization of GFP (Figure 4C). These results indicate that translation initiation occurs within the interval of in-frame amino acids 5 to 15 upstream of the annotated Met to produce dual targeting. Interruption by a stop codon in this interval apparently results in translation initiation only at the annotated initiator Met, producing plastid targeting, as shown in Figure 2.
Figure 4.
Analysis of the POLγ2 UTR Sequence in Protein Targeting.
Particle bombardment results are shown for single leaf epidermal cells expressing the transgenic constructs CP2 ([A]; 294-nucleotide UTR), CP2*-16 ([B]; 294-nucleotide UTR with a stop codon 16 amino acids upstream of the initiator Met), CP2*-5 ([C]; 294-nucleotide UTR with a stop codon 5 amino acids upstream of the initiator Met), CP4M ([D]; 87-nucleotide segment of the UTR with an added Met), CPU4M ([E]; 87-nucleotide segment is shown with an added Met, no targeting presequence), CP*4M ([F]; 294-nucleotide UTR, substituting Leu for two Mets at the annotated translation start site), CP2L-7L ([G]; 294-nucleotide UTR, substituting CTC for the CTG at codon −7), and CP2L-7M ([H]; 294-nucleotide UTR, substituting ATG for the CTG at codon −7). To visualize unusually low GFP fluorescence levels in (G), the confocal microscope settings required significant adjustment, accounting for the spectral overlap in autofluorescing plastids in neighboring cells. A control for chloroplast targeting (ribulose-1,5-bisphosphate carboxylase/oxygenase [Rubisco] targeting peptide fused with GFP) and a control for mitochondrial targeting (F1-ATPase γ-subunit N terminus fused with GFP) are shown in (I). White arrows indicate mitochondria, and yellow arrows indicate plastids. Unaffected plastids in neighboring nontransformed cells autofluoresce red under the conditions used. All images are at magnification ×600.
The sequence upstream of the annotated initiator Met is as follows: −16 −15 −14 −13 −12 −11 −10 −9 −8 −7 −6 −5 −4 −3 −2 −1 +1 +2 +3, CGA CTC TGT CGT CAC TTC TCC TTC AAG CTG GGT AGA TTA AGT TCG CTT ATG GCC ATG, with the most probable plant non-AUG start codon underlined (Gordon et al., 1992). This CTG resides favorably relative to Kozak consensus predictions, with a purine (A) at position −3 and a G at position +4 (Kozak, 1986).
Translation of the Predicted UTR Confers Mitochondrial Targeting Capacity
To confirm that the upstream interval in question had the potential to confer dual targeting, three additional constructs were developed (Figure 3). The first, designated CP4M, truncated the upstream region up to 87 nucleotides (29 codons) immediately upstream of the annotated initiator ATG, making up approximately half of the UTR sequence and converting the ATA at codon −30 to an ATG. The addition of the ATG ensures translation of this interval. This construct, placed under the control of the CaMV 35S promoter, truncates the UTR to the interval that shows alignment between POLγ1 and POLγ2. The second construct, CPU4M, contained only the 87-nucleotide UTR sequence plus an artificially added ATG, fused to GFP under the control of the CaMV 35S promoter, but without the predicted targeting presequence located downstream of the annotated initiation site. The third construct, CP4*M, was identical to CP4M but contained a stop codon (TGA) introduced 15 nucleotides (5 codons) upstream of the annotated initiator ATG. This construct was again designed to assess the translational activity of the truncated UTR sequence and involved the identical nucleotide substitution as clone CP2*-5.
Protein targeting results from clone CP4M indicated dual targeting of GFP (Figure 4D), confirming that the dual-targeting capacity that we observed previously is conferred by translation of the sequence immediately upstream of the annotated initiator Met. Bombardment with clone CPU4M produced mitochondrial targeting of GFP, suggesting that the upstream UTR sequence encodes a mitochondrial targeting sequence (Figure 4E). No GFP expression was detected with clone CP4*M, based on four independent experiments of five leaves each (data not shown). This observation is consistent with the known negative effects of introducing short open reading frames upstream of coding regions (Hanfrey et al., 2003; Wiese et al., 2004), indicating that most or all translation initiates at the artificially introduced Met codon.
Upstream Translation Initiation Occurs but Requires the Presence of the Downstream ATG
Because all of our results suggested that the annotated Met codon might not serve as a primary site for the initiation of translation, we were interested to learn whether this Met codon was dispensable for translation of the gene. We postulated that the Met codon might participate in alternative translation initiation, whereby an upstream non-ATG initiator gives rise to a mitochondrial product, whereas initiation at the ATG results in a plastid product. We also wished to learn whether disruption of the ATG would affect the production of the upstream-derived mitochondrial product.
The annotated translation initiation site contains two Met (ATG) codons separated by a single Ala (GCC). We developed construct CP2-M1,3L by altering CP2 to substitute Leu (TTG) for the two Met (ATG) codons located at the annotated start of the gene (Figure 3). The results from this construct were intriguing. Bombardment with clone CP2-M1,3L resulted in extremely low-efficiency (<1%) GFP production, with only three expressing cells detected from four independent bombardment experiments of at least 12 leaves each, and GFP expression levels were markedly lower than normal. Within the three GFP-expressing cells, protein localization was mitochondrial (Figure 4F). This observation, confirmed with three independent clones, is important to our study in two respects. From these results we assume that dual targeting in POLγ2 arises by alternative translation initiation, so that amino acid substitution of the initiator Met eliminates plastid targeting and results in a mitochondrially targeted protein. Of particular interest, however, is the additional observation that the annotated initiator Met seems to be important for efficient translation overall. Our results suggest that the efficiency of upstream non-ATG translation for a mitochondrially targeted product was also affected by disruption of these downstream ATG codons. Whether this change in translation efficiency is attributable to altered secondary structure caused by the introduced mutations has not yet been determined. It is also possible that mRNA stability is reduced as a result of this change. When both the +1 and +3 ATG codons are eliminated, there are no other ATG codons in the first exon, and the first ATG codon in the second exon is out of frame, leading to a premature stop codon. Although upstream non-ATG initiation appears to be quite efficient, the elimination of these two in-frame ATG codons in exon 1 may lead to nonsense-mediated decay. It is known that in-frame nonsense codons are not required for nonsense-mediated decay (Alonso, 2005) and that leaky scanning resulting in initiation at a downstream out-of-frame ATG can also result in nonsense-mediated decay (Culbertson and Leeds, 2003).
Mutation of the −7 CTG Codon Suggests That Translation Initiates at That Site
With the introduction of stop codons, we were able to delimit at least one translation initiation site to the −5 to −15 codon interval upstream of the annotated ATG. To more directly test the CTG at the −7 codon position for translation initiation activity, we modified the CP2 clone to produce two additional gene constructs. The first, CP2L-7L, converted the CTG to CTC (both code for Leu), and the second, CP2L-7M, converted the CTG to ATG. Particle bombardment experiments with these two constructs suggested that the CTG at position −7 is the likely site of translation initiation. The CP2L-7L clone, which changes the CTG to CTC, a less suitable non-AUG initiation codon, showed only chloroplast targeting (Figure 4G). Eliminating the initiation of translation at the CTG at codon −7 results in the same targeting pattern as allowing initiation to occur at the annotated ATG by deleting the UTR. Changing the CTG codon at −7 to an ATG still produced dual targeting (Figure 4H), indicating that translation initiation likely occurs at this site, whether a CTG or an ATG is present.
The Upstream UTR Does Not Undergo Transcript Processing
One alternative explanation for the observed translation initiation at a non-ATG start codon is transcript splicing within the UTR. Such splicing could introduce an ATG within the transcript that is not detected by genomic sequence analysis. Sequences from three independent cDNA clones (see Methods) and RT-PCR amplification of the UTR interval confirmed that this region does not undergo transcript splicing, and the mRNA sequence was identical to the genomic sequence for the 294-bp interval preceding the annotated ATG (data not shown). The CTG-to-CTC result also rules out the hypothesis that an alternative ATG codon is spliced to the message, because this change would have no effect on translation initiating at an upstream ATG.
Similar UTR Influence on Protein Targeting Is Evident in Other Genes
With a dual-domain, dual-targeting presequence in POLγ2, and the apparent similarity in sequences upstream of the annotated ATG start codons for the rice homologs (Figure 1), we surmise that one means of protein dual-targeting evolution might be the translational incorporation of sequences upstream of an original translation initiation site. If this is the case, additional genes might exist encoding organellar proteins that display in-frame sequences upstream of the annotated start codon. These sequences, when artificially appended to the open reading frame, would be expected to alter protein targeting features. To date, aside from the genes mentioned in rice (Figure 1) and an organellar RNA polymerase (Hedtke et al., 2002; Kobayashi et al., 2002), we have identified additional candidates likely to initiate at non-ATG start codons to confer protein targeting specificity. Three of these genes are listed in Table 1.
Table 1.
Plant Loci Identified in Silico as Likely Candidates for Protein Targeting Influenced by Sequences Upstream of the Annotated Met
| Predotar
|
TargetP
|
iPSORT
|
|||||
|---|---|---|---|---|---|---|---|
| Gene Identifier | Annotation | +UTR | −UTR | +UTR | −UTR | +UTR | −UTR |
| At3g20540 | γ-Like DNA polymerase POLγ1 | 0.953 (M) | 0.776 (M) | 0.830 (M) | 0.741 (M) | Mitochondrial | Mitochondrial |
| 0.028 (P) | 0.640 (P) | 0.165 (P) | 0.588 (P) | ||||
| At1g50840 | γ-Like DNA polymerase POLγ2 | 0.962 (M) | 0.005 (M) | 0.298 (M) | 0.314 (M) | Plastid | Plastid |
| 0.048 (P) | 0.957 (P) | 0.466 (P) | 0.928 (P) | ||||
| Ap005164 copy I | Putative DNA polymerase in rice | 0.993 (M) | 0.180 (M) | 0.574 (M) | 0.738 (M) | Mitochondrial | Plastid |
| 0.018 (P) | 0.922 (P) | 0.105 (P) | 0.441 (P) | ||||
| Ap005164 copy II | Putative DNA polymerase in rice | 0.991 (M) | 0.366 (M) | 0.677 (M) | 0.425 (M) | Mitochondrial | Plastid |
| 0.034 (P) | 0.682 (P) | 0.168 (P) | 0.760 (P) | ||||
| At3g23780 | DNA-directed RNA polymerase subunit | 0.105 (M) | 0.920 (M) | 0.424 (M) | 0.410 (M) | Plastid | Mitochondrial |
| 0.892 (P) | 0.000 (P) | 0.100 (P) | 0.393 (P) | ||||
| At3g10270 | Putative DNA gyrase subunit B | 0.990 (M) | 0.000 (M) | 0.042 (M) | 0.115 (M) | Mitochondrial | Neither |
| 0.001 (P) | 0.002 (P) | 0.644 (P) | 0.197 (P) | ||||
| At3g51690 | DNA helicase homolog PIF1 | 0.702 (M) | 0.002 (M) | 0.042 (M) | 0.202 (M) | Mitochondrial | Plastid |
| 0.026 (P) | 0.040 (P) | 0.644 (P) | 0.405 (P) | ||||
| At3g24320 | MutS homolog 1 AtMsh1 | 0.767 (M) | 0.943 (M) | 0.512 (M) | 0.856 (M) | Mito | Mito |
| 0.367 (P) | 0.222 (P) | 0.102 (P) | 0.215 (P) | ||||
M, mitochondria; P, plastid. Discrimination values in boldface are those we considered significant (P > 0.35) in our analysis.
In the case of Arabidopsis MutS Homolog 1 (MSH1) (previously designated CHM), we tested whether the gene likely initiates translation from a codon upstream of the annotated ATG by developing a gene construct that incorporates only the predicted targeting presequence starting with the annotated ATG as well as a gene construct that includes additional upstream sequence. GFP was used as the reporter gene; the constructs were under the control of the CaMV 35S promoter, and particle bombardment was used for transgene delivery. Results of these experiments are shown in Figure 5.
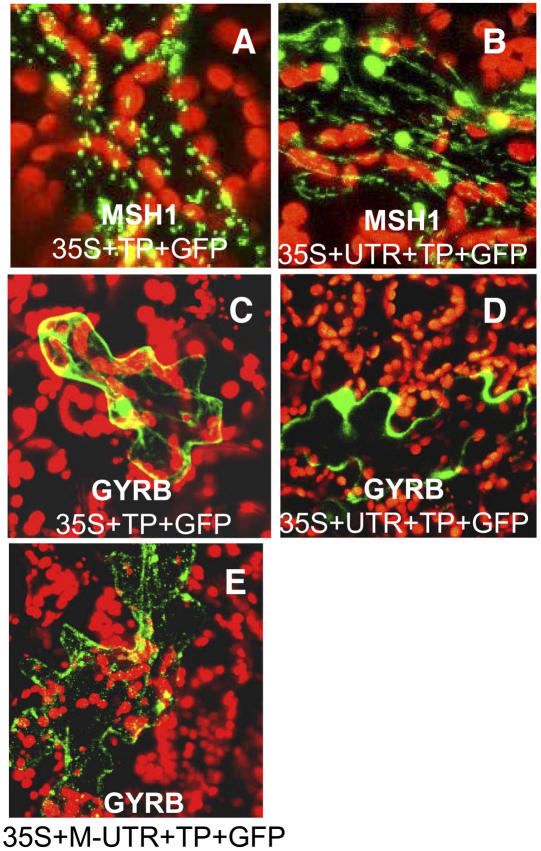
Figure 5.
Influence of the UTR on GFP Localizations for the MSH1 and DNA Gyrase B (GYRB; At3g10270) Arabidopsis Genes.
Translational fusions were made to GFP of the Arabidopsis MSH1 (At3g24320) targeting presequence (A) and the targeting presequence plus the UTR (B), followed by particle bombardment and analysis by confocal laser scanning microscopy. No artificial Met was added. Similar experiments conducted with DNA Gyrase B (GYRB) included the targeting presequence (C), the targeting presequence plus the UTR (D), and the targeting presequence plus the UTR with added ATG at the promoter–UTR junction (E). All images are at magnification ×600.
AtMSH1 translation initiation at the annotated ATG resulted in predominantly mitochondrial targeting of the protein, although a few plastids were evident in some experiments (Figure 5A). Inclusion of the predicted UTR sequence resulted in dual targeting of the protein (Figure 5B). These results suggest that the identified upstream region likely influences targeting features of this protein. Mutation of the MSH1 gene results in both a mitochondrial and a plastid phenotype (Redei, 1973; Abdelnoor et al., 2003).
In the case of accession At3g10270, encoding a DNA gyrase subunit B, protein targeting predictions suggest that translation initiation of this gene at the annotated start codon provides no targeting specificity (Table 1). This was confirmed by GFP targeting experiments (Figure 5C). Interestingly, addition of upstream UTR sequence, together with an artificially introduced ATG, resulted in mitochondrial targeting, with some level of plastid targeting evident (Figure 5E). Without the addition of the ATG at the 5′ end of the UTR, cytoplasmic localization resulted (Figure 5D). Similar experiments conducted with the DNA helicase gene (At3g51690) resulted in identical results to those of gyrase B (data not shown). We interpret this outcome as an indication that upstream sequences may be important to protein localization. However, in the case of gyrase B and helicase, we likely included an upstream region that was too limited, providing inadequate leader sequence for proper initiation and necessitating the introduction of an artificial start codon in both cases (Van Etten and Janssen, 1998). Our observations may also relate to those reported by Ambard-Bretteville et al. (2003), in which the omission or substitution of even one or two codons at the N terminus of a targeting presequence could render an organelle-targeted protein cytoplasmic.
DISCUSSION
We identified a number of nuclear genes encoding organellar proteins with at least some portion of the protein targeting information lying within the region upstream of the annotated start codon. Although others have identified a chloroplast-targeted RNA polymerase initiated at a non-AUG codon (Hedtke et al., 2002; Kobayashi et al., 2002), we have shown non-AUG initiation of a dual-targeted protein and suggest that this phenomenon may regulate the localization of a number of dual-targeted proteins. Genome sequences are annotated mechanically to indicate only open reading frames beginning with an AUG; however, it is clear from our data that information relevant to targeting is located upstream of the AUG and is translated. An intriguing possible explanation for the observations reported here is that the endosymbiotic events encompassing two subcellular organelles, the mitochondrion and the plastid, were accompanied by nuclear genetic phenomena that facilitated protein targeting. One of these strategies perhaps involved the translational incorporation of in-frame sequence residing upstream of the initiation codon, permitting the expansion of protein targeting capacity.
This hypothesis relies on several premises. If protein targeting capacity can be influenced by the addition of presumably random UTR sequences at the N terminus of the protein, one must assume that codon usage favors those amino acids known to be important to protein targeting properties. In fact, the amino acids Ser (six codons), Arg (six codons), Leu (six codons), and Ala (four codons) are among the amino acids with highest frequency in random sequences and represent amino acids shown to be important to protein targeting (Peeters and Small, 2001). Those amino acids found in lowest frequencies in plant targeting presequences, Asp (two codons) and Glu (two codons), correspond to low-frequency amino acids encoded by random sequence.
Computer-based randomization of 1000 sequences, each 40 amino acids in length, and their subsequent analysis (Predotar program) for predicted organellar targeting capacity indicated that 43.7% of these random sequences would likely target to organelles (24.5% to mitochondria, 18.6% to plastids, and 0.6% dual; data not shown). This observation appears consistent with the report that incorporation of random 20- to 70-bp Escherichia coli DNA sequences, or sequences from a eukaryotic gene, identifies 2.7 or 5.0% of the sequences, respectively, that are capable of directing protein transport to mitochondria in yeast (Baker and Schatz, 1987). These results are also consistent with the suggestion that N-terminal sequences of many bacterial genes already appear to have protein targeting potential (Lucattini et al., 2004).
The translation of upstream UTR sequence would also likely necessitate a relaxation of translation initiation controls in the system. Hashimoto et al. (2002) have shown in yeast that a single amino acid substitution in the β-subunit of eukaryotic translation initiation factor 2 can result in enhanced translation initiation at non-ATG codons. Moreover, the identification of internal ribosome entry sites within eukaryotic UTRs has shown several of these to involve CUG rather than AUG sites (Komar and Hatzoglou, 2005). In yeast, translation of the mitochondrial and cytoplasmic glycyl-tRNA synthetase originates from a single locus. Whereas the cytoplasmic form of the protein initiates at the annotated AUG, the mitochondrial form, encoding a 23-amino acid N-terminal extension, initiates at an upstream, in-frame UUG (Chang and Wang, 2004). A very similar situation also exists for the yeast alanyl-tRNA synthetase, using upstream ACG as a translation initiation codon for the mitochondrial form (Tang et al., 2004). These observations appear to further support our assumption that the relaxation of certain translation initiation controls occurred to expand the targeting capacity of proteins within the eukaryotic cell.
In some genes, alternative translation initiation sites appear to exist, with only one predominating in activity (reviewed in Silva-Filho, 2003). The zinc metalloprotease that degrades signal peptides in Arabidopsis contains two putative initiator Met codons, but only one appears functional (Bhushan et al., 2003). Translation initiation at the first Met results in a dual-targeted protein, whereas artificial truncation to the second Met results in a plastid-targeted protein. In this case, a dual-domain, dual-targeting presequence functions with an initiator Met at the start of translation, although the second Met might have once functioned as the initiator codon for a protein targeted exclusively to plastids.
Similarly, the RNA polymerase gene of the moss Physcomitrella patens contains two in-frame Met codons, with translation initiation at the upstream site conferring dual targeting and initiation at the second site conferring mitochondrial targeting. When one research group assayed targeting by including the coding sequence beginning with the upstream Met but excluding untranslated 5′ leader sequence, targeting was dual (Richter et al., 2002). When a second group tested the same gene but incorporated the 5′ untranslated leader into their reporter gene constructs, targeting was mitochondrial, and intriguingly, initiation occurred at the second Met only (Kabeya and Sato, 2005). These results suggest that gene context plays a role in the selection of the translation initiation site and, in this case, that the protein localizes exclusively to mitochondria. If this is the case, however, certain details remain unclear. If only the second Met functions in translation initiation, why would the two Met codons be conserved not only in this gene but in the corresponding locus of Arabidopsis (Hedtke et al., 2002)? The THI1 gene of Arabidopsis also relies on two different in-frame AUG codons to direct the protein to chloroplasts and mitochondria. In this case, the second AUG predominates, suggesting that leaky scanning, reinitiation, or internal entry of ribosomes may be required to achieve mitochondrial targeting of this protein (Chabregas et al., 2002).
Inclusion of the entire 294-nucleotide upstream region of the POLγ2 gene, together with the annotated targeting presequence, resulted in dual targeting of the reporter protein. Inclusion of only the targeting presequence, initiating at the annotated Met, resulted in plastid targeting. From these results, we assume that at least one additional non-ATG translation initiation site exists within the 294-nucleotide sequence. UTR sequence alignment between the POLγ1 and POLγ2 genes and their homologs in rice suggests that translation might initiate ∼87 nucleotides (29 codons) upstream of the annotated start site. However, introduction of stop codons in this region appeared to indicate translation initiation much closer to the annotated initiator Met, at least six codons upstream.
Forcing initiation at the −30 codon position resulted in dual targeting of the protein under our experimental conditions. Within this upstream region, introduced stop codons suggested initiation between codons −5 and −15, and changing the −7 CTG codon to CTC eliminated the mitochondrial component of the protein localization, presumably because the only remaining initiation site is the annotated ATG. These observations suggest that the −7 codon is a major site of translation initiation, resulting in dual targeting. Interestingly, conversion of the −7 CTG codon to an ATG still showed dual targeting. There are two possible interpretations for this result. One possibility is that initiation at codon −7 results in mitochondrial localization, initiation at codons +1 or +3 results in chloroplast localization, and the observed dual targeting is attributable to initiation at both sites, even when the upstream site is an ATG. The other possibility is that initiation at the −7 codon results in dual targeting, and the efficiency of initiation at this position determines whether dual targeting or chloroplast targeting will occur. That an ATG gives the same result as a CTG suggests that the non-ATG initiation of this gene is remarkably efficient, perhaps because of the sequence context of the UTR. The influence of upstream, untranslated leader sequence on the selection of an ATG versus a non-ATG translation initiation site has been postulated in other systems as well (Botto et al., 1997). Whether additional non-ATG sites for translation initiation exist within the upstream region, thus accounting for the upstream sequence conservation between POLγ1 and POLγ2, remains to be determined. This upstream sequence could instead enhance the efficiency of translation of the CTG and could explain why there has apparently been no selection for an ATG at this site.
Some evidence suggests that the relative efficiency of the +1 Met site versus the upstream non-ATG site for translation initiation, observed as plastid versus mitochondrial products, is influenced by trans-acting cellular or developmental cues. Stable transformants containing GFP reporter constructs with the entire native promoter and UTR from POLγ2 showed dual targeting in young tissues but largely mitochondrial targeting in mature tissues. This transition could be in response to reduced plastid demands for DNA replication at these stages (Oldenburg and Bendich, 2004). However, this result suggests that the non-ATG site for translation initiation becomes predominant in mature tissues of the plant.
Artificial addition of an ATG at the promoter–UTR junction was found to be required for organellar targeting of DNA gyrase and helicase-GFP fusions. These results appear to further support the importance of the local sequence environment on translation initiation, and it is likely that we included insufficient upstream sequence to permit proper translation initiation. Alternatively, one must assume that the DNA gyrase and helicase loci used in this study encode cytoplasmic proteins. Given the prokaryotic origins of DNA gyrases as bacterial DNA topoisomerases (Maxwell, 1999) and their physiological functions, this seems highly unlikely.
A recent report described the organellar targeting properties of the DNA gyrase encoded by accession At3g10270 (Wall et al., 2004). This study reported plastid targeting of the gene, whereas we demonstrate mitochondrial or dual targeting and the necessity of upstream UTR sequences for this targeting capacity. The discrepancies between these two studies appear to be because the previous authors were working with At5g04130 rather than At3g10270. This misidentification in the Wall et al. (2004) study is likely the result of sequence similarities that exist between duplicate organellar DNA gyrase genes present in the Arabidopsis genome. Although At3g10270 requires UTR sequence for targeting, At5g04130 (misidentified as At3g10270 in the previous study) contains alternative translation start sites.
The in vivo assay used in our studies offers important advantages for the investigation of protein targeting determinants, but limitations of the system should be mentioned. Given the clear influence of gene context and upstream sequences on translation initiation, this effect can confound experimental design. In this study, most clones were developed using the pCAMBIA1302 (http://www.cambia.org) vector that includes a multiple cloning site between the CaMV 35S promoter and GFP. This cloning site includes an NcoI restriction site designed to artificially create an ATG at the 5′ end of any clone inserted. This was taken into account in our experiments, requiring that most analyses use the full 294-nucleotide upstream region rather than truncations. When identical clones were developed in the pK7FWG2 destination vector (Plant Systems Biology, Ghent University) designed for protein targeting studies, surprising differences in protein targeting were observed. The pK7FWG2 vector includes additional intervening sequence between the CaMV 35S promoter and the 5′ end of the inserted clone, including part of the ω leader from Tobacco mosaic virus (Karimi et al., 2005), which is known to affect translational efficiency (Gallie, 2002), and this sequence appeared to alter targeting results in all cases in which a truncated leader was used rather than the 294-nucleotide leader sequence (data not shown).
In Figure 1, we show the unusual features within the putative UTR of duplicate Arabidopsis organellar DNA polymerase genes that suggested that the UTR sequence was translated. This figure also shows similar features in the corresponding rice genes. These rice loci were previously described by another group, with somewhat different interpretations of gene structure and predicted protein targeting (Kimura et al., 2002). The apparent contradiction in these two reports is accounted for by advancements in sequence quality and refinement in splice site annotation within the public database that followed the original study.
Within an evolutionary context, the data presented here and in previous detailed studies of dual targeting by others suggest that protein targeting capacity may have evolved in stages. This interpretation appears consistent with the frequent observation of functionally distinct mitochondrial and plastid targeting domains within dual-targeting presequences. It was originally thought that when two targeting signals are arranged in tandem, the most N-terminal sequence determines the final localization (Silva-Filho et al., 1996). However, this does not appear to be the case in naturally occurring dual-targeting configurations (Rudhe et al., 2002). Targeting presequence acquisition by nuclear genes may have involved a combination of intergenic recombination together with limited relaxation of translation initiation constraints. This process likely led to the misannotation of some dual-targeted protein coding genes by focusing on predicted features at the most prominent Met initiator codon and omitting essential upstream sequences that may have been secondarily acquired. The importance of upstream regions of a gene to translational regulation and response to cellular change has become increasingly evident (Komar and Hatzoglou, 2005). The possibility of this type of regulation in nuclear–organellar coordination presents intriguing opportunities for further study.
METHODS
Development of Gene Constructs
Preparation of gene constructs containing the targeting presequence for both POLγ1 and POLγ2, in association with the CaMV 35S promoter, was performed as described by Elo et al. (2003). The corresponding native promoter constructs were prepared by amplifying 330 bp of upstream sequence for POLγ1 and 1022 bp of upstream sequence for POLγ2 together with the entire predicted targeting presequence of each gene. PCR amplifications used forward primer 5′-CCCTGCAGGAGAGTTTTCGTGTTCCCCAT-3′ and reverse primer 5′-GTTCCGCCAACTGTGAAACAAGTCATGACC-3′ for POLγ1 and forward primer 5′-CCGTCGACATCACAGAGACGGAGAAACC-3′ and reverse primer 5′-GGTCATGACTACCTCCGTCTGATTTCCAAC-3′ for POLγ2.
Constructs involving truncated versions of the POLγ2 native promoter were prepared using the following PCR primer combinations: CP2 (−294bp+TP), forward primer 5′-CCTCATGATTTGACAGAGCAATGCAATCT-3′ and reverse primer 5′-GGTCATGACTACCTCCGTCTGATTTCCAAC-3′; CP4M (−87bp+TP), forward primer 5′-CCTCATGACGACAAAATTCGCCACCATA-3′ and reverse primer 5′-GGTCATGACTACCTCCGTCTGATTTCCAAC-3′; CPU4M (−87bp-TP), forward primer 5′-CCCCATGGCGACAAAATTCGCCACCATA-3′ and reverse primer 5′-CCCCATGGCCATAAGCGAACTT-3′.
PCR products were ligated to the pGEM cloning vector (Promega) for DNA sequence confirmation and then transferred to vector pCAMBIA1302 or modified pCAMBIA1302 (minus the CaMV 35S promoter) (http://www.cambia.org).
Introduction of a stop codon to clone CP4M to create CP4*M was performed using a one-step overlap extension PCR method described by Urban et al. (1997). The modification of clone CP4M to create CP4*M was performed by introducing an A-to-T substitution 15 nucleotides upstream of the ATG.
Constructs of the mutant derivatives of CP2 were made using overlapping PCR as described by Urban et al. (1997). The primers CP2-F1 (5′-TCGGTACCCGGGGATCCTCTAGAGT-3′) and CP2-R1 (5′-GTCAGATCTACCATGACTACCTCCGTCTG-3′) were used in each case to amplify the region from upstream of the PstI site in pCAMBIA1302 to downstream of the BglII site, which encompassed the entire Polγ2 insert. Mutants of this region were constructed using overlapping PCR. In each case, the region between CP2-F1 and the mutagenic reverse primer, and the region between CP2-R1 and the mutagenic forward primer, were amplified. The amplification products were gel-purified, mixed together, and used as template for reamplification with CP2-F1 and CP2-R1 only. The products of this reaction were gel-purified, digested with PstI and BglII, and ligated to PstI-BglII–digested pCAMBIA1302. DNA sequences for all clones were verified. The mutagenic primers were as follows, with the nucleotide change noted in boldface. To change the Lys at codon position −27 to a stop, we used K-27* forward (5′-GGAACGATAACGACATAATTCGCCACC-3′) and K-27* reverse (5′-GGTGGCGAATTATGTCGTTATCGTTCC-3′). To change the Arg at codon position −16 to a stop, we used R-16* forward (5′-CCTATTTTTAATCTCTGACTCTGTCGTC-3′) and R-16* reverse (5′-GACGACAGAGTCAGAGATTAAAAATAGG-3′). To change the Arg at codon position −5 to a stop, we used R-5* forward (5′-CTTCAAGCTGGGTTGATTAAGTTCGC-3′) and R-5* reverse (5′-GCGAACTTAATCAACCCAGCTTGAAG-3′). To change the ATG codons at positions +1 and +3 to Leu codons, we used M1LM3L forward (5′-GATTAAGTTCGCTTTTGGCCTTGGGGGTTTC-3′) and M1LM3L reverse (5′-GAAACCCCCAAGGCCAAAAGCGAACTTAATC-3′). To change the CTG Leu codon at position −7 to a CTC codon, we used L-7L forward (5′-CGTCACTTCTCCTTCAAGCTCGGTAGATTAAGTTCGC-3′) and L-7L reverse (5′-GCGAACTTAATCTACCGAGCTTGAAGGAGAAGTGACG-3′). To change the CTG Leu codon at position −7 to an ATG codon, we used L-7M forward (5′-CGTCACTTCTCCTTCAAGATGGGTAGATTAAGTTCGC-3′) and L-7M reverse (5′-GCGAACTTAATCTACCCATCTTGAAGGAGAAGTGACG-3′).
Constructs for accessions At3g24323 (AtMSH1) and At3g10270 (DNA gyrase subunit B) were created using the same methods described above with the following primers. For AtMSH1 UTR and targeting presequence, forward primer 5′-GGCCATGGTGTGAATTGCATAGTGGTCG-3′ and reverse primer 5′-GGCCATGGAAACATCACTTGACGTCTTC-3′; for AtMSH1 annotated start ATG and targeting presequence, forward primer 5′-CACCATGCATTGGATTGCTACCAG-3′ and reverse primer 5′-AGTGAGAACATCACTTGACGT-3′; for Gyrase UTR+M, forward primer 5′-CACCATGCCATTATTCACATTTGGTTTCAGG-3′; for Gyrase UTR, forward primer 5′-CACCCCATTATTCACATTTGGTTTCAGG-3′; for Gyrase annotated start ATG, forward primer 5′-CACCATGGAGTCTCTCCAAGAGAGCTCT-3′; the reverse primer used in all gyrase constructs was 5′-TGAAGCAAAACCAGCTTGGGCCT-3′.
Control constructs for mitochondrial (targeting presequence of the F0F1 ATPase γ-subunit gene AtpG) and plastid (transit peptide sequence of the Rubisco small subunit gene RbcS) targeting were graciously provided by D. Stern and L. Allison (Beardslee et al., 2002).
Transient and Stable Transformations
In separate experiments, 7 μg of DNA from individual constructs was delivered into 4-week-old leaves of Arabidopsis thaliana (ecotype Columbia) using tungsten particles and the Biolistic PDS-1000/He system (Bio-Rad). Particles were bombarded into Arabidopsis leaves using 900 p.s.i. rupture discs under a vacuum of 26 inches of Hg. After bombardment, Arabidopsis leaves were allowed to recover for 18 to 22 h on Murashige and Skoog medium plates at 22°C in 16 h of light before assaying GFP expression. Stable transformants were produced using the floral dip method (Clough and Bent, 1998).
GFP Expression Assay
Localization of GFP expression was conducted by laser scanning confocal microscopy using 488- and 633-nm excitation and two-channel measurement of emission, 522 nm (green/GFP) and 680 nm (red/chlorophyll).
Protein Gel Blot Analysis
Plant extracts were prepared by grinding in liquid nitrogen and adding the powder to 1× SDS sample buffer without dyes (62.5 mM Tris, pH 6.8, 2% SDS, 5% 2-mercaptoethanol, and 10% glycerol); 0.5 mL of this was added per gram of wet weight. The sample was boiled for 15 min and centrifuged, and the supernatant was subjected to gel electrophoresis. The gel was 12.5% polyacrylamide, run at 120 V for 15 min, then at 140 V until the 29-kD marker was near the bottom. Protein transfer to membrane was in Tris-Gly-15% methanol to polyvinylidene difluoride membranes. Blocking was in Tris-buffered saline, 4% milk, and 0.05% Tween 20 at 4°C overnight. Primary antibody was a mixture of two mouse monoclonal anti-GFP antibodies (clones 7.1 and 13.1; Roche Diagnostics). The secondary antibody was goat anti-mouse (IgG + IgM) peroxidase-conjugated antibody. Detection was with the Pierce SuperSignal West Pico Chemiluminescent substrate.
Bioinformatics Analysis
The search for N-terminal and amino acid sequence similarities was performed with BLAST (Altschul et al., 1990, 1997) through GenBank and MatDB (http://mips.gsf.de/proj/thal/db/index.html) (Schoof et al., 2002). Rice (Oryza sativa) sequence information was obtained by BLASTX analysis of The Institute for Genomic Research rice genome database. Presequence/transit peptide predictions were performed using Predotar (http://www.inra.fr/predotar/), TargetP (http://www.cbsdtu.dk/services/TargetP/), PSORT (http://psort.nibb.ac.jp/form.html), and iPSORT (http://hypothesiscreator.net/iPSORT/).
A computer program was developed to generate random sequences 40 amino acids in length. Care was taken to conserve the relative amino acid frequencies within the sequences according to the genetic code without consideration of Arabidopsis codon usage bias. The output of the program was directed to the faceless version of Predotar (http://www.inra.fr/predotar/) for targeting predictions.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At3g20540 (POLγ1) and At1g50840 (POLγ2). The three independent POLγ2 cDNA sequences used in this study were accessions AY091072, AF46286, and AV831550.
Acknowledgments
We thank the University of Nebraska Core Facility for Microscopy for technical assistance in GFP imaging. We gratefully acknowledge Chris Wittgren for technical assistance with construct development. This work was funded by grants to S.A.M. from the National Science Foundation and the Department of Energy. This research was also supported in part by funds provided through the Hatch Act. This is a contribution of the University of Nebraska Agricultural Research Division, Journal Series number 14503.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sally A. Mackenzie (smackenzie2@unl.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035287.
References
- Abdelnoor, R.V., Yule, R., Elo, A., Christensen, A., Meyer-Gauen, G., and Mackenzie, S. (2003). Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 100, 5968–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L., Daley, D.O., Whelan, J., and Palmer, J.D. (2002). Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell 14, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, C.R. (2005). Nonsense-mediated RNA decay: A molecular system micromanaging individual gene activities and suppressing genomic noise. Bioessays 27, 463–466. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambard-Bretteville, F., Small, I., Grandjean, O., Colas des Francs-Small, C. (2003). Discrete mutations in the presequence of potato formate dehydrogenase inhibit the in vivo targeting of GFP fusions into mitochondria. Biochem. Biophys. Res. Commun. 311, 966–971. [DOI] [PubMed] [Google Scholar]
- Baker, A., and Schatz, G. (1987). Sequences from a prokaryotic genome or the mouse dihydrofolate reductase gene can restore the import of a truncated precursor protein into yeast mitochondria. Proc. Natl. Acad. Sci. USA 84, 3117–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardslee, T.A., Roy-Chowdhury, S., Jaiswal, P., Buhot, L., Lerbs-Mache, S., Stern, D.B., and Allison, L.A. (2002). A nucleus-encoded maize protein with sigma factor activity accumulates in mitochondria and chloroplasts. Plant J. 31, 199–209. [DOI] [PubMed] [Google Scholar]
- Bhushan, S., Lefebvre, B., Stahl, A., Wright, S.J., Bruce, B.D., Boutry, M., and Glaser, E. (2003). Dual targeting and function of a protease in mitochondria and chloroplasts. EMBO Rep. 4, 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto, J.M., Vincent, J.P., and Mazella, J. (1997). Existence of two translation initiation sites leading to the expression of two proteins from the rat high-affinity neurotensin-receptor cDNA: Possible regulation by the 5′ end non-coding region. Biochem. J. 324, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennicke, A., Grohmann, L., Hiesel, R., Knoop, V., and Schuster, W. (1993). The mitochondrial genome on its way to the nucleus: Different stages of gene transfer in higher plants. FEBS Lett. 325, 140–145. [DOI] [PubMed] [Google Scholar]
- Chabregas, S.M., Luche, D.D., Van Sluys, M.-A., Menck, C.F.M., and Silva-Filho, M.C. (2002). Differential usage of two in-frame translational start codons regulates subcellular localization of Arabidopsis thaliana THI1. J. Cell Sci. 116, 285–291. [DOI] [PubMed] [Google Scholar]
- Chang, K.-J., and Wang, C.-C. (2004). Translation initiation from a naturally occurring non-AUG codon in Saccharomyces cerevisiae. J. Biol. Chem. 279, 13778–13785. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Culbertson, M.R., and Leeds, P.F. (2003). Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13, 207–214. [DOI] [PubMed] [Google Scholar]
- Elo, A., Lyznik, A., Gonzalez, D.O., Kachman, S.D., and Mackenzie, S. (2003). Nuclear genes encoding mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 15, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie, D.R. (2002). The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 30, 3401–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois, J.-L., Achard, P., Green, G., and Mache, R. (2001). The Arabidopsis chloroplast ribosomal protein L21 is encoded by a nuclear gene of mitochondrial origin. Gene 274, 179–185. [DOI] [PubMed] [Google Scholar]
- Gordon, K., Futterer, J., and Hohn, T.E. (1992). Efficient initiation of translation at non-AUG triplets in plant cells. Plant J. 2, 809–813. [PubMed] [Google Scholar]
- Hanfrey, C., Franceschetti, M., Mayer, M.J., Illingworth, C., Elliott, K., Collier, M., Thompson, B., Perry, B., and Michael, A.J. (2003). Translational regulation of the plant S-adenosylmethionine decarboxylase. Biochem. Soc. Trans. 31, 424–427. [DOI] [PubMed] [Google Scholar]
- Hashimoto, N.N., Carnevalli, L.S., and Castilho, B.A. (2002). Translation initiation at non-AUG codons mediated by weakened association of eukaryotic initiation factor (eIF) 2 subunits. Biochem. J. 15, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke, B., Legen, J., Weihe, A., Herrmann, R.G., and Borner, T. (2002). Six active phage-type RNA polymerase genes in Nicotiana tabacum. Plant J. 30, 625–637. [DOI] [PubMed] [Google Scholar]
- Kabeya, Y., and Sato, N. (2005). Unique translation initiation at the second AUG codon determines mitochondrial localization of the phage-type RNA polymerases in the moss Physcomitrella patens. Plant Physiol. 138, 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, K., Kubo, N., Ozawa, K., and Hirai, A. (1996). Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J. 15, 6652–6661. [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., De Meyer, B., and Hilson, P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10, 103–105. [DOI] [PubMed] [Google Scholar]
- Kimura, S., et al. (2002). A novel DNA polymerase homologous to Escherichia coli DNA polymerase I from a higher plant, rice (Oryza sativa L.). Nucleic Acids Res. 30, 1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Dokiya, Y., Kumazawa, Y., and Sugita, M. (2002). Non-AUG translation initiation of mRNA encoding plastid-targeted phage-type RNA polymerase in Nicotiana sylvestris. Biochem. Biophys. Res. Commun. 299, 57–61. [DOI] [PubMed] [Google Scholar]
- Komar, A.A., and Hatzoglou, M. (2005). Internal ribosome entry sites in cellular mRNAs: The mystery of their existence. J. Biol. Chem. 280, 23425–23428. [DOI] [PubMed] [Google Scholar]
- Kozak, M. (1986). Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Lucattini, R., Likic, V.A., and Lithgow, T. (2004). Bacterial proteins predisposed for targeting to mitochondria. Mol. Biol. Evol. 21, 652–658. [DOI] [PubMed] [Google Scholar]
- Lurin, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., and Herrmann, R.G. (1998). Gene transfer from organelles to the nucleus: How much, what happens and why? Plant Physiol. 118, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, A. (1999). DNA gyrase as a drug target. Biochem. Soc. Trans. 27, 48–53. [DOI] [PubMed] [Google Scholar]
- Oldenburg, D.J., and Bendich, A.J. (2004). Changes in the structure of DNA molecules and the amount of DNA per plastid during chloroplast development in maize. J. Mol. Biol. 344, 1311–1330. [DOI] [PubMed] [Google Scholar]
- Peeters, N., and Small, I. (2001). Dual targeting to mitochondria and chloroplasts. Biochim. Biophys. Acta 1541, 54–63. [DOI] [PubMed] [Google Scholar]
- Redei, G.P. (1973). Extra-chromosomal mutability determined by a nuclear gene locus in Arabidopsis. Mutat. Res. 18, 149–162. [Google Scholar]
- Richter, U., Kiessling, J., Hedtke, B., Decker, E., Reski, R., Borner, T., and Weihe, A. (2002). Two RpoT genes of Physcomitrella patens encode phage-type RNA polymerases with dual targeting to mitochondria and plastids. Gene 290, 95–105. [DOI] [PubMed] [Google Scholar]
- Rudhe, C., Clifton, R., Whelan, J., and Glaser, E. (2002). N-terminal domain of the dual-targeted pea glutathione reductase signal peptide controls organellar targeting efficiency. J. Mol. Biol. 324, 577–585. [DOI] [PubMed] [Google Scholar]
- Sakai, A., Takano, H., and Kuroiwa, T. (2004). Organelle nuclei in higher plants: Structure, composition, function, and evolution. Int. Rev. Cytol. 238, 59–118. [DOI] [PubMed] [Google Scholar]
- Schoof, H., Zaccaria, P., Gundlach, H., Lemcke, K., Rudd, S., Kolesov, G., Arnold, R., Mewes, H.W., and Mayer, K.F. (2002). MIPS Arabidopsis thaliana Database (MAtDB): An integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res. 30, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Filho, M.C. (2003). One ticket for multiple destinations: Dual targeting of proteins to distinct subcellular locations. Curr. Opin. Plant Biol. 6, 589–595. [DOI] [PubMed] [Google Scholar]
- Silva-Filho, M.C., Chaumont, F., Leterme, S., and Boutry, M. (1996). Mitochondrial and chloroplast targeting sequences in tandem modify protein import specificity in plant organelles. Plant Mol. Biol. 30, 769–780. [DOI] [PubMed] [Google Scholar]
- Small, I., Wintz, H., Akashi, K., and Mireau, H. (1998). Two birds with one stone: Genes that encode products targeted to two or more compartments. Plant Mol. Biol. 38, 265–277. [PubMed] [Google Scholar]
- Tang, H.-L., Yeh, L.-S., Chen, N.-K., Ripmaster, T., Schimmel, P., and Wang, C.-C. (2004). Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J. Biol. Chem. 26, 49656–49663. [DOI] [PubMed] [Google Scholar]
- Urban, A., Neukirchen, S., and Jaeger, K.-E. (1997). A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res. 25, 2227–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten, W.J., and Janssen, G.R. (1998). An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol. Microbiol. 27, 987–1001. [DOI] [PubMed] [Google Scholar]
- Wall, M.K., Mitchenall, L.A., and Maxwell, A. (2004). Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA 101, 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, A., Etzinga, N., Wobbes, B., and Smeekens, S. (2004). A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16, 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.-P., and Glaser, E. (2002). Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci. 7, 14–21. [DOI] [PubMed] [Google Scholar]