Abstract
The Arabidopsis thaliana secretome was analyzed by the proteomic approach, which led to the identification of secreted proteins implicated in many aspects of cell biology. We then investigated the change in the Arabidopsis secretome in response to salicylic acid and identified several proteins involved in pathogen response. One of these, a secreted lipase with a GDSL-like motif designated GDSL LIPASE1 (GLIP1), was further characterized for its function in disease resistance. glip1 plants were markedly more susceptible to infection by the necrotrophic fungus Alternaria brassicicola compared with the parental wild-type plants. The recombinant GLIP1 protein possessed lipase and antimicrobial activities that directly disrupt fungal spore integrity. Furthermore, GLIP1 appeared to trigger systemic resistance signaling in plants when challenged with A. brassicicola, because pretreatment of the glip1 mutant with recombinant GLIP1 protein inhibited A. brassicicola–induced cell death in both peripheral and distal leaves. Moreover, glip1 showed altered expression of defense- and ethylene-related genes. GLIP1 transcription was increased by ethephon, the ethylene releaser, but not by salicylic acid or jasmonic acid. These results suggest that GLIP1, in association with ethylene signaling, may be a critical component in plant resistance to A. brassicicola.
INTRODUCTION
The cell wall or extracellular matrix (ECM) is a dynamic zone harboring active components that regulate cell–cell interactions. The ECM contains secreted proteins that play crucial roles in a variety of physiological processes, such as growth, development, and defense responses (Showalter, 1993; Pennell, 1998). The most abundant and well-studied cell wall proteins are structural proteins, including extensins, Gly-rich proteins, Pro-rich proteins, solanaceous lectins, and arabinogalactan proteins (Showalter, 1993). However, the ECM also contains a large number of nonstructural proteins, including peroxidases, invertases, proteases, mannosidases, galactosidases, and glucanases.
ECM peptide or protein participation in the plant defense process has been reported previously. For example, plants have been found to contain families of antimicrobial peptides (Garcia-Olmedo et al., 1998), which include the thionins, defensins, lipid transfer proteins, hevein- and knottin-like peptides, maize (Zea mays) basic peptide 1, Impatiens balsamina antimicrobial peptide, snakins, and systemins. Systemins are a unique class of 18–amino acid peptides derived from the precursor polypeptide prosystemin and have been functionally well defined (Ryan et al., 2002). They activate the defense signaling process by interacting with the plasma membrane receptor kinase SR160. Plants also bear polygalacturonase-inhibiting proteins, which are ubiquitous cell wall proteins that protect plants from fungal invasion (De Lorenzo and Ferrari, 2002). Polygalacturonase-inhibiting proteins directly inhibit the polygalacturonases secreted by pathogenic fungi and at the same time enhance the accumulation of oligogalacturonides, which in turn activates the defense response.
Proteomics is the global analysis of gene products in various tissues and physiological states of cells (Pandey and Mann, 2000) and has become very important in the functional genomics field now that several genomes have been sequenced completely and analytical methods for protein characterization have been developed. Progress in plant proteomics has largely been made possible by two-dimensional (2D) gel electrophoresis–based proteomic approaches (van Wijk, 2001; Kersten et al., 2002), and several major plant proteomics studies have focused on subcellular proteomes and protein complexes (Rouquie et al., 1997; Peltier et al., 2000; Prime et al., 2000; Kruft et al., 2001; Millar et al., 2001; Chivasa et al., 2002; Bae et al., 2003).
Despite technical difficulties in preparing pure cell walls, the proteomic approach has also been used to study plant cell wall proteins (Okushima et al., 2000; Chivasa et al., 2002; Ndimba et al., 2003). In one of these studies, isolated cell walls were subjected to sequential protein extractions using CaCl2 and a urea buffer, resulting in fractions that included classical cell wall proteins such as glucanases, expansins, glucosidases, pectinesterases, and peroxidases (Chivasa et al., 2002). In the cell wall proteomics study of Okushima et al. (2000), cultured BY2 cells were used to examine the extracellular proteins of tobacco (Nicotiana tabacum) cells. Six proteins were found by N-terminal amino acid sequencing to be similar to wheat (Triticum aestivum) chemically induced 5, ascorbate oxidase, peroxidase, and exoglucanase. In the proteomics study of Ndimba et al. (2003), Arabidopsis thaliana cell suspension cultures were investigated for the proteomic changes induced by fungal elicitors, chitosan, and extracts of Fusarium moniliforme. Proteins whose abundance was changed by these treatments were identified in the cell wall fraction and in the culture filtrate.
In this work, we conducted a comprehensive analysis of the secretome of cultured Arabidopsis cells. We collected proteins secreted into the culture medium and analyzed them by 2D gel electrophoresis and matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). To identify the secreted proteins involved in the plant pathogen response, we evaluated salicylic acid (SA)–induced changes in the isolated secretome. 2D gel analysis revealed that 18 protein spots corresponding to 13 different protein species were either upregulated or downregulated by SA treatment. Of these, GDSL LIPASE1 (GLIP1), a secreted lipase with a GDSL motif, was selected for further characterization. We found that GLIP1 plays critical roles in plant resistance to the fungus Alternaria brassicicola. This disease resistance is regulated by two different mechanisms: direct disruption of fungal spores, and systemic resistance signaling associated with ethylene. As a result, we conclude that the proteomics approach can be an effective means of identifying novel proteins implicated in signaling processes.
RESULTS
Isolation of Secreted Proteins from Arabidopsis Cell Cultures
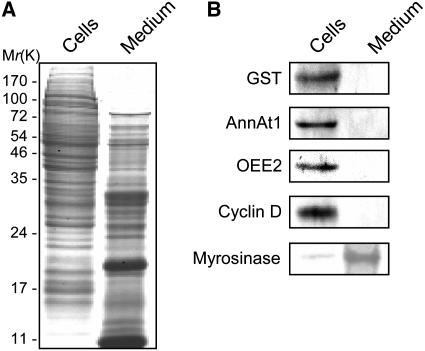
Cell cultures derived from Arabidopsis ecotype Columbia (Col-0) were established to isolate proteins secreted into the medium. The culture medium was harvested on d 4, at which point the cells were actively proliferating at the mid-log phase. The isolated secreted proteins were tested for contamination with nonsecretory proteins by protein gel blot analysis using antibodies specific for the cell wall protein myrosinase (Husebye et al., 2002), the cytosol proteins glutathione S-transferase (GST) and cyclin D, the cytosol and membrane protein annexin1 (AnnAt1), and the chloroplast protein oxygen-evolving enhancer protein2 (OEE2) (Figure 1). The extracellular protein myrosinase was found mostly in the secreted fraction, whereas the others were detected exclusively in the cultured cells (i.e., the nonsecreted fraction).
Figure 1.
Isolation of the Secreted Proteins from Arabidopsis Cell Cultures.
Arabidopsis suspension cultures were separated into cell and culture medium fractions and subjected to gel electrophoresis. Each lane was loaded with the same amount of protein (10 μg).
(A) Fractionated proteins separated by SDS-gel electrophoresis and visualized by Coomassie blue staining.
(B) Protein gel blot analysis of fractionated proteins with anti-GST, anti-AnnAt1, anti-OEE2, anti-cyclin D, and anti-myrosinase antibodies.
To confirm the lack of contamination of the culture medium, we estimated cytosolic marker enzyme activities, namely, those of alcohol dehydrogenase (ADH) and glucose-6-phosphate dehydrogenase (G6PDH), in the separated medium and callus fractions (Table 1). The enzyme activities were high in the cells but not detectable in the medium. These data suggest that the secretome prepared from the culture medium was essentially free of contamination by nonsecreted proteins. However, we cannot rule out the possibility that some proteins in the secretome were leaking from dead or damaged cells. The instability of contaminating cytosolic marker proteins accumulating in the culture medium may explain this.
Table 1.
Cytosolic Marker Enzyme Activities in the Cell and Culture Medium Fractions
| Fraction | ADH Activity (units/mg) | G6PDH Activity (units/mg) |
|---|---|---|
| Nonea | 0.002 ± 0.003 | 0.001 ± 0.002 |
| Medium | 0.002 ± 0.004 | 0.001 ± 0.003 |
| Cells | 2.193 ± 0.047 | 0.033 ± 0.004 |
One unit of activity was defined as the amount of enzyme catalyzing the reduction of 1 μmol of NAD+ or NADP+ per minute under the specific assay conditions described in Methods. The values shown are means ± sd (n = 10).
No fractions added.
Identification of the Arabidopsis Secreted Proteins
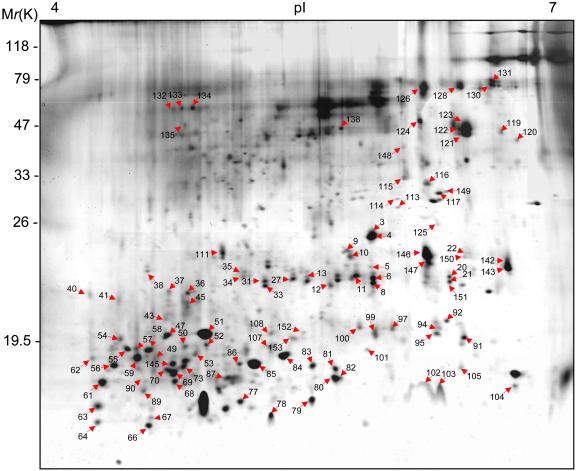
For proteomic analysis, the isolated secreted proteins were subjected to 2D gel electrophoresis (Figure 2). Approximately 400 protein spots were detected on a silver-stained 2D gel. The secreted proteins were relatively small and mostly distributed in the 10- to 100-kD range. Analysis of the protein spots by MALDI-TOF MS led to the identification of 107 spots that correspond to 91 different proteins (Figure 2; see Supplemental Table 1 online).
Figure 2.
Silver-Stained 2D Gel of Secreted Proteins.
Isolated secreted proteins were resolved in the range of pH 4 to 7. Protein spots identified by MALDI-TOF MS are numbered and listed in Supplemental Table 1 online.
Of the proteins identified, 60 (66%) have known functions or sequences similar to those of known proteins, whereas 31 (34%) are novel and have not been assigned any functions. All proteins were categorized into classes based on their functions. The largest class consisted of the novel or uncharacterized proteins (34%). The next largest class contained proteins involved in signaling and regulation (29.6%). The other functions included catalysis (17.6%), protein degradation (6.7%), protein protection (5.5%), structure (3.3%), and retrotransposition (3.3%). These results show that secreted proteins are implicated in a variety of cellular functions.
Secreted proteins are known to bear a short stretch of amino acids called a signal sequence at their N terminus (Nielsen et al., 1997). To determine whether the secreted proteins we identified contain such a signal sequence, we analyzed them with SignalP software (http://www.cbs.dtu.dk/services/SignalP), which searches for signal sequences and their cleavage sites (Nielsen et al., 1997). Of the proteins analyzed, 49 (54%) appear to contain signal sequences with mean S scores > 0.48 (see Supplemental Table 1 online).
Identification of SA-Responsive Secreted Proteins
To identify secreted proteins involved in the pathogen response, we analyzed the change in the Arabidopsis secretome in response to SA, a plant hormone that regulates defense signaling (Dürner et al., 1997). Thus, Arabidopsis cell cultures were left untreated or were treated with 0.5 mM SA for 2 and 6 h, after which the culture medium was collected and analyzed by comparative 2D gel electrophoresis. Protein spots whose intensity in the SA-treated medium was altered relative to the untreated control medium were quantitatively analyzed. We performed three independent experiments and selected only those spots whose abundance varied reproducibly by greater than twofold. Eighteen protein spots were selected and subjected to MALDI-TOF MS analysis for protein identification (Table 2).
Table 2.
Identification of SA-Responsive Proteins Secreted by Arabidopsis Cell Cultures
| Match MM (kD)/pIb
|
Induction Factore
|
||||
|---|---|---|---|---|---|
| Spot No.a | Accession No.c | Protein Named | 2 h | 6 h | |
| 6 | 17.15/5.8 | 18403909 | Ubiquitin-like UBQ7/AtRUB2 | 0.63 | 0.21 |
| 8 | 17.15/5.8 | 18403909 | Ubiquitin-like UBQ7/AtRUB2 | 0.82 | 0.43 |
| 9 | 34.05/5.3 | 26452648 | Jacalin-related protein | 2.91 | 2.51 |
| 10 | 34.05/5.3 | 26452648 | Jacalin-related protein | 1.56 | 2.10 |
| 20 | 22.83/6.4 | 24417448 | Leu-rich repeat, plant specific | 0.81 | 0.50 |
| 21 | 22.83/6.4 | 24417448 | Leu-rich repeat, plant specific | 0.90 | 0.50 |
| 151 | 22.83/6.4 | 24417448 | Leu-rich repeat, plant specific | 0.88 | 0.37 |
| 92 | 19.03/5.6 | 11288845 | Hypothetical protein | 0.38 | 0.29 |
| 100 | 23.43/5.7 | 26451897 | Unknown protein | 0.18 | 0.11 |
| 116 | 27.22/5.8 | 15221583 | Expressed protein | 2.82 | 2.77 |
| 117 | 32.43/6.3 | 21553514 | Unknown protein | 2.25 | 3.54 |
| 119 | 41.71/6.6 | 15237530 | GDSL motif lipase/hydrolase–like protein | 3.04 | 3.03 |
| 143 | 20.81/5.7 | 15226149 | Athila retroelement open reading frame1–like protein | 2.04 | 1.06 |
| 148 | 37.74/6.1 | 15228497 | Esterase/lipase/thioesterase, active site | 1.79 | 2.00 |
| 149 | 32.32/5.4 | 15234160 | Expressed protein | 1.34 | 2.19 |
| 150 | 25.50/6.0 | 15239633 | Unknown protein | 3.59 | 2.31 |
| 152 | 18.56/5.4 | 15223328 | Hypothetical protein | 0.36 | 0.49 |
| 153 | 18.56/5.4 | 15223328 | Hypothetical protein | 0.18 | 0.11 |
Number of the spot.
Predicted molecular mass (MM) and pI of the matched sequence.
Accession number in the National Center for Biotechnology Information nonredundant database.
Entry name according to the National Center for Biotechnology Information database.
Fold change in protein expression upon SA treatment as calculated by ImageMaster analysis software.
The identified protein spots corresponded to 13 different proteins, as some were present in multiple spots, which suggests that they undergo posttranslational modifications. Of the 13 proteins, 3 had protein names or known functions but 10 were novel or unknown proteins in the Arabidopsis genome databases. These unknown proteins were then scanned against Prosite (http://ca.expasy.org/tools/#pattern), a database of protein families and domains, and additionally assigned functional motifs (i.e., a jacalin-related protein, a Leu-rich repeat, and the active site of esterase/lipase/thioesterase; Table 2).
Sequence Comparison of GLIP1 with Other GLIPs
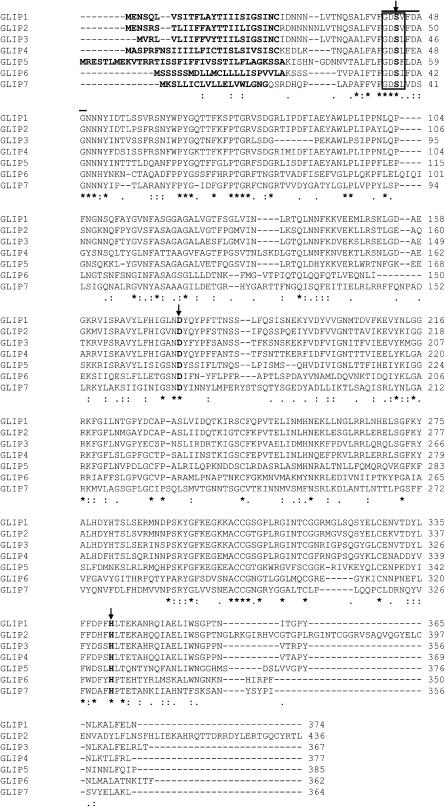
A GDSL motif lipase/hydrolase–like protein (spot 119), designated GLIP1, was selected for further analysis because plant-derived secreted lipases have not been implicated previously in the pathogen response. In a BLAST search of the Arabidopsis genome database, six additional GLIP genes were identified and designated GLIP2 to GLIP7 (Figure 3). A comparison of the amino acid sequences reveals that GLIP1 is 68 to 78% identical to GLIP2, GLIP3, and GLIP4, whereas its similarity to the other GLIPs is lower (32 to 52% identical to GLIP5, GLIP6, and GLIP7). The GLIPs represent a novel lipase gene family with a GDSL-like motif (Brick et al., 1995) and a signal peptide as predicted by SignalP (Nielsen et al., 1997). All seven GLIPs contain a catalytic triad of Ser, Asp, and His conserved among the lipase and esterase families. Another characteristic feature of the GLIPs is that the classical lipase signature sequence GxSxG is replaced by GxSxxxxG, which overlaps with the GDSL motif (Brick et al., 1995).
Figure 3.
Alignment of the Deduced Amino Acid Sequence of GLIP1 with Those of Other GLIPs.
Sequences were aligned using ClustalW. The signal sequences are shown in boldface at the N termini, and the residues of the catalytic triad (Ser, Asp, and His) are indicated in boldface and by arrows. The lipase signature sequence is shown by the solid line, and the GDSL-like motif is boxed. Consensus symbols under the alignment are as follows: asterisks indicate identical residues in all sequences; colons and periods indicate conserved and semiconserved substitutions, respectively.
Subcellular Localization of GLIP1
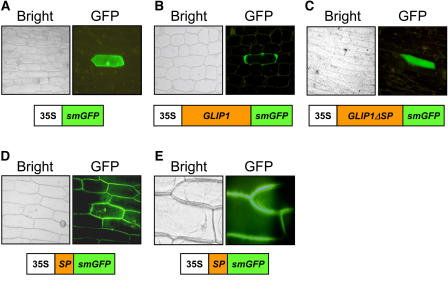
To investigate the subcellular localization of GLIP1, we fused soluble, modified green fluorescent protein (smGFP) to GLIP1 expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The smGFP fusion constructs of full-length GLIP1, GLIP1 lacking the signal peptide (GLIP1ΔSP), and the signal peptide alone (SP) were introduced into onion (Allium cepa) epidermal cells. After incubation for 24 h, fluorescence from smGFP was examined by fluorescence microscopy (Figure 4). The smGFP control and GLIP1ΔSP were expressed in the cytosol. However, GLIP1 and SP fluorescence signals were clearly located in the intercellular space, indicating that the signal sequence leads to the secretion of GLIP1. The fluorescence of GLIP1 appeared to be confined in the intercellular space, unlike the diffuse pattern of SP. The intercellular localization was clarified in the presence of mannitol, which induces plasmolysis, pulling the cytosol away from the cell wall, and thus allowing the two to be distinguished from one another (Figure 4E). These results are consistent with the proteomic data demonstrating that GLIP1 is secreted into the cell wall or extracellular space.
Figure 4.
Subcellular Localization of GLIP1 Proteins.
GFP fusion constructs were introduced into onion epidermal cells by particle bombardment. The expression of the fusion proteins was driven by the CaMV 35S promoter. Bright field (left) and fluorescence (right) images are shown. Schemes of the fusion constructs are shown at the bottom of each panel: 35S, CaMV 35S promoter.
(A) Expression of a GFP control.
(B) Expression of the full-length GLIP1-GFP fusion protein.
(C) Expression of the GLIP1ΔSP-GFP fusion protein.
(D) Expression of the SP-GFP fusion protein.
(E) Expression of SP-GFP in the presence of 1 M mannitol. Plasmolysis was induced in cells that express GFP fusion proteins.
Isolation of GLIP1 T-DNA Insertion Mutants
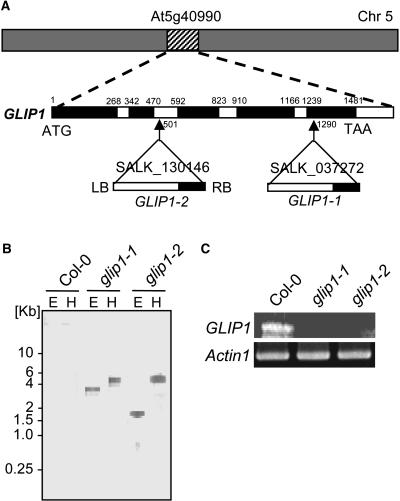
To assess the in vivo function of GLIP1, we searched the Salk Institute insertion sequence database for T-DNA insertion mutants and obtained two alleles, glip1-1 and glip1-2, in which the T-DNA inserts are in the fifth exon and second intron, respectively (Figure 5A). The mutant seeds were originally heterozygous for a single T-DNA insertion with a 3:1 segregation ratio. Homozygous lines were identified in the next generation. To determine the exact position of the T-DNA insertion, genomic DNA fragments of the GLIP1 gene and T-DNA junction were amplified and sequenced (Figure 5A). DNA gel blot analysis determined that these lines contained a single T-DNA insertion (Figure 5B). It was also demonstrated that the homozygous mutants did not express GLIP1 transcripts (Figure 5C). Under normal growth conditions, glip1 plants showed growth similar to Col-0 (data not shown).
Figure 5.
T-DNA Insertion Mutants of GLIP1.
(A) Genomic organization of GLIP1 and location of the T-DNA insertion in the glip1 mutants. The arrows indicate the positions of the T-DNA insertions (triangles). Genomic GLIP1 DNA is represented by exons (black) and introns (white). The T-DNA orientation is indicated by left (LB) and right (RB) borders. The numbers refer to nucleotides in the genomic GLIP1 DNA. Chr 5, chromosome 5.
(B) DNA gel blot analysis of the T-DNA insertion in Col-0 and glip1 plants. The genomic DNA was digested with EcoRI (E) and HindIII (H), and the blot was probed with the T-DNA left border–specific DNA.
(C) RNA analysis of GLIP1 gene expression in Col-0 and glip1 plants. GLIP1 expression was analyzed by RT-PCR. Total RNA (0.1 μg) was used to detect GLIP1 and Actin1 (loading control) expression.
Function of GLIP1 in Disease Resistance
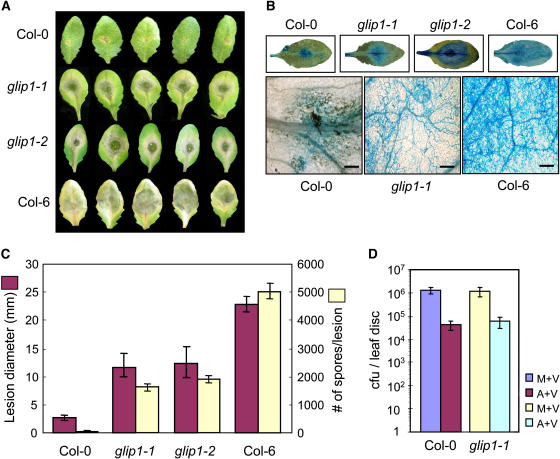
Because GLIP1 was isolated in a screen for SA-responsive secreted proteins, it was first assessed whether glip1 mutants exhibit altered responses to infection by the bacterial pathogen Pseudomonas syringae, which is closely associated with SA signaling (Crute et al., 1994). The bacterial growth and disease symptom development after inoculation with virulent Pseudomonas syringae pv tomato DC3000 (Pst DC3000) did not differ significantly between Col-0 and glip1 plants (Figure 6D). The development of systemic acquired resistance (SAR) was also observed in both Col-0 and glip1 plants, as examined by treatment of primary leaves with avirulent Pst DC3000 carrying avrRpt2 [Pst DC3000 (avrRpt2)] before challenge of secondary leaves with Pst DC3000 (Figure 6D). These results suggest that GLIP1 is not involved in resistance to P. syringae in Arabidopsis.
Figure 6.
Disease Development in glip1 Mutants Inoculated with P. syringae and A. brassicicola.
Four-week-old plants were challenged with either 20 μL of P. syringae bacterial suspensions (106 colony-forming units [cfu]/mL) for both primary and secondary inoculations (D) or 10 μL of fungal spore suspensions (5 × 105 spores/mL) ([A] to [C]). The inoculated leaves were detached 4 d after inoculation for analysis. This experiment was repeated three times with similar results.
(A) Necrotic lesion phenotypes.
(B) Necrotic lesions stained with lactophenol–aniline blue. The formation of fungal mycelium on glip1 and Col-6 leaves was visualized with a microscope. Bars = 25 μm.
(C) Measurement of lesion diameter and number of newly formed spores. The bars and error bars represent averages and sd, respectively, of three independent experiments each with 10 leaves.
(D) Bacterial growth determined in secondary leaves challenged with Pst DC3000 1 d after the inoculation of primary leaves with either 10 mM MgCl2 or Pst DC3000 (avrRpt2). M+V, primary and secondary inoculations with MgCl2 and Pst DC3000, respectively; A+V, primary and secondary inoculations with Pst DC3000 (avrRpt2) and Pst DC3000, respectively. The bars and error bars represent averages and sd, respectively, of three independent experiments.
We then examined the resistance phenotypes of glip1-1, glip1-2, Col-0, and a different Columbia ecotype, Col-6, after inoculation with the necrotrophic fungus A. brassicicola. In the infection cycle, spore germination is followed by the extension of the germ tube over the leaf surface. At the tip of hyphae, fungi develop appressoria for direct penetration into the host and eventually colonize the tissue (McRoberts and Lennard, 1996). Col-0 and Col-6 were used as natural wild-type ecotypes that are resistant and susceptible, respectively, to A. brassicicola (Kagan and Hammerschmidt, 2002). It was previously shown that inoculation of Col-0 wild-type plants with A. brassicicola induces the hypersensitive response, resulting in small necrotic lesions restricted to the infection sites (Thomma et al., 1998, 1999). We also observed that Col-0 inoculated with A. brassicicola formed small brown necrotic lesions (Figure 6A). By contrast, the glip1-1, glip1-2, and Col-6 plants developed spreading lesions upon inoculation (Figures 6A and 6C). As visualized by lactophenol–aniline blue staining, those spreading lesions were heavily colonized by fungal hyphae (Figure 6B). Furthermore, determination of spore number produced de novo at the inoculation site revealed that new spores were abundantly produced on glip1 and Col-6 leaves, unlike Col-0, on which few, if any, new spores were formed (Figure 6C). No difference between glip1-1 and glip1-2 was observed in disease resistance, indicating that the observed phenotypes are the consequence of a T-DNA insertion into the GLIP1 gene.
Antimicrobial Activity of Recombinant GLIP1 Proteins
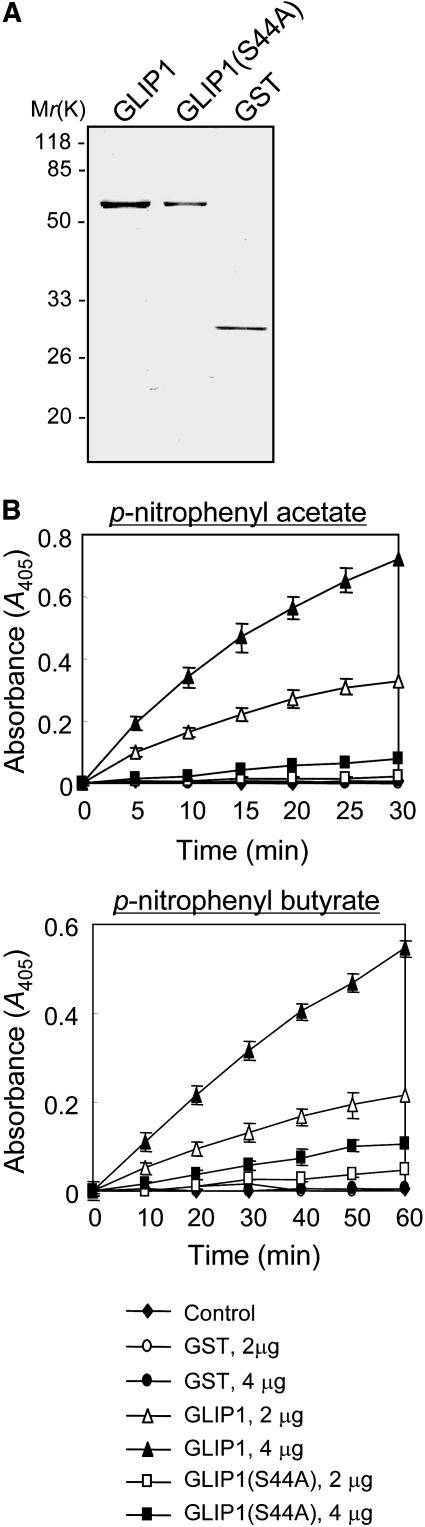
To determine whether GLIP1 has a lipase activity that is required for resistance to A. brassicicola, the recombinant GLIP1 protein was expressed in Escherichia coli (Figure 7A). Along with wild-type protein, we prepared a GLIP1 mutant whose Ser-44 of the catalytic triad was replaced by Ala (S44A). These proteins were assayed for lipase activity using p-nitrophenyl acetate and p-nitrophenyl butyrate as substrates (Figure 7B). The wild-type GLIP1 exhibited high activity, but the GLIP1(S44A) mutant did not, demonstrating that GLIP1 is a lipase and that Ser-44 in the catalytic triad participates in the hydrolytic activity.
Figure 7.
Lipase Activity of Recombinant GLIP1 Proteins.
(A) Coomassie blue–stained gel of purified recombinant GST-GLIP1 proteins.
(B) Lipase activities of the recombinant GLIP1 and GLIP1(S44A) proteins and the GST control. In the assays, 2 μg (open symbols) and 4 μg (closed symbols) of proteins were incubated with two substrates, p-nitrophenyl acetate and p-nitrophenyl butyrate. Each data point represents the average and sd of five independent measurements.
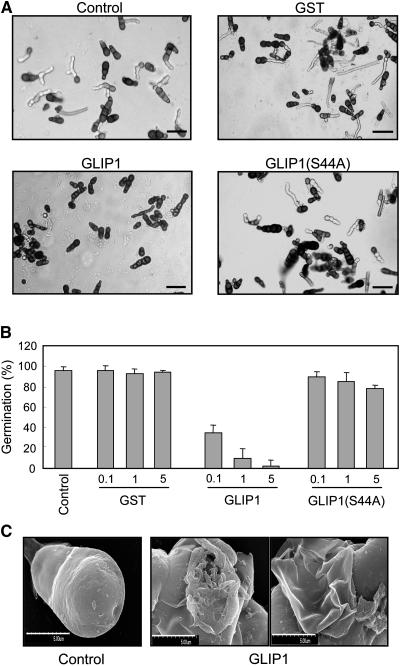
Because GLIP1 is a secreted protein, it is conceivable that GLIP1 directly inhibits the pathogenic activity of A. brassicicola. The antimicrobial activity of GLIP1 against A. brassicicola was thus assessed. When fungal spores were incubated with recombinant wild-type and mutant proteins, spore germination was inhibited by the active form of GLIP1 (Figures 8A and 8B). GLIP1(S44A) had little effect on the fungal growth. Furthermore, scanning electron microscopy showed that GLIP1 caused severe morphological changes in the fungal spores (Figure 8C). These results suggest that GLIP1 may directly disrupt the structure of the fungal cell wall and/or membrane in a specific manner.
Figure 8.
Antimicrobial Activity of Recombinant GLIP1 Proteins against A. brassicicola.
Recombinant proteins were added to 10 μL of spore suspension (5 × 105 spores/mL). The samples were incubated for 24 h at 23°C and observed with a microscope. PBS was used as a mock control.
(A) The inhibitory effect of recombinant proteins (5 μg) on the germination of fungal spores as observed by light microscopy. Bars = 50 μm.
(B) Quantitative analysis of germination rates. The percentage of germinated spores shown in (A) was determined by light microscopy. Bars and error bars represent averages and sd, respectively, of three independent experiments.
(C) Scanning electron microscopy images of fungal spores incubated with GLIP1 proteins (5 μg). Five spores for each condition were observed with similar results. PBS was used as a mock control. Bars = 500 μm.
Function of GLIP1 in Systemic Defense Response
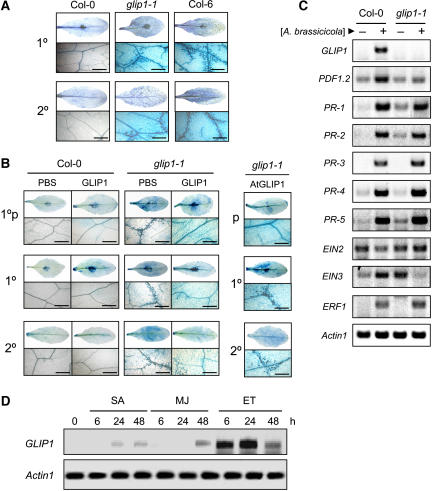
When inoculated with A. brassicicola, Col-0 activates the SAR response that leads to the induction of defense-related genes such as PDF1.2, PR-3, and PR-4 (Penninckx et al., 1996; Thomma et al., 1998). We thus investigated whether GLIP1 is implicated in local and systemic defense responses that are induced in plants by pathogen challenge. When the leaves of A. brassicicola–challenged plants were stained with trypan blue, extensive cell death associated with the systemic spreading of this fungus was observed in both the peripheral and distant leaves in glip1-1 but not in Col-0 (Figure 9A). In Col-0 with SAR induced, cell death was restricted to the inoculated site. Therefore, the expression of SAR could be determined by monitoring the presence of trypan blue–stained cells. The glip1-1 plants were pretreated with recombinant GLIP1 protein, followed by localized A. brassicicola inoculation (1°p) (Figure 9B). This localized inoculation on the pretreated leaves (1°p) was then followed by the application of A. brassicicola to the other distal leaves (1°). There was a significant decrease in both local (1°p) and systemic (1° and 2°) cell death, reflecting the induction of SAR in leaves remote to the infection (Figure 9B). It was noticeable that pretreatment of glip1-1 with GLIP1 protein in local leaves (1°p) systemically inhibited cell death in distant leaves (1°) inoculated with A. brassicicola, in which cell death would be otherwise strongly induced. By contrast, pretreatment with GLIP1 in the absence of localized A. brassicicola inoculation (p) failed to inhibit cell death in both 1° and 2° leaves (Figure 9B, right). Here, primary (1°) and secondary (2°) indicate inoculated and uninoculated leaves, respectively. These data suggest that GLIP1 activates systemic resistance signaling only when challenged with A. brassicicola.
Figure 9.
Function of GLIP1 in the Systemic Defense Response.
Four-week-old plants were inoculated with a 10-μL drop of A. brassicicola spore suspension (5 × 105 spores/mL). In (A) and (B), primary (1°) and secondary (2°) indicate inoculated and uninoculated leaves, respectively. The term 1°p indicates local leaves that were pretreated with PBS or GLIP1 proteins and then inoculated with A. brassicicola. Bars = 50 μm. High magnification (×400) micrographs at the bottom show cell death at a distance from the inoculation site in the case of the primary leaves (1°p and 1°). This experiment ([A] to [D]) was repeated more than three times with similar results.
(A) Cell death, as shown by staining with lactophenol–trypan blue. The staining of primary and secondary leaves was performed 4 d after the primary leaves were treated with A. brassicicola.
(B) Inhibition of the A. brassicicola–induced local and systemic cell death in the glip1-1 mutant by pretreatment with GLIP1 proteins. For the pretreatment, plants were infiltrated into one site on the abaxial surface of each leaf with PBS (control) or GLIP1 proteins (1 μg) as indicated, incubated for 1 d, and then inoculated with A. brassicicola. This localized inoculation on the pretreated leaves (1°p) was followed by the application of A. brassicicola to the other distal leaves 1 d later. As a control (right), leaves were only pretreated with GLIP1 proteins (p) without local inoculation of A. brassicicola. Cell death was monitored by trypan blue staining over 4 d.
(C) Induction of pathogenesis- and ethylene-related genes in Col-0 and glip1-1 plants infected with A. brassicicola. Four-week-old plants were inoculated with a 10-μL drop of spore suspension (5 × 105 spores/mL), harvested 1 d after treatment, and used to extract total RNA. RNA levels were determined by RT-PCR analysis. Actin1 was used as a control.
(D) RNA analysis of the induction of GLIP1 in response to hormone treatments. SA (1 mM), methyl jasmonate (MJ; 50 μM), and the ethylene releaser ethephon (ET; 1.5 mM) were dissolved in water and applied to 4-week-old Col-0 plants. Total RNAs were isolated at the indicated times after treatment, and RNA levels were determined by RT-PCR analysis. Actin1 was used as a control.
Signaling molecules such as SA, jasmonic acid (JA), and ethylene accumulate during defense and, in turn, trigger the coordinated expression of defense genes. Resistance to A. brassicicola involves the cooperative actions of JA and ethylene (Dong, 1998; Penninckx et al., 1998). PDF1.2, PR-3, and PR-4 require JA and ethylene for activation, whereas PR-1, PR-2, and PR-5 are induced by SA (Kunkel and Brooks, 2002). In addition, many genes (e.g., ERF1, EIN2, and EIN3) have been identified to function in ethylene signaling (Wang et al., 2002). We thus analyzed the expression levels of these genes, including GLIP1, in Col-0 and glip1-1 upon A. brassicicola treatment. GLIP1 transcription was strongly induced in Col-0 (Figure 9C). Whereas PR gene expression did not differ, PDF1.2 was less induced in glip1-1. Furthermore, the basal RNA levels of the SA-dependent PR genes, PR-1, PR-2, and PR-5, were much higher in glip1-1 than in Col-0. Of the ethylene-related genes, EIN2 and EIN3 also exhibited altered expression in glip1-1. The expression of EIN2 was repressed in Col-0 but somewhat induced in glip1-1 in response to A. brassicicola. An increase of EIN3 in Col-0 was reversed in glip1-1 with reduced expression. These results suggest that glip1 may be defective in ethylene signaling. In agreement with this, the high basal expression level of SA-related PR genes in glip1-1 is consistent with the suggestions that the SA and ethylene pathways interact in an antagonistic manner (Wang et al., 2002). Although A. brassicicola infection induces PR-1, it has been shown that the resistance to this pathogen is dependent on the JA/ethylene signaling pathway (Penninckx et al., 1996). Noticeably, GLIP1 was strongly induced by ethephon over 24 h (Figure 9D). A slight increase in GLIP1 expression was observed upon treatment with SA and JA. These results demonstrate that the increase of GLIP1 protein in the secretome is regulated posttranslationally (i.e., by secretion) and/or transcriptionally. Together, these results suggest that GLIP1 may be a critical component in the resistance response in association with ethylene signaling.
DISCUSSION
In this work, we present a comprehensive analysis of the Arabidopsis secretome in the medium of cultured cells. Subsequently, to identify secreted proteins that participate in plant defense, we measured increased or decreased protein accumulation in the secretome in response to SA treatment. This revealed several intriguing proteins. In particular, our studies highlight the functional importance of a secreted lipase, GLIP1, in the defense response against A. brassicicola. These results suggest that GLIP1 protects plants from A. brassicicola attack in two distinct ways: by directly disrupting fungal spore integrity, and by activating defense signaling in plants.
Proteomic Analysis of Secreted Proteins
In our analysis of the secretome, we focused on the culture medium, which would contain diffusible nonsticky proteins in the cell wall. We were able to isolate and identify proteins involved in a variety of cellular functions, such as signaling and regulation, catalysis, protein degradation, protein protection, and structure. These results indicate that complicated biological events take place in the extracellular environment. The purity of the isolated secretome was evaluated by immunoblotting with antibodies for proteins in different subcellular compartments. Two cytoplasmic proteins, GST and cyclin D, the chloroplast protein OEE2, and the cytosol and membrane protein AnnAt1 were not detected in the secretome, although they were found in the medium-free callus. By contrast, the secreted protein myrosinase was mostly detected in the secretome. The cytoplasmic contamination of the secreted fraction was also assessed by enzyme assays. These data together indicated that the secretome prepared from the culture medium was enriched in secreted proteins with no detectable contamination.
SignalP analysis predicted that 54% of the secreted proteins possess a signal peptide that would target them for secretion. The rest of the proteins lack an identifiable signal peptide, suggesting the presence of nonclassical endoplasmic reticulum– and Golgi-independent secretory pathways. Consistent with this finding, several proteins lacking a signal sequence in bacteria and yeast are secreted through an alternative secretory machinery such as ATP binding cassette transporters (Kuchler, 1993). Accumulating evidence also suggests that eukaryotic cells secrete proteins lacking signal peptides, including interleukins (MacKenzie et al., 2001), fibroblast growth factors (Bryckaert et al., 2000), annexin II (Siever and Erickson, 1997), and galectins (Barondes et al., 1994). These proteins are extracellularly translocated by the alternative secretory pathways, which may also involve ATP binding cassette transporters (Lottaz et al., 2001). Moreover, research in the last few years has revealed the cell-to-cell movement of plant regulatory proteins through plasmodesmata as a novel mechanism that controls developmental processes (Heinlein, 2002; Oparka, 2004). Those non-cell-autonomous proteins include transcription factors such as LEAFY (Sessions et al., 2000), KNOTTED1 (Lucas et al., 1995), and SHORTROOT (Nakajima et al., 2001). In addition, a wide range of molecules are released into the phloem through plasmodesmata for long-distance transport, suggesting the apoplastic movement of these molecules (Oparka and Cruz, 2000). Such alternative secretory processes may facilitate the extracellular secretion of proteins that lack a classical signal sequence.
ECM Proteins in Defense Response
In our search for secreted proteins involved in the defense process, we isolated 13 proteins whose levels are changed in the secretome by SA treatment. Of the eight SA-induced proteins, four share sequences with known proteins. Jacalin-related proteins are known to be inducible defense proteins (Mann et al., 2001). They share the domain structure of plant lectins and are upregulated by several low molecular mass regulators, such as JA, SA, and ethylene, or by altered ambient conditions, such as salt stress (Zhang et al., 2000). Another enhanced secreted protein is an Athila retroelement open reading frame1 (ORF1)–like protein. The Athila retroelement is a Ty3-gypsy group retrotransposon with an envelope-like ORF (Wright and Voytas, 1998). Plant retrotransposons are activated by stresses, including wounding, pathogen attack, freezing, UV light, JA, and SA (Kimura et al., 2001). This may be either a survival strategy or a stress-induced generator of genomic diversity (Wessler, 1996).
The other two secreted proteins with increased expression have sequence similarity to lipases or esterases but belong to different gene families, such as the GDSL motif lipase and Ser hydrolase families (eukaryotic lipases and esterases). That lipases are implicated in disease resistance in plants has been reported previously. Mutational analyses of Arabidopsis to screen for genes required for R gene–mediated resistance or SAR identified two lipase-encoding genes, enhanced disease susceptibility1 (EDS1) and phytoalexin deficient4 (PAD4), that have been well characterized (Falk et al., 1999; Jirage et al., 1999). Mutations in both EDS1 and PAD4 suppressed disease resistance conferred by R proteins. EDS1 functions upstream of SA-dependent PR1 mRNA accumulation and is not required for JA-induced PDF1.2 expression (Falk et al., 1999). PAD4 also participates in a positive regulatory loop that increases SA levels, thus activating SA-dependent defense responses (Jirage et al., 1999). A recent study demonstrated that SA binding protein2 (SABP2) in tobacco binds SA with high affinity and belongs to the α/β-Ser hydrolase superfamily (Kumar and Klessig, 2003). SABP2 retains SA-stimulated lipase activity and is required for full local and systemic resistance to pathogen infection. In a differential screening study, a different lipase gene family of Arabidopsis was also identified (Jakab et al., 2003). It contains nine genes encoding pathogenesis-related lipase-like proteins (PRLIP1 to PRLIP9), which are induced by treatment with SA, JA, or ethylene. All of these lipases, including EDS1 and PAD4 in Arabidopsis and tobacco SABP2, belong to the α/β-Ser hydrolase superfamily. They share little sequence homology but contain the catalytic triad (Ser, Asp, His) and the lipase signature sequence GxSxG. The two lipases that we isolated in this work also contain the catalytic triad but have different features (i.e., the GxSxxxxG signature sequence instead of GxSxG and a signal sequence at the N terminus). Thus, these two proteins are members of distinct and unique families of secreted lipases.
Secreted Lipases in Pathogen Response
Of the two putative secreted lipases we isolated, we further characterized the GDSL motif lipase/hydrolase GLIP1. It possesses a typical signal peptide at its N terminus, which allowed secretion to the cell wall, as verified by expression analysis. Analysis of the Arabidopsis genomic database revealed six more proteins that share 50 to 85% similarity (32 to 78% identity) to the sequence of GLIP1, and all have a signal sequence as predicted by SignalP.
The extracellular localization of GLIP1 led to the notion that it may directly inhibit fungal infection and/or produce a lipid-derived defense signaling molecule. Indeed, we found that fungal spores were disrupted when treated with recombinant GLIP1 protein, suggesting that GLIP1 directly interferes with the fungal infection process. It has been reported previously that fungal plant pathogens secrete hydrolyzing enzymes such as cutinases or esterases to degrade components of the plant cell wall during their invasion of the host. Moreover, extracellular lipases have been considered to be virulence factors of pathogenic bacterial and fungal species, including A. brassicicola (Berto et al., 1999; Eddine et al., 2001). We show here that the fungal spores may actually be counterattacked by the plant-derived lipase GLIP1. Fungal cells have significant internal turgor pressure and thus undergo lysis when their cell walls are even slightly perturbed (Selitrennikoff, 2001). The lipase activity of GLIP1 was required for its function, because the alteration of a Ser residue at the active site (S44A) eliminated both the enzymatic activity of GLIP1 and its ability to disrupt the fungal spores. Thus, we have demonstrated here a plant-derived secreted lipase that can directly disrupt invading fungi. Notably, GLIP1 appears to have substrate specificity toward fungal cell walls that are composed of components different from those of plant hosts (Selitrennikoff, 2001).
It was previously shown that inoculation of Col-0 wild-type plants with A. brassicicola induced a hypersensitive response resulting in small necrotic lesions restricted to the infection sites (Thomma et al., 1998, 1999). This initial response activated a SAR response that led to the induction of defense-related genes such as PDF1.2, PR-3, and PR-4 (Penninckx et al., 1996; Thomma et al., 1998). Based on our observation, GLIP1 may play dual roles in the defense response, as it not only directly interacts with the fungal pathogen but may also influence the defense signaling in the plant itself. This possibility is suggested by the results that the expression of defense- and ethylene-related genes was partially defective in glip1 plants and that pretreatment of glip1 with GLIP1 recombinant protein systemically inhibited the cell death induced by fungal infection. The defense signaling may be activated by a lipid-derived molecule generated by the lipid-hydrolyzing activity of GLIP1. Alternatively, materials or chemicals released by the disrupted fungal spores may function as elicitors that trigger the systemic resistance signaling. The existence of a lipid-derived mobile signal that promotes long-distance signaling has been proposed previously (Maldonado et al., 2002). It was shown that defective in induced resistance1 (dir1) was defective in developing SAR to virulent Pseudomonas or Peronospora parasitica. DIR1 is an apoplastic lipid transfer protein (LTP) and thus may function in the generation of an essential mobile signal or its transmission from the induced leaf. It is conceivable that a GLIP1-derived mobile signal moves around as an LTP-bound form, which remains to be examined. We found that GLIP1 may be implicated in both short- and long-distance defense responses, because glip1 failed to develop local and systemic resistance responses to A. brassicicola, but these responses were recovered upon pretreatment with recombinant GLIP1.
We originally identified GLIP1 as an SA-stimulated secreted protein. However, it turned out that GLIP1 is actually involved in ethylene signaling, unlike the previously characterized SA-dependent lipases, such as EDS1 and PAD4. Whereas SA has been recognized as a key modulator for the activation of both the hypersensitive response and SAR, its role in the defense response triggered by necrotrophic fungal pathogens is not yet clear (Schenk et al., 2003). Accumulating data indicate that resistance to A. brassicicola is induced via the activation of the JA and ethylene signaling pathways (Penninckx et al., 1998; Pieterse and van Loon, 1999). Furthermore, the ethylene and JA pathways cooperate with each other, as supported by the fact that the induction of PDF1.2 requires both JA and ethylene (Wang et al., 2002). Here, we propose that GLIP1 is a critical component for both local and systemic resistance responses in the incompatible interaction with A. brassicicola in the ethylene-dependent pathway.
METHODS
Plant Materials, Growth Conditions, and Treatments
Arabidopsis thaliana plants were grown in a growth room at 23°C under long-day conditions (16-h light/8-h dark cycle). Plants were also grown under short-day conditions (8-h light/16-h dark cycle) for infection with Pseudomonas syringae, because Arabidopsis displays age-related resistance to P. syringae (Kus et al., 2002). Arabidopsis ecotype Col-0 (catalog no. WT-02) was purchased from Lehle Seeds. Arabidopsis ecotype Col-6 (catalog no. CS8155) and the T-DNA insertion mutants glip1-1 (SALK_037272) and glip1-2 (SALK_130146) were obtained from the ABRC.
Callus was generated from the cotyledons of young seedlings and induced on induction medium (Murashige and Skoog [MS] medium, pH 5.8, 3% sucrose, 0.8% agar, and 2 mg/L 2,4-D). A cell suspension culture was initiated by inoculating 1 to 2 g of callus into 50 mL of MS medium supplemented with 3% sucrose and 2 mg/L 2,4-D as described (May and Leaver, 1993). The suspension cells (10 mL) were subcultured into 50 mL of fresh medium once every week and maintained with gentle agitation (120 rpm).
For hormone treatments, SA (1 mM), methyl jasmonate (50 μM), and ethephon (1.5 mM) were dissolved in water and applied as a spray to 4-week-old plants. The treated plants were kept at 100% RH and then harvested at the indicated times.
Infection with P. syringae was performed as described (Alvarez et al., 1998). Pst DC3000 (avrRpt2) and Pst DC3000 were grown overnight in King's B medium containing 100 μg/mL rifampicin with and without 50 μg/mL kanamycin, respectively, washed twice, and resuspended in 10 mM MgCl2. Four-week-old plants were infiltrated at two sites of two primary leaves with 20-μL aliquots of 10 mM MgCl2 or Pst DC3000 (avrRpt2) (106 colony-forming units/mL), which was followed by infiltration at two sites of secondary leaves with 20-μL aliquots of Pst DC3000 (106 colony-forming units/mL) the next day.
Inoculation with Alternaria brassicicola was performed by applying a 10-μL drop of spore suspension (5 × 105 spores/mL) on each leaf of 4-week-old plants. For pretreatment with GLIP1 proteins, plants were infiltrated into one site on the abaxial surface of each leaf with PBS (control) or GLIP1 proteins (1 μg) in 10-μL aliquots and incubated for 24 h before pathogen inoculation. Inoculated plants were kept at 100% RH in a growth chamber for the indicated times. To count the number of spores, 10 leaves were collected and vigorously shaken in 5 mL of 0.1% Tween 20 in a test tube. Leaves were removed, and the spore-containing suspension was centrifuged at 5000g for 15 min. The spores were resuspended in 200 μL of 0.1% Tween 20, serially diluted, and counted with a microscope.
Preparation of Secreted Proteins
The suspension cultures were left untreated or treated with 0.5 mM SA 4 d after subculture. At the indicated times, the cultures were passed twice through a membrane filter (0.2 μm) in a vacuum apparatus to remove the cells from the culture medium. The collected medium (secreted fraction) was then freeze-dried in a vacuum lyophilizer and dialyzed against buffer (10 mM MES, pH 5.8). The prepared secreted proteins were stored at −70°C.
2D Gel Electrophoresis
The 2D gel electrophoresis and gel analysis were performed as described (Lee et al., 2004). Secreted proteins were resuspended in isoelectric focusing sample buffer (7 M urea, 2 M thiourea, 4% 3-[(3-cholamido propyl)-dimethylammonio]-1-propane sulfate, 1% carrier ampholytes, 20 mM DTT, and 0.001% bromophenol blue) and used to rehydrate immobilized pH gradient strips (240 mm, pH 4 to 7; Amersham Biosciences). Isoelectric focusing was performed for a total of 320,000 to 350,000 voltage hours on the IPGphor system (Amersham Biosciences). To run gels in the second dimension, 12% SDS polyacrylamide gels were used. The 2D gels were stained with silver nitrate according to the manufacturer's instructions (Amersham Biosciences). To create the reference map, 2D gels were scanned and analyzed by ImageMaster 2D Elite software (Amersham Biosciences). For each condition analyzed, three to five gels were prepared from three different protein extractions. The volumes of silver-stained spots were normalized according to the volumes of internal standards. The SA-induced changes were subjected to statistical analysis with Student's t test, and those spots with P < 0.05 were considered for identification by MALDI-TOF MS.
MALDI-TOF MS and Database Searching
Peptide samples were prepared as described previously (Lee et al., 2004). Peptide masses were measured on a MALDI-TOF MS system (Voyager-DE STR; Applied Biosystems). Peptide mass fingerprint data were matched to the National Center for Biotechnology Information nonredundant database entries using the MS-Fit program (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm). The following search parameters were applied. Mass tolerance was set to 50 ppm [(experimental mass – theoretical mass)/theoretical mass in daltons], and one incomplete cleavage was allowed. Acetylation of the N terminus, alkylation of Cys by carbamidomethylation, oxidation of Met, and pyroGlu formation of N-terminal Gln were set as possible modifications. Molecular mass and pI ranges were set to 10 to 200 kD and 4 to 7, respectively. The database search disclosed matching proteins ranked according to peptide number matches, sequence coverage, and the Molecular Weight Search (MOWSE) score. Although the candidate ranked at the top was considered a positive identification, protein identification was assigned when the following criteria were met: at least five matching peptides, >15% sequence coverage, and a MOWSE score > 103.
Protein Analysis
For immunoblotting, proteins were separated on 12% SDS-polyacrylamide gels and electrotransferred onto nitrocellulose membranes. Membranes were incubated with anti-GST, anti-AnnAt1 (Lee et al., 2004), anti-OEE2 (Yang et al., 2003), anti-cyclin D (Cho et al., 2004), and anti-myrosinase (Husebye et al., 2002) antibodies. Anti-myrosinase antibody was provided by Atle Bones, and the other antibodies were prepared in the Kumho Life and Environmental Science Laboratory. Antibody-bound proteins were detected by incubation with secondary antibodies conjugated to horseradish peroxidase using the enhanced chemiluminescence system (Amersham Biosciences).
DNA and RNA Analysis
The genomic DNA was digested, separated, and blotted onto a Hybond N+ nylon membrane (Amersham Biosciences). The T-DNA–specific probe for DNA analysis contained the partial sequence of the left border of T-DNA and was generated by PCR using primers 5′-GCTGCCTGTATCGAGTGGTGATTTTGT-3′ and 5′-TCCTTTCGCTTTCTTCCCTTCCTTTCTC-3′. Hybridization was performed for 16 to 24 h at 60°C. Membranes were then washed with 1× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate)/0.1% SDS, then with 0.5× SSC/0.1% SDS, and finally with 0.1% SSC/0.1% SDS. The bands were visualized by autoradiography.
RT-PCR was performed with 0.1 μg of total RNA using the Access RT-PCR system (Promega). The primers used in the RT-PCR are as follows: for GLIP1, 5′-CGATTGTGCACCAGCCTCATTGGTT-3′ and 5′-CAGCGCTTTGAGATTATAGGGTCC-3′; for PR-1, 5′-TCGTCTTTGTAGCTCTTGTAGGTG-3′ and 5′-TAGATTCTCGTAATCTCAGCTCT-3′; for PR-2, 5′-CGTTGTGGCTCTTTACAAACAACAAAAC-3′ and 5′-GAAATTAACTTCATACTTAGACTGTCGAT-3′; for PR-3, 5′-CGGTGGTACTCCTCCTGGACCCACCGGC-3′ and 5′-CGGCGGCACGGTCGGCGTCTGAAGGCTG-3′; for PR-4, 5′-GACAACAATGCGGTCGTCAAGG-3′ and 5′-AGCATGTTTCTGGAATCAGGCTGCC-3′; for PR-5, 5′-ATGGCAAATATCTCCAGTATTCACA-3′ and 5′-ATGTCGGGGCAAGCCGCGTTGAGG-3′; for PDF1.2, 5′-GCTAAGTTTGCTTCCATCATCACCCTT-3′ and 5′-AACATGGGACGTAACAGATACACTTGTG-3′; for EIN2, 5′-TGCAGCTCGCATAAGCGTTGTGACTGGTA-3′ and 5′-CGCTCTCTCCATTTAACCGAGTTAACAC-3′; for EIN3, 5′-GATGTTGATGAATTGGAGAGGAGGATG-3′ and 5′-ACGTCTCTGAGGAGGATCACAGTGT-3′; for ERF1, 5′-CGGCTTCTCACCGGAATATTCTATCG-3′ and 5′-TCTCCGAAAGCGACTCTTGAACTCTCT-3′; and for Actin1, 5′-GGCGATGAAGCTCAATCCAAACG-3′ and 5′-GGTCACGACCAGCAAGATCAAGACG-3′.
Assays of Endogenous Enzyme Activities
Endogenous ADH and G6PDH activities were measured as described (Ranieri et al., 1996; Chung and Ferl, 1999). Proteins were extracted in extraction buffer (50 mM Tris, pH 8.0, and 15 mM DTT) and centrifuged at 12,000g for 15 min at 4°C. The reaction mixture for ADH contained 50 mM Tris, pH 9.0, 0.867 mM NAD+, 20% ethanol, and aliquots of protein extracts. The reaction for G6PDH was performed in 86.3 mM triethanolamine, pH 7.6, 6.7 mM MgCl2, 0.37 M NADP+, 12 mM glucose-6-phosphate, and aliquots of protein extracts. The increase in absorbance at 340 nm was monitored every 15 s for 60 s. The activities were expressed as micromoles of NAD+ or NADP+ reduced per minute per milligram of protein.
Subcellular Localization of GLIP1
The DNAs of GLIP1, GLIP1ΔSP, and SP were generated by PCR using the following primers with the sequences for cloning sites XbaI and XhoI (underlined): for GLIP1, 5′-GATCTCTAGAATGGAAAACTCTCAATTAG-3′ and 5′-GATCCTCGAGATTAAGTTCAAACAGCGC-3′; for GLIP1ΔSP, 5′-GATCTCTAGAATGGACAACAATAATCTTGTA-3′ and 5′-GATCCTCGAGATTAAGTTCAAACAGCGC-3′; and for SP, 5′-GATCTCTAGAATGGAAAACTCTCAATTAG-3′ and 5′-CCGCTCGAGGACACTAGAGAGAGTATC-3′. The amplified DNAs were cloned into the XbaI and XhoI sites of the pBI221 vector (Clontech) so that smGFP was fused in frame to the C termini of the DNA products. The resulting constructs were introduced into onion (Allium cepa) epidermal cells using a helium biolistic particle delivery system (Bio-Rad) as described by Shieh et al. (1993). After incubation for 12 to 48 h at 23°C, the subcellular distribution of the GFP fusion proteins was examined with a fluorescence microscope (Olympus BX-51TRF).
Purification of GLIP1 and Lipase Assay
The full-length coding region of GLIP1 was cloned into the pGEX 6P-1 vector (Amersham Biosciences). The GLIP1(S44A) mutant was generated by mutagenic PCR using the following primers with one mismatch (lowercase): for sense, 5′-TCGTGTTTGGAGATgCTGTGTTCGATGCTGGA-3′; for antisense, 5′-TCCAGCATCGAACACAGcATCTCCAAACACGA-3′. Escherichia coli BL21(DE3) cells transformed with constructs were cultured, and the expression of the recombinant proteins was induced by the addition of 0.1 mM isopropyl β-d-thiogalactopyranoside for 30 min. The GST-GLIP1 fusion proteins were purified according to the manufacturer's instructions, dialyzed against PBS, and stored at −70°C before use. Lipase activity was measured as described (Baudouin et al., 1997). The enzyme reaction mixture contained 0.5 M HEPES, pH 6.5, 1 mM substrate (p-nitrophenyl acetate or p-nitrophenyl butyrate), and aliquots of recombinant proteins (2 to 4 μg) and was incubated for 60 min at 30°C. Absorbance was measured at 405 nm every 5 min for 60 min.
Microscopic Analyses
The lesions and fungal hyphae were visualized by staining the infected leaves according to a previously described method (Kumar et al., 2004). Leaves detached 4 d after inoculation were stained with lactophenol–aniline blue. Lactophenol was prepared by mixing phenol, acetic acid, distilled water, and glycerol at a ratio of 1:1:1:2. Aniline blue (0.1%) and lactophenol were then mixed at a ratio of 1:3. Cell death was detected by lactophenol–trypan blue staining followed by destaining in saturated chloral hydrate as described (Koch and Slusarenko, 1990). Stained samples were mounted in 60% glycerol and examined using a light microscope (Olympus BX-51TRF).
The antimicrobial activity of GLIP1 was determined by adding the recombinant proteins (0.1 to 5 μg) to 10 μL of spore suspension (5 × 105 spores/mL). The samples were placed on glass slides, incubated for 24 h at 23°C, and observed by light microscopy. Fungi samples were prepared for scanning electron microscopy by fixation in fixing solution (0.1 M sodium phosphate, pH 7.0, 1% sucrose, 3% paraformaldehyde, and 0.2% glutaraldehyde) under vacuum for 6 h, dehydration through a graded ethanol series, and critical point drying. The samples were sputter-coated with gold and viewed using a S-2500 scanning electron microscope (Hitachi).
Accession Numbers
Arabidopsis Genome Initiative locus numbers for the genes described in this work are as follows: GLIP1 (At5g40990), GLIP2 (At1g53940), GLIP3 (At1g53990), GLIP4 (At3g14225), GLIP5 (At1g53920), GLIP6 (At1g71120), and GLIP7 (At5g15720). Other accession numbers are listed in Table 2.
Supplementary Material
Acknowledgments
We are grateful to Atle Bones for generously providing the anti-myrosinase antibody. We also thank Eun Young Choi, Eun Ju Cho, Won Hyun Song, and Mi-Ok Han for technical assistance. This research was supported by grants from the Plant Signaling Network Research Center funded by the Korea Science and Engineering Foundation and the BioGreen 21 Program funded by the Rural Development Administration, Republic of Korea. This article is Kumho Life and Environmental Science Laboratory Publication 81.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ohkmae K. Park (omkim@korea.ac.kr).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.034819.
References
- Alvarez, M.E., Pennell, R.I., Meijer, P.J., Ishikawa, A., Dixon, R.A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. [DOI] [PubMed] [Google Scholar]
- Bae, M.S., Cho, E.J., Choi, E.-Y., and Park, O.K. (2003). Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 36, 652–663. [DOI] [PubMed] [Google Scholar]
- Barondes, S.H., Cooper, D.N., Gitt, M.A., and Leffler, H. (1994). Galectins: Structure and function of a large family of animal lectins. J. Biol. Chem. 269, 20807–20810. [PubMed] [Google Scholar]
- Baudouin, E., Charpenteau, M., Roby, D., Marco, Y., Ranjeva, R., and Ranty, B. (1997). Functional expression of a tobacco gene related to the serine hydrolase family—Esterase activity towards short-chain dinitrophenyl acylesters. Eur. J. Biochem. 248, 700–706. [DOI] [PubMed] [Google Scholar]
- Berto, P., Commenil, P., Belingheri, L., and Dehorter, B. (1999). Occurrence of a lipase in spores of Alternaria brassicicola with a crucial role in the infection of cauliflower leaves. FEMS Microbiol. Lett. 180, 183–189. [DOI] [PubMed] [Google Scholar]
- Brick, D.J., Brumlik, M.J., Buckley, J.T., Cao, J.X., Davies, P.C., Misra, S., Tranbarger, T.J., and Upton, C. (1995). A new family of lipolytic plant enzymes with members in rice, Arabidopsis and maize. FEBS Lett. 377, 475–480. [DOI] [PubMed] [Google Scholar]
- Bryckaert, M., Guillonneau, X., Hecquet, C., Perani, P., Courtois, Y., and Mascarelli, F. (2000). Regulation of proliferation-survival decisions is controlled by FGF1 secretion in retinal pigmented epithelial cells. Oncogene 19, 4917–4929. [DOI] [PubMed] [Google Scholar]
- Chivasa, S., Ndimba, B.K., Simon, W.J., Robertson, D., Yu, X.L., Knox, J.P., Bolwell, P., and Slabas, A.R. (2002). Proteomic analysis of the Arabidopsis thaliana cell wall. Electrophoresis 23, 1754–1765. [DOI] [PubMed] [Google Scholar]
- Cho, J.W., Park, S.C., Shin, E.A., Kim, C.K., Han, W., Sohn, S.I., Song, P.S., and Wang, M.H. (2004). Cyclin D1 and p22ack1 play opposite roles in plant growth and development. Biochem. Biophys. Res. Commun. 324, 52–57. [DOI] [PubMed] [Google Scholar]
- Chung, H.-J., and Ferl, R.J. (1999). Arabidopsis alcohol dehydrogenase expression in both shoots and roots is conditioned by root growth environment. Plant Physiol. 121, 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute, I., Beynon, J., Dangl, J., Holub, E., Mauch-Mani, B., Slusarenko, A., Staskawicz, B., and Ausubel, F. (1994). Microbial pathogenesis of Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 705–747.
- De Lorenzo, G., and Ferrari, S. (2002). Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 5, 295–299. [DOI] [PubMed] [Google Scholar]
- Dong, X. (1998). SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1, 316–323. [DOI] [PubMed] [Google Scholar]
- Dürner, J., Shah, J., and Klessig, D.F. (1997). Salicylic acid and disease resistance in plants. Trends Plant Sci. 2, 266–274. [Google Scholar]
- Eddine, A.N., Hannemann, F., and Schafer, W. (2001). Cloning and expression analysis of NhL1, a gene encoding an extracellular lipase from the fungal pea pathogen Nectria haematococca MP VI (Fusarium solani f. sp. pisi) that is expressed in planta. Mol. Genet. Genomics 265, 215–224. [DOI] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Olmedo, F., Molina, A., Alamillo, J.M., and Rodriguez-Palenzuela, P. (1998). Plant defense peptides. Biopolymers 47, 479–491. [DOI] [PubMed] [Google Scholar]
- Heinlein, M. (2002). Plasmodesmata: Dynamic regulation and role in macromolecular cell-to-cell signaling. Curr. Opin. Plant Biol. 5, 543–552. [DOI] [PubMed] [Google Scholar]
- Husebye, H., Chadchawan, S., Winge, P., Thangstad, O.P., and Bones, A.M. (2002). Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128, 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab, G., Manrique, A., Zimmerli, L., Metraux, J.P., and Mauch-Mani, B. (2003). Molecular characterization of a novel lipase-like pathogen-inducible gene family of Arabidopsis. Plant Physiol. 132, 2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan, I.A., and Hammerschmidt, R. (2002). Arabidopsis ecotype variability in camalexin production and reaction to infection by Alternaria brassicicola. J. Chem. Ecol. 28, 2121–2140. [DOI] [PubMed] [Google Scholar]
- Kersten, B., Burkle, L., Kuhn, E.J., Giavalisco, P., Konthur, Z., Lueking, A., Walter, G., Eickhoff, H., and Schneider, U. (2002). Large-scale plant proteomics. Plant Mol. Biol. 48, 133–141. [PubMed] [Google Scholar]
- Kimura, Y., Tosa, Y., Shimada, S., Sogo, R., Kusaba, M., Sunaga, T., Betsuyaku, S., Eto, Y., Nakayashiki, H., and Mayama, S. (2001). OARE-1, a Ty1-copia retrotransposon in oat activated by abiotic and biotic stresses. Plant Cell Physiol. 42, 1345–1354. [DOI] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A.J. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruft, V., Eubel, H., Jansch, L., Werhahn, W., and Braun, H.P. (2001). Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Kuchler, K. (1993). Unusual routes of protein secretion: The easy way out. Trends Cell Biol. 3, 421–426. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Tripathi, K., Rana, M., Purwar, S., and Garg, G.R. (2004). Dibutyryl c-AMP as an inducer of sporidia formation: Biochemical and antigenic changes during morphological differentiation of Karnal bunt (Tilletia indica) pathogen in axenic culture. J. Biosci. 29, 23–31. [DOI] [PubMed] [Google Scholar]
- Kumar, D., and Klessig, D.F. (2003). High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc. Natl. Acad. Sci. USA 100, 16101–16106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Kus, J.V., Zaton, K., Sarkar, R., and Cameron, R.K. (2002). Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. Plant Cell 14, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Lee, E.J., Yang, E.J., Lee, J.E., Park, A.R., Song, W.H., and Park, O.K. (2004). Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 16, 1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottaz, D., Beleznay, Z., and Bickel, M. (2001). Inhibition of ATP-binding cassette transporter downregulates interleukin-1beta-mediated autocrine activation of human dermal fibroblasts. J. Invest. Dermatol. 117, 871–876. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1980–1983. [DOI] [PubMed] [Google Scholar]
- MacKenzie, A., Wilson, H.L., Kiss-Toth, E., Dower, S.K., North, R.A., and Surprenant, A. (2001). Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity 15, 825–835. [DOI] [PubMed] [Google Scholar]
- Maldonado, A.M., Doerner, P., Dixon, R.A., Lamb, C.J., and Cameron, R.K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399–403. [DOI] [PubMed] [Google Scholar]
- Mann, K., Farias, C.M., Del Sol, F.G., Santos, C.F., Grangeiro, T.B., Nagano, C.S., Cavada, B.S., and Calvete, J.J. (2001). The amino-acid sequence of the glucose/mannose-specific lectin isolated from Parkia platycephala seeds reveals three tandemly arranged jacalin-related domains. Eur. J. Biochem. 268, 4414–4422. [DOI] [PubMed] [Google Scholar]
- May, M.J., and Leaver, C.J. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRoberts, N., and Lennard, J.H. (1996). Pathogen behaviour and plant cell reactions in interactions between Alternaria species and leaves of host and nonhost plants. Plant Pathol. 45, 742–752. [Google Scholar]
- Millar, A.H., Sweetlove, L.J., Giege, P., and Leaver, C.J. (2001). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127, 1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Nakajima, K., Sena, G., Nawy, T., and Benfey, P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311. [DOI] [PubMed] [Google Scholar]
- Ndimba, B.K., Chivasa, S., Hamilton, J.M., Simon, W.J., and Slabas, A.R. (2003). Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 3, 1047–1059. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Okushima, Y., Koizumi, N., Kusano, T., and Sano, H. (2000). Secreted proteins of tobacco cultured BY2 cells: Identification of a new member of pathogenesis-related proteins. Plant Mol. Biol. 42, 479–488. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J. (2004). Getting the message across: How do plant cells exchange macromolecular complexes? Trends Plant Sci. 9, 33–41. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., and Cruz, S.S. (2000). The great escape: Phloem transport and unloading of macromolecules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 323–347. [DOI] [PubMed] [Google Scholar]
- Pandey, A., and Mann, M. (2000). Proteomics to study genes and genomes. Nature 405, 837–846. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Friso, G., Kalume, D.E., Roepstorff, P., Nilsson, F., Adamska, I., and van Wijk, K.J. (2000). Proteomics of the chloroplast: Systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12, 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell, R. (1998). Cell walls: Structures and signals. Curr. Opin. Plant Biol. 1, 504–510. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A., Eggermont, K., Terras, F.R., Thomma, B.P., De Samblanx, G.W., Buchala, A., Metraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Thomma, B.P., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M., and van Loon, L.C. (1999). Salicylic acid-independent plant defence pathways. Trends Plant Sci. 4, 52–58. [DOI] [PubMed] [Google Scholar]
- Prime, T., Sherrier, D., Mahon, P., Packman, L., and Dupree, P. (2000). A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21, 3488–3499. [DOI] [PubMed] [Google Scholar]
- Ranieri, A., D'Urso, G., Nali, C., Lorenzini, G., and Soldatini, G.F. (1996). Ozone stimulates apoplastic antioxidant systems in pumpkin leaves. Physiol. Plant. 97, 381–387. [Google Scholar]
- Rouquie, D., Peltier, J.B., Marquis-Mansion, M., Tournaire, C., Doumas, P., and Rossignol, M. (1997). Construction of a directory of tobacco plasma membrane proteins by combined two-dimensional gel electrophoresis and protein sequencing. Electrophoresis 18, 654–660. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A., Pearce, G., Scheer, J., and Moura, D.S. (2002). Polypeptide hormones. Plant Cell 14 (suppl.), S251–S264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Manners, J.M., Anderson, J.P., Simpson, R.S., Wilson, I.W., Somerville, S.C., and Maclean, D.J. (2003). Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. 132, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selitrennikoff, C.P. (2001). Antifungal proteins. Appl. Environ. Microbiol. 67, 2883–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., Yanofsky, M.F., and Weigel, D. (2000). Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289, 779–782. [DOI] [PubMed] [Google Scholar]
- Shieh, M.W., Wessler, S.R., and Raikhel, N.V. (1993). Nuclear targeting of the maize R protein requires two nuclear localization sequences. Plant Physiol. 101, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter, A.M. (1993). Structure and function of plant cell wall proteins. Plant Cell 5, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siever, D.A., and Erickson, H.P. (1997). Extracellular annexin II. Int. J. Biochem. Cell Biol. 29, 1219–1223. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P., Nelissen, I., Eggermont, K., and Broekaert, W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19, 163–171. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Penninckx, I.A.M.A., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk, K.J. (2001). Challenges and prospects of plant proteomics. Plant Physiol. 126, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K.L., Li, H., and Ecker, J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 (suppl.), S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S.R. (1996). Turned on by stress: Plant retrotransposons. Curr. Biol. 6, 959–961. [DOI] [PubMed] [Google Scholar]
- Wright, D.A., and Voytas, D.F. (1998). Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics 149, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, E.J., Oh, Y.A., Lee, E.S., Park, A.R., Cho, S.K., Yoo, Y.J., and Park, O.K. (2003). Oxygen-evolving enhancer protein 2 is phosphorylated by glycine-rich protein 3/wall-associated kinase 1 in Arabidopsis. Biochem. Biophys. Res. Commun. 305, 862–868. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Peumans, W.J., Barre, A., Houles-Astoul, C., Rovira, P., Rougé, P., Proost, P., Truffa-Bachi, P., Jalali, A.A.H., and Van Damme, E.J.M. (2000). Isolation and characterization of jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta 210, 970–978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.