Abstract
The transmembrane protein Notch is cleaved by γ-secretase to yield an active form, Notch intracellular domain (Notch-IC), in response to the binding of ligands, such as Jagged. Notch-IC contributes to the regulation of a variety of cellular events, including cell fate determination during embryonic development as well as cell growth, differentiation, and survival. We now show that Notch1-IC suppresses the scaffold activity of c-Jun N-terminal kinase (JNK)-interacting protein 1 (JIP1) in the JNK signaling pathway. Notch1-IC physically associated with the JNK binding domain of JIP1 and thereby interfered with the interaction between JIP1 and JNK. JIP1 mediated the activation of JNK1 induced by glucose deprivation in mouse embryonic fibroblasts, and ectopic expression of Notch1-IC inhibited JNK activation and apoptosis triggered by glucose deprivation. Taken together, these findings suggest that Notch1-IC negatively regulates the JNK pathway by disrupting the scaffold function of JIP1.
Keywords: scaffold, apoptosis, glucose deprivation
Signaling by Notch is associated with a variety of cellular events, including cell fate control during embryonic development, differentiation, cell growth, and apoptosis (1). Presenilin (PS) facilitates Notch signaling (2-7). PS-activated γ-secretase thus mediates the processing of Notch1 into an active form, Notch1 intracellular domain (Notch1-IC), and the generation of Notch1-IC is inhibited in PS1-deficient cells (5). Evidence has suggested that the biological function of Notch is completely dependent on its cleavage to Notch-IC, which then translocates to the nucleus and, together with the transcription factor CBF1/RBP-Jκ, activates the transcription of target genes (1, 8). The existence of a transcription-independent function of Notch in the cytoplasm has also been suggested, however, although the cytoplasmic targets of Notch remain unclear (9-12).
Mitogen-activated protein kinase (MAPK) pathways also play important roles in a variety of cellular processes, including cell proliferation, differentiation, and death (13-15). Mammalian MAPKs are classified into three subfamilies: extracellular signal-regulated kinase, c-Jun N-terminal kinase (JNK), or stress-activated protein kinase (SAPK), and p38 MAPK (16-21). Each MAPK pathway includes three distinct components: a MAPK, a MAPK kinase (MAP2K), and a MAPK kinase kinase (MAP3K). MAP3Ks phosphorylate and activate MAP2Ks, which in turn phosphorylate and activate MAPKs. Activated MAPKs phosphorylate various substrate proteins, including transcription factors, and thereby regulate gene expression and other cellular functions. The JNK/SAPK signaling pathway is activated in response to exposure of cells to a variety of cellular stresses or to proinflammatory cytokines, such as TNF-α (13, 14). This pathway consists of JNK/SAPK, a MAP2K, such as SEK1 (also known as MKK4 or JNKK1) or MKK7, and a MAP3K, such as mixed lineage kinase (MLK)3 or MEKK1. Although these components of the JNK pathway transmit their signals through a series of binary interactions, some of them exist in signaling complexes whose formation is mediated by scaffold proteins, such as JNK-interacting protein 1 (JIP1) (22-26). JIP1 physically interacts with JNK, MKK7, and members of the MLK group of MAP3Ks (24), and it enhances the selectivity and effectiveness of kinase activation during JNK signaling (27).
We have shown previously that PS1 inhibits the JNK pathway in a manner dependent on its γ-secretase-activating function (28), suggesting that a processing product of a cellular substrate of γ-secretase might suppress activation of this pathway. In this regard, previous studies have proposed that Notch might negatively regulate the JNK pathway (11, 12). We have now investigated the mechanism for the regulation of the JNK pathway by Notch1. Our data show that Notch inhibits this pathway and that this effect is mediated through physical interaction of Notch1-IC with JIP1 and consequent prevention of JIP1-mediated activation of JNK.
Materials and Methods
DNA Constructs. Complementary DNAs encoding deletion mutants of murine Notch1 (mNotch1) were generated by PCR and include mNotch1 containing the extracellular cysteine-rich LIN-12/Notch/GLP repeats (mNotch1-LNG) (amino acids 1444-2480), mNotch1 lacking the extracellular domain (mNotch1-ΔE) (amino acids 1701-2480), or mNotch1-IC (amino acids 1747-2480). The amplification products were digested with SmaI and EcoRV and then cloned into the EcoRV site of the pEF-BOS-myc vector. Vector constructs encoding the kinase components of the JNK pathway are described in refs. 28-31.
Cell Culture and Apoptosis Assay. HEK293 cells, Jurkat T cells, and mouse embryonic fibroblasts (MEFs) from PS1+/+ or PS1-/- mice (32) or from JIP1+/+ or JIP1-/- mice (33) were maintained at 37°C in DMEM supplemented with 10% FBS. MEFPS1(+/+) and MEFPS1(-/-) cells at the passage from four to six cells and MEFJIP1(+/+) and MEFJIP1(-/-) cells at the passage from six to 10 cells were used, and each pair of the MEF cells were in the same passage at the experiments. Cultured cells were transfected by the calcium phosphate method or with the use of a GenePorter II system (Gene Therapy Systems, San Diego). For measurement of apoptosis, cells were transfected for 48 h with the indicated vector constructs plus pEGFP (BD Biosciences, Franklin Lakes, NJ). The transfected cells were incubated for 4 h in serum-free DMEM or serum/glucose-free DMEM and then for an additional 24 h in complete medium. The cells were then fixed with 4% formaldehyde, stained with 10 μg/ml DAPI, and examined for apoptotic nuclear morphology with the use of a Zeiss Axiovert fluorescence microscope. The proportion of apoptotic nuclei among cells expressing GFP was determined. More than 200 cells were counted in each experiment, and results from three independent experiments were analyzed. Alternatively, cells were analyzed for apoptosis by TUNEL staining with in situ cell death detection kit (Roche Applied Science, Indianapolis).
For induction of Notch1 signaling by Jagged1, a soluble form of Myc-tagged Jagged1 was immobilized on culture dishes that had been coated with a mouse monoclonal antibody to Myc as described in ref. 34. The dishes were then used for studies of Jagged1-induced Notch1 signaling.
Immune Complex Kinase Assay. Cultured cells were lysed in a lysis buffer (35), and the cell lysates were subjected to immunoprecipitation with appropriate antibodies (30, 36). The resulting precipitates were assayed for JNK activity with the use of GST-c-Jun (1-79) as substrate (36). The phosphorylated substrate was resolved by SDS/PAGE on a 10% gel and analyzed with a Fuji BAS-2500 phosphorimager. The cell lysates were also subjected directly to immunoblot analysis probed with the indicated antibodies. Immunoreactive bands were visualized with the use of enhanced chemiluminescence reagents (Amersham Biosciences).
Coimmunoprecipitation and in Vitro Binding Analyses. Coimmunoprecipitation analysis was performed as described in ref. 37. For in vitro binding experiments, HEK293 cells that had been transfected for 48 h with pcDNA3-FLAG or pcDNA3-JIP1-FLAG were subjected to immunoprecipitation with antibodies to FLAG. mNotch1-IC and its deletion variants were produced by in vitro translation in the presence of [35S]methionine with the use of a Quick Coupled TnT kit (Promega). The 35S-labeled mNotch-IC variants were then incubated for 1 h at 4°C with JIP1-FLAG or control immunoprecipitates in a binding solution (37). The precipitates were recovered and then analyzed by SDS/PAGE and autoradiography.
Luciferase Activity Assay. HEK293 cells were cotransfected for 24 h with pGL2-3X-CBF1-luciferase and pcDNA3-β-galactosidase together with the indicated vector constructs. Cells were then unexposed or exposed to UV (80 J/m2), incubated for 6 h at 37°C, and lysed. Cells lysates were assayed for luciferase activity with the use of a luciferase assay kit (Promega). Luciferase activity in each sample was normalized relative to the β-galactosidase activity in the same sample.
Results
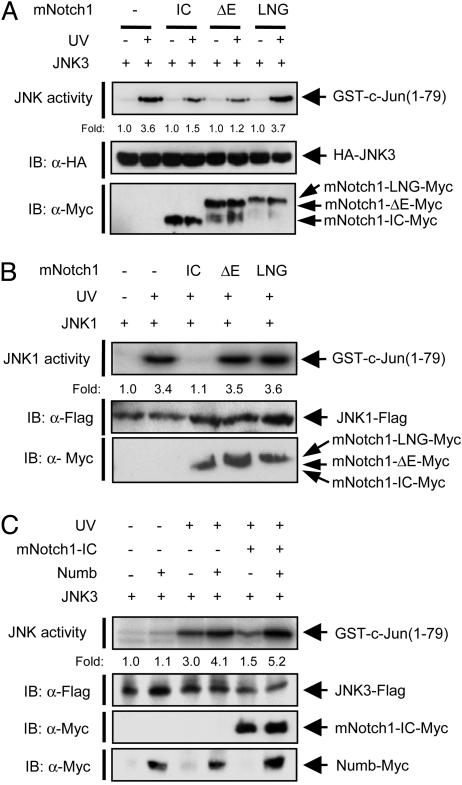
Notch1 Inhibits Activation of the JNK Pathway. To investigate the possible effect of Notch1 on the JNK pathway, we transfected HEK293 cells with an expression vector encoding hemagglutinin epitope (HA)-tagged JNK3/SAPKβ alone or together with vectors for various Myc epitope-tagged mNotch1 variants. Exposure of the cells to UV light induced activation of JNK3 (Fig. 1A). Coexpression of mNotch1-IC resulted in inhibition of UV-induced JNK3 activation. A form of mNotch1 lacking the extracellular domain (mNotch1-ΔE) also inhibited this effect of UV irradiation. Immunoblot analysis showed that a proportion of the expressed mNotch1-ΔE was processed to mNotch1-IC, presumably by an endogenous γ-secretase activity in the cells (Fig. 1 A), as suggested in ref. 38. In contrast, a form of mNotch1 containing the extracellular cysteine-rich LIN-12/Notch/GLP repeats (mNotch1-LNG), which is not converted to mNotch1-IC by γ-secretase (38), failed to inhibit UV-induced JNK3 activation (Fig. 1 A).
Fig. 1.
Notch1 inhibits activation of the JNK signaling pathway. (A) HEK293 cells were transfected for 50 h with an expression vector encoding HA-JNK3/SAPKβ alone or together with vectors for Myc-epitope-tagged mNotch1 variants (mNotch1-IC, mNotch1-ΔE, and mNotch1-LNG), as indicated. The cells were then unexposed or exposed to UV irradiation (80 J/m2), incubated for an additional 30 min, and lysed. Cell lysates were subjected to immunoprecipitation with antibodies to HA, and the resulting precipitates were examined for JNK activity by an immune complex kinase assay. The numbers below each lane indicate fold increase in JNK activity relative to the value for the corresponding nonirradiated cells. The amounts of the recombinant proteins in cell lysates were examined by immunoblot (IB) analysis with the indicated antibodies. (B) MEFs from PS1-/- mice were transfected for 50 h with an expression vector encoding JNK1-FLAG alone or together with vectors for Myc-epitope-tagged mNotch1-IC variants as indicated, exposed to UV, and analyzed as in A with the exception that immunoprecipitation was performed with antibodies to FLAG. (C) HEK293 cells were transfected for 50 h with a vector encoding JNK3-FLAG alone or together with vectors for mNotch1-IC-Myc and Numb-Myc as indicated, exposed to UV, and analyzed as in B.
We next examined the effects of these mNotch1 variants on UV-induced JNK activation in MEFs from PS1 knockout mice, in which the γ-secretase-mediated processing of endogenous Notch1 is impaired compared with that in MEFs from PS1+/+ mice (5). Ectopic expression of mNotch1-IC in PS1-/- MEFs resulted in inhibition of UV-induced activation of JNK1 (Fig. 1B). In contrast, neither mNotch1-ΔE nor mNotch1-LNG affected JNK activation. Taken together, these results suggested that mNotch1-IC, an active form of mNotch1, inhibits activation of the JNK pathway.
We then investigated the effect of Numb, a negative regulator of Notch1, on Notch1-mediated JNK inhibition. Numb antagonizes Notch1 signaling through direct interaction with Notch1-IC (39). The inhibitory effect of mNotch1-IC on UV-induced activation of JNK3 (Fig. 1C) or JNK1 (data not shown) was prevented by coexpression of Numb.
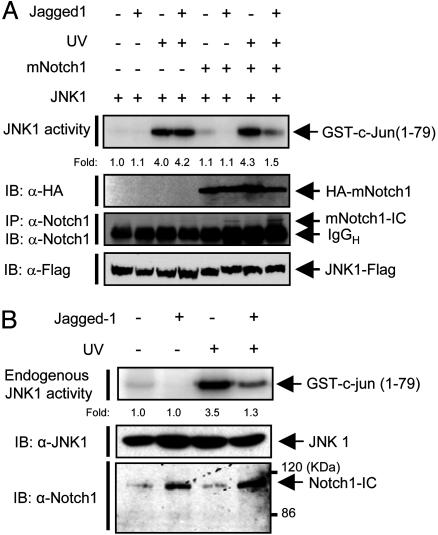
Jagged1-Induced Processing of Notch1 to Notch1-IC Results in Inhibition of JNK Activation. The γ-secretase-mediated Notch processing depends on the binding of Notch ligands, such as Jagged (40-42). We therefore examined the effect of Jagged1-induced processing of Notch1 on JNK activation. HEK293 cells were transfected with a vector for JNK1-FLAG alone or together with a vector for mNotch1, and the cells were then unexposed or exposed to Jagged1. Jagged1 induced processing of mNotch1 to mNotch1-IC as well as inhibition of UV-stimulated JNK1 activity in the mNotch1-transfected cells (Fig. 2A). In comparison, Jagged1 did not affect UV-stimulated JNK1 activity in cells that had not been transfected with the mNotch1 vector. We next examined whether the Jagged1-induced activation of endogenous Notch1 by cell-cell interaction prevents JNK activation (Fig. 2B). Jurkat T cells were incubated for 12 h with HEK293 cells that had been transfected with a vector for Jagged1 or with an empty control vector. The interaction between Jurkat T cells and Jagged1-expressing HEK293 cells induced the processing of endogenous Notch1 to Notch1-IC as well as inhibition of the UV-stimulated activity of endogenous JNK1 in Jurkat T cells.
Fig. 2.
Jagged1 induces the inhibitory action of Notch1 on JNK activation. (A) HEK293 cells were transfected for 24 h with a vector encoding JNK1-FLAG alone or along with a vector for HA-tagged full-length mNotch-1. The cells were then transferred to control culture dishes or to dishes containing immobilized Jagged1. After culture for 24 h, the cells were unexposed or exposed to UV light (80 J/m2), incubated for an additional 30 min at 37°C, and lysed. Cell lysates were subjected to immunoprecipitation with antibodies to FLAG, and the resulting immunoprecipitates were assayed for JNK1 activity with an immune complex kinase assay. Cell lysates were also subjected to immunoblot analysis with antibodies to HA and to FLAG. The processing of full-length Notch1 into mNotch1-IC was monitored by immunoprecipitation (IP) and immunoblot (IB) analysis of cell lysates with antibodies to Notch1. (B) HEK293 cells were transfected for 40 h with a vector for Jagged1 or an empty vector. The transfected HEK293 cells were then incubated with Jurkat T cells. Jurkat T cells in suspension were collected, transferred to new culture dishes, and then untreated or treated with UV light (80 J/m2). The lysates of Jurkat cells were subjected to immunoprecipitation with anti-JNK1 antibody, and the resulting precipitates were assayed for JNK1 activity. Cell lysates were also subjected directly to immunoblot analysis with antibodies to JNK1 and to Notch1 (Santa Cruz Biotechnology).
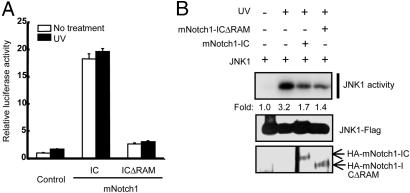
CBF1-Dependent Transcription Is Not Required for Inhibition of JNK Signaling by Notch1. Notch1-IC associates with CBF1 and translocates to the nucleus to activate transcription of target genes (1, 40, 43). We therefore examined whether transcriptional activation by Notch1-IC is required for the inhibitory action of Notch1-IC on the JNK pathway. The CBF1-dependent expression of a luciferase reporter gene was enhanced by mNotch1-IC but not by a mNotch1-IC mutant (mNotch1-ICΔRAM) that lacks the CBF1-interacting RAM domain (Fig. 3A). In contrast, mNotch1-IC and mNotch1-ICΔRAM inhibited the UV-induced activation of JNK1 (Fig. 3B). These results suggested that mNotch1-IC inhibits JNK activation by a pathway independent of CBF1-mediated signaling.
Fig. 3.
Inhibition of JNK activation by Notch1-IC is independent of CBF1-mediated signaling. (A) HEK293 cells were transfected for 24 h with pGL2-3X-CBF1-luciferase and pcDNA3-β-galactosidase together with vectors for HA-mNotch1-IC or HA-mNotch1-ICΔRAM, as indicated. The cells were then unexposed or exposed to UV light (80 J/m2), incubated for an additional 6 h, and lysed. Cell lysates were assayed for luciferase activity, which was normalized by the corresponding β-galactosidase activity. Data are means ± average deviation of triplicates from a representative experiment. (B) HEK293 cells were transfected for 48 h with expression vectors for JNK1-Flag, HA-mNotch1-IC, and HA-mNotch1-ICΔRAM, as indicated. The cells were then unexposed or exposed to UV light (80 J/m2), incubated for an additional 30 min at 37°C, and lysed. Cell lysates were subjected to immunoprecipitation with anti-FLAG antibody, and the resulting precipitates were assayed for JNK1 activity with an immune complex kinase assay. Cell lysates were also subjected to immunoblot analysis with antibodies to FLAG and to HA.
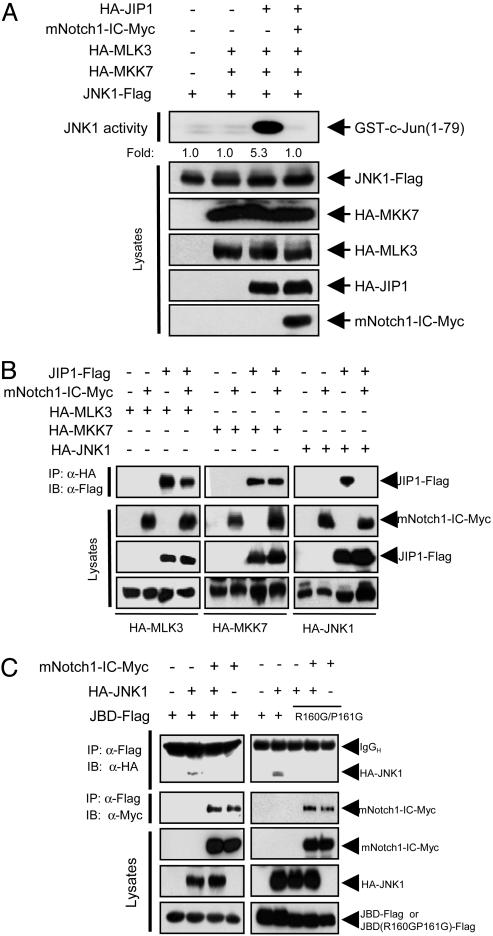
Notch1-IC Interferes with the Scaffold Function of JIP1 in the JNK Pathway. To characterize further the mechanism underlying the inhibitory action of mNotch1-IC on activation of the JNK pathway, we examined whether mNotch1-IC affects the function of JIP1. JIP1 physically associates with MLK3, MKK7, and JNK and, thus, functions as a scaffold protein that facilitates JNK activation by MLK3-MKK7-JNK signaling (22, 26, 44). Coexpression of JIP1 increased the kinase activity of ectopic JNK1 in HEK293 cells that had been cotransfected with vectors for MLK3, MKK7, and JNK1 (Fig. 4A). This scaffold function of JIP1 was inhibited by mNotch1-IC. We then examined whether mNotch1-IC could alter the binding of JIP1 to MLK3, MKK7, or JNK1. Coimmunoprecipitation analysis revealed that mNotch1-IC did not affect the interaction between JIP1 and MKK7 and only slightly inhibited that between JIP1 and MLK3 (Fig. 4B). In contrast, mNotch1-IC markedly inhibited the association between JIP1 and JNK1. JIP1 binds to JNK through its JNK-binding domain (JBD) (22). We therefore examined the effect of mNotch1-IC on the interaction between JNK1 and the JBD of JIP1 in transfected HEK293 cells. Coimmunoprecipitation analysis showed that the JBD of JIP1 physically associated with JNK1 in the transfected cells and that this interaction was inhibited by mNotch1-IC (Fig. 4C). Furthermore, mNotch1-IC coprecipitated with the JBD of JIP1. Introduction of point mutations in JBD of JIP1 at Arg-160 and Pro-161 has been shown to disrupt the interaction between JIP1 and JNK (45). Indeed, JNK1 coimmunoprecipitated with JBD but not with JBD(R160G/P161G) (Fig. 4C). Interestingly, this JBD mutant still coimmunoprecitated with mNotch1-IC, and this interaction was not affected by coexpression of JNK1.
Fig. 4.
Inhibition of the scaffold activity of JIP1 by Notch1-IC. (A) HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors for HA-MLK3, HA-MKK7, JNK1-FLAG, HA-JIP1, and mNotch1-IC-Myc. Cell lysates were then subjected to immunoprecipitation with anti-FLAG antibody, and the resulting precipitates were assayed for JNK1 activity with an immune complex kinase assay. Cell lysates were also subjected to immunoblot analysis with the indicated antibodies. (B) HEK239 cells were transfected for 48 h with the indicated combinations of expression vectors for HA-MLK3, HA-MKK7, HA-JNK1, JIP1-FLAG, and mNotch1-IC-Myc. Cell lysates were then subjected to immunoprecipitation (IP) with anti-HA antibody, and the resulting precipitates were subjected to immunoblot (IB) analysis with anti-FLAG antibody. Cell lysates were also subjected directly to immunoblot analysis with antibodies to HA, FLAG, and Myc. (C) HEK293 cells were transfected for 50 h with a vector encoding JBD-FLAG or JBD(R160G/P161G) together with vectors for HA-JNK1 and mNotch1-IC-Myc, as indicated. Cell lysates were subjected to immunoprecipitation with anti-FLAG antibody, and the resulting precipitates were subjected to immunoblot analysis with antibodies to HA or to Myc. Cell lysates were also subjected directly to immunoblot analysis with antibodies to Myc, HA, or FLAG. IgGH, heavy chain of IgG.
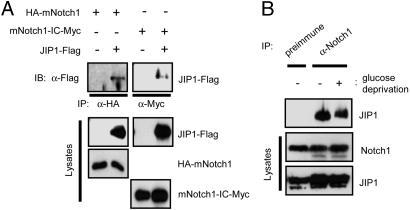
Notch1-IC Physically Interacts with JIP1 in Vitro and in Intact Cells. Next, we further investigated the physical interaction between mNotch1-IC and JIP1. In vitro binding analysis with in vitro-translated S35-labeled mNotch1-IC variants revealed that mNotch1-IC indeed interacted with JIP1 and that a central region (amino acids 1898-2197) of mNotch1-IC was important for this binding to JIP1 (data not shown). Physical interaction between mNotch1 and JIP1 in intact cells was also examined by coimmunoprecipitation analysis of HEK293 cells transfected with various combinations of expression vectors for JIP1-FLAG, HA-mNotch1, and mNotch1-IC-Myc. Immunoblot analysis of immunoprecipitates prepared with antibodies to HA or to Myc revealed that JIP1 physically associated with mNotch1 and also with mNotch1-IC (Fig. 5A). We did not detect interaction between mNotch1-IC and MLK3, MKK7, or JNK1 in similar experiments (data not shown). Physical interaction between endogenous JIP1 and Notch1 was also apparent in MEFs, and the extent of this interaction was not affected by exposure of the cells to cellular stresses, including glucose deprivation (Fig. 5B) and UV (data not shown).
Fig. 5.
Notch1 physically interacts with JIP1 in intact cells. (A) HEK293 cells were transfected for 48 h with the indicated combinations of vectors for JIP1-FLAG, HA-mNotch1, and mNotch1-IC-Myc. Cell lysates were then subjected to immunoprecipitation (IP) with antibodies to HA or Myc, as indicated. The resulting immunoprecipitates were subjected to immunoblot (IB) analysis with antibodies to FLAG. Cell lysates were also subjected directly to immunoblot analysis with antibodies to FLAG, HA, or Myc. (B) MEFs were incubated for 4 h at 37°C in DMEM (serum-free) or glucose-free DMEM (serum-free) and then for an additional 12 h in complete medium. Cell lysates were subjected to immunoprecipitation with antibodies to Notch1 (Upstate Biotechnology, Lake Placid, NY) or with rabbit preimmune IgG. The resulting precipitates were examined by immunoblot analysis with goat anti-JIP1 antibody (Santa Cruz Biotechnology). Cell lysates were also subjected directly to immunoblot analysis with antibodies to Notch1 or to JIP1.
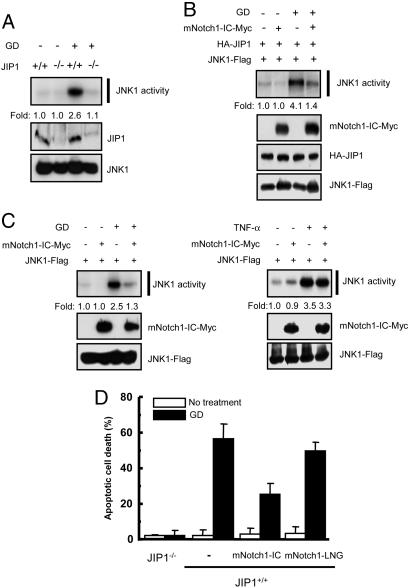
Notch1-IC Inhibits Glucose Deprivation-Induced JNK Activation and Apoptosis. Deprivation of glucose and oxygen has been shown to induce activation of the JNK pathway and consequent apoptosis, and these effects are mediated by JIP1 in hippocampal neurons (26). Glucose deprivation thus induced JNK1 activation in JIP1+/+ MEFs but not in JIP1-/- MEFs (Fig. 6A), and ectopic expression of JIP1 in the JIP1-/- MEFs resulted in glucose deprivation-induced JNK1 activation (Fig. 6B). Ectopic mNotch1-IC inhibited the activation of JNK1 induced by glucose deprivation in JIP1-transfected JIP1-/- MEFs (Fig. 6B) as well as in JIP1+/+ MEFs (Fig. 6C). In contrast, mNotch1-IC did not affect the activation of JNK1 induced by TNF-α (Fig. 6C) or sorbitol (data not shown), both of which induce JNK activation in a JIP1-independent manner (data not shown). Furthermore, mNotch1-IC, but not mNotch1-LNG, inhibited glucose deprivation-induced apoptosis in JIP1+/+ MEFs as shown by DAPI staining (Fig. 6C) as well as TUNEL staining (data not shown). Glucose deprivation did not induce apoptosis in JIP1-/- MEFs. Taken together, these results suggest that Notch1-IC inhibits the activation of JNK and apoptosis mediated by JIP1.
Fig. 6.
Notch1-IC inhibits glucose deprivation-induced JNK activation and apoptosis in MEFs. (A) JIP1+/+ or JIP1-/- MEFs were incubated first for 4 h in DMEM (serum-free) or glucose-free DMEM (serum-free) and then for an additional 12 h in complete medium. Cell lysates were subjected to immunoprecipitation with anti-JNK1 antibody, and the resulting precipitates were assayed for JNK1 activity with an immune complex kinase assay. Cell lysates were also subjected directly to immunoblot analysis with antibodies to JIP1 or to JNK1. (B) JIP1-/- MEFs were transfected for 48 h with plasmid vectors for HA-JIP1 and JNK1-FLAG in the absence or presence of a vector for mNotch1-IC-Myc. The transfected cells were incubated first for 4 h in DMEM (serum-free) or glucose-free DMEM (serum-free) and then for an additional 12 h in complete medium. Cell lysates were then subjected to immunoprecipitation with anti-FLAG antibody, and the resulting precipitates were assayed for JNK activity. Cell lysates were also examined by immunoblot analysis with antibodies to Myc, HA, or FLAG. (C) JIP1+/+ MEFs were transfected for 48 h with a vector for JNK1-FLAG alone or together with a vector for mNotch1-IC-Myc. For the glucose deprivation experiment, the transfected cells were treated as described for B. For the TNF-α experiment, the transfected cells were untreated or treated with 20 ng/ml TNF-α for 15 min. Cell lysates were then subjected to immunoprecipitation with anti-FLAG antibody, and the immunoprecipitates were assayed for JNK activity. Cell lysates were also subjected directly to immunoblot analysis with antibodies to Myc or to FLAG. (D) JIP1+/+ or JIP1-/- MEFs were transfected for 48 h with pEGFP alone or together with an expression vector for mNotch1-IC or for mNotch1-LNG. The transfected cells were incubated first for 4 h in DMEM (serum-free) or glucose-free DMEM (serum-free) and then for an additional 24 h in complete medium. The cells were then stained with DAPI, and GFP-positive cells were scored for apoptotic nuclei with a fluorescence microscope. Data are means ± average deviation of values from three independent experiments. GD, glucose deprivation.
Discussion
We have previously shown that PS1 negatively regulates the JNK signaling pathway and JNK-mediated apoptosis (28). The PS1-mediated inhibition of JNK signaling appears to require the activation of γ-secretase, given that functionally inactive mutants of PS1 did not inhibit JNK activation (28). γ-Secretase mediates the processing of its target proteins, including β-amyloid precursor protein (APP) and Notch (5, 46-48). Cleavage of APP by γ-secretase generates β-amyloid peptide and cytoplasmic APP-C99 (49). The observation that neither of these products inhibited the JNK pathway (data not shown) suggests that PS1-mediated inhibition of this pathway is not attributable to γ-secretase-mediated cleavage of APP. In contrast, we now show that Notch1-IC, an active form of Notch1 generated by the action of γ-secretase, functions as a negative regulator of JNK signaling. It was previously proposed that Notch might modulate the JNK signaling pathway (11, 12). Notch and its cytoplasmic regulator Deltex were thus found to inhibit the activation of transcription by c-Jun (11), which is one of the substrates of JNK. Furthermore, JNK activity was found to be higher in Notch mutant Drosophila embryo than in wild-type embryo (12). These findings are consistent with our present data showing that Notch1-IC inhibits the activation of JNK.
Notch-IC exerts many of its biological actions by activating the transcription factor Su(H) (Suppressor of Hairless) in Drosophila or CBF1/RBP-Jκ in mammals (8, 50). Notch-IC interacts with CBF1 and translocates to the nucleus, resulting in the elevated expression of target genes (8, 10, 50). Our data now indicate that activation of the transcription factor CBF1 is not required for inhibition of the JNK pathway by Notch1-IC. Moreover, Numb, a cytoplasmic negative regulator of Notch1 (1, 39), blocked the inhibitory action of Notch1-IC on JNK activation. Instead, we found that Notch1-IC physically interacts with JIP1, which, by associating with MLK3, MKK7, and JNK, functions as a scaffold protein that facilitates JNK activation (26). Our data show that Notch1-IC, by binding to JIP1, interferes with the physical interaction between JIP1 and JNK, thereby antagonizing the JIP1-mediated activation of JNK signaling. Akt1 has been demonstrated to physically associate with JIP1 and inhibit JIP1-mediated JNK activation in primary neurons, thereby preventing excitototic neuronal death (51). Given that Notch1-IC and Akt1 bind to the distinct regions of JIP1 (Fig. 4C) (51) and that both of the proteins have antiapoptotic function, it would be worthwhile to examine the possibility of the functional cooperation between Notch1 and Akt1 for regulating JIP1-mediated JNK activation and apoptosis. It would be also of considerable interest to determine whether Notch1, like Akt1, has a protective role in excitotoxic neuronal injury. Inhibition by Notch of JIP1-facilitated signaling by the MLK3-MKK7-JNK pathway may thus contribute to the mechanism by which Notch plays a pivotal role in a variety of the cellular events, such as embryonic development, cell growth, cell differentiation, and apoptosis.
Acknowledgments
We thank Roger J. Davis (University of Massachusetts Medical School, Worcester), J. R. Woodgett (Ontario Cancer Institute, Toronto), Jeffrey S. Nye (Johnson & Johnson, New Brunswick, NJ),Lawrence B. Holzman (University of Michigan, Ann Arbor), and Thomas Maciag (Maine Medical Center Research Institute, Scarborough) for providing JNK1, SAPKβ, Notch1, JIP1(R160G/P161G), and Jagged1 cDNA clones, respectively. We also thank Jie Shen (Harvard Medical School, Boston) for MEFPS1(+/+) and MEFPS1(-/-) cells. This work was supported by the Creative Research Initiatives of the Korean Ministry of Science and Technology (E.-J.C.) and by grants from the Functional Proteomics Center (to T.-S.C.) and the Brain Research Center of the 21st Century Frontier Research Program of the Korean Ministry of Science and Technology (to H.-S.P.).
Author contributions: J.W.K., M.J.K., and E.-J.C. designed research; J.W.K., M.J.K., K.J.K., H.J.Y., J.S.C., S.G.H., and T.-S.C. performed research; H.-S.P., K.-W.L., P.-L.H., S.-G.C., and T.-W.K. contributed new reagents/analytic tools; J.W.K., M.J.K., and E.-J.C. analyzed data; and J.W.K., M.J.K., and E.-J.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: APP, amyloid β-protein precursor; HA, hemagglutinin; MAPK, mitogen-activated protein kinase; MAP2K, MAPK kinase; MAP3K, MAPK kinase kinase; MEF, mouse embryonic fibroblast; MLK, mixed lineage kinase; JNK, c-Jun N-terminal kinase; PS, presenilin; SAPK, stress-activated protein kinase; JBD, JNK-binding domain; mNotch1, murine Notch1; Notch1-IC, Notch1 intracellular domain.
References
- 1.Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. (1999) Science 284, 770-776. [DOI] [PubMed] [Google Scholar]
- 2.Levitan, D. & Greenwald, I. (1995) Nature 377, 351-354. [DOI] [PubMed] [Google Scholar]
- 3.Li, X. & Greenwald, I. (1997) Proc. Natl. Acad. Sci. USA 94, 12204-12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, Y. M. & Jan, Y. N. (1999) Neuron 23, 201-204. [DOI] [PubMed] [Google Scholar]
- 5.De Strooper, B., Annaert, W., Cupers, P., Saftig, P., Craessaerts, K., Mumm, J. S., Schroeter, E. H., Schrijvers, V., Wolfe, M. S. & Ray, W. J. (1999) Nature 398, 518-522. [DOI] [PubMed] [Google Scholar]
- 6.Ray, W. J., Yao, M., Nowotny, P., Mumm, J., Zhang, W., Wu, J. Y., Kopan, R. & Goate, A. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3263-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li, Y. M., Xu, M., Lai, M. T., Huang, Q., Casto, J. L., DiMuzio-Mower, J., Harrison, T., Lellis, C., Nadin, A. & Neduvelil, J. G. (2000) Nature 405, 689-694. [DOI] [PubMed] [Google Scholar]
- 8.Fortini, M. E. & Artavanis-Tsakonas, S. (1994) Cell 79, 273-282. [DOI] [PubMed] [Google Scholar]
- 9.Busseau, I., Diederich, R. J., Xu, T. & Artavanis-Tsakonas, S. (1994) Genetics 136, 585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuno, K., Go, M. J., Sun, X., Eastman, D. S. & Artavanis-Tsakonas, S. (1997) Development (Cambridge, U.K.) 124, 4265-4273. [DOI] [PubMed] [Google Scholar]
- 11.Ordentlich, P., Lin, A., Shen, C. P., Blaumueller, C., Matsuno, K., Artavanis-Tsakonas, S. & Kadesch, T. (1998) Mol. Cell. Biol. 18, 2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zecchini, V., Brennan, K. & Martinez-Arias, A. (1999) Curr. Biol. 9, 460-469. [DOI] [PubMed] [Google Scholar]
- 13.Minden, A. & Karin, M. (1997) Biochim. Biophys. Acta. 1333, F85-F104. [DOI] [PubMed] [Google Scholar]
- 14.Ip, Y. T. & Davis, R. J. (1998) Curr. Opin. Cell Biol. 10, 205-219. [DOI] [PubMed] [Google Scholar]
- 15.Schaeffer, H. J. & Weber, M. J. (1999) Mol. Cell. Biol. 19, 2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boulton, T. G., Nye, S. H., Robbins, D. J., Ip, N. Y., Radziejewska, E., Morgenbesser, S. D., DePinho, R. A., Panayotatos, N., Cobb, M. H. & Yancopoulos, G. D. (1991) Cell 65, 663-675. [DOI] [PubMed] [Google Scholar]
- 17.Han, J., Lee, J. D., Bibbs, L. & Ulevitch, R. J. (1994) Science 265, 808-811. [DOI] [PubMed] [Google Scholar]
- 18.Cobb, M. H. & Goldsmith, E. J. (1995) J. Biol. Chem. 270, 14843-14846. [DOI] [PubMed] [Google Scholar]
- 19.Kyriakis, J. M. & Avruch, J. (1996) BioEssays 18, 567-577. [DOI] [PubMed] [Google Scholar]
- 20.Su, B. & Karin, M. (1996) Curr. Opin. Immunol. 8, 402-411. [DOI] [PubMed] [Google Scholar]
- 21.Fanger, G. R., Gerwins, P., Widmann, C., Jarpe, M. B. & Johnson, G. L. (1997) Curr. Opin. Genet. Dev. 7, 67-74. [DOI] [PubMed] [Google Scholar]
- 22.Dickens, M., Rogers, J. S., Cavanagh, J., Raitano, A., Xia, Z., Halpern, J. R., Greenberg, M. E., Sawyers, C. L. & Davis, R. J. (1997) Science 277, 693-696. [DOI] [PubMed] [Google Scholar]
- 23.Bonny, C., Nicod, P. & Waeber, G. (1998) J. Biol. Chem. 273, 1843-1846. [DOI] [PubMed] [Google Scholar]
- 24.Whitmarsh, A. J., Cavanagh, J., Tournier, C., Yasuda, J. & Davis, R. J. (1998) Science 281, 1671-1674. [DOI] [PubMed] [Google Scholar]
- 25.Kim, I. J., Lee, K. W., Park, B. Y., Lee, J. K., Park, J., Choi, I. Y., Eom, S. J., Chang, T. S., Kim, M. J., Yeom, Y. I., et al. (1999) J. Neurochem. 72, 1335-1343. [DOI] [PubMed] [Google Scholar]
- 26.Whitmarsh, A. J., Kuan, C. Y., Kennedy, N. J., Kelkar, N., Haydar, T. F., Mordes, J. P., Appel, M., Rossini, A. A., Jones, S. N. & Flavell, R. A. (2001) Genes Dev. 15, 2421-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison, D. K. & Davis, R. J. (2003) Annu. Rev. Cell Dev. Biol. 19, 91-118. [DOI] [PubMed] [Google Scholar]
- 28.Kim, J. W., Chang, T. S., Lee, J. E., Huh, S. H., Yeon, S. W., Yang, W. S., Joe, C. O., Mook-Jung, I., Tanzi, R. E., Kim, T. W. & Choi, E. J. (2001) J. Cell Biol. 153, 457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shim, J., Lee, H., Park, J., Kim, H. & Choi, E. J. (1996) Nature 381, 804-806. [DOI] [PubMed] [Google Scholar]
- 30.Shim, J., Park, H. S., Kim, M. J., Park, J., Park, E., Cho, S. G., Eom, S. J., Lee, H. W., Joe, C. O. & Choi, E. J. (2000) J. Biol. Chem. 275, 14107-14111. [DOI] [PubMed] [Google Scholar]
- 31.Kim, J. W., Joe, C. O. & Choi, E. J. (2001) J. Biol. Chem. 276, 27064-27070. [DOI] [PubMed] [Google Scholar]
- 32.Shen, J., Bronson, R. T., Chen, D. F., Xia, W., Selkoe, D. J. & Tonegawa, S. (1997) Cell 89, 629-639. [DOI] [PubMed] [Google Scholar]
- 33.Im, J. Y., Lee, K. W., Kim, M. H., Lee, S. H., Ha, H. Y., Cho, I. H., Kim, D., Yu, M. S., Kim, J. B., Lee, J. K., et al. (2003) J. Neurosci. Res. 74, 326-332. [DOI] [PubMed] [Google Scholar]
- 34.Ross, D. A., Rao, P. K. & Kadesch, T. (2004) Mol. Cell. Biol. 24, 3505-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang, T. S., Kim, M. J., Ryoo, K., Park, J., Eom, S. J., Shim, J., Nakayama, K. I., Nakayama, K., Tomita, M., Takahashi, K., et al. (2003) J. Biol. Chem. 278, 48092-48098. [DOI] [PubMed] [Google Scholar]
- 36.Park, H. S., Park, E., Kim, M. S., Ahn, K., Kim, I. Y. & Choi, E. J. (2000) J. Biol. Chem. 275, 2527-2531. [DOI] [PubMed] [Google Scholar]
- 37.Cho, S. G., Kim, J. W., Lee, Y. H., Hwang, H. S., Kim, M. S., Ryoo, K., Kim, M. J., Noh, K. T., Kim, E. K., Cho, J. H., et al. (2003) J. Cell Biol. 163, 71-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopan, R., Schroeter, E. H., Weintraub, H. & Nye, J. S. (1996) Proc. Natl. Acad. Sci. USA 93, 1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhyu, M. S., Jan, L. Y. & Jan, Y. N. (1994) Cell 76, 477-491. [DOI] [PubMed] [Google Scholar]
- 40.Kidd, S., Lieber, T. & Young, M. W. (1998) Genes Dev. 12, 3728-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi, H., Rand, M. D., Wu, X., Sestan, N., Wang, W., Rakic, P., Xu, T. & Artavanis-Tsakonas, S. (1999) Science 283, 91-94. [DOI] [PubMed] [Google Scholar]
- 42.Mumm, J. S., Schroeter, E. H., Saxena, M. T., Griesemer, A., Tian, X., Pan, D. J., Ray, W. J. & Kopan, R. (2000) Mol. Cell 5, 197-206. [DOI] [PubMed] [Google Scholar]
- 43.Struhl, G. & Greenwald, I. (1999) Nature 398, 522-525. [DOI] [PubMed] [Google Scholar]
- 44.Davis, R. J. (2000) Cell 103, 239-252. [DOI] [PubMed] [Google Scholar]
- 45.Nihalani, D., Wong, H. N. & Holzman, L. B. (2003) J. Biol. Chem. 278, 28694-28702. [DOI] [PubMed] [Google Scholar]
- 46.De Strooper, B., Saftig, P., Craessaerts, K., Vanderstichele, H., Guhde, G., Annaert, W., Von Figura, K. & Van Leuven, F. (1998) Nature 391, 387-390. [DOI] [PubMed] [Google Scholar]
- 47.Ni, C. Y., Murphy, M. P., Golde, T. E. & Carpenter, G. (2001) Science 294, 2179-2181. [DOI] [PubMed] [Google Scholar]
- 48.Lee, H. J., Jung, K. M., Huang, Y. Z., Bennett, L. B., Lee, J. S., Mei, L. & Kim, T. W. (2002) J. Biol. Chem. 277, 6318-6323. [DOI] [PubMed] [Google Scholar]
- 49.Selkoe, E. J. (1998) Trends Cell Biol. 8, 447-453. [DOI] [PubMed] [Google Scholar]
- 50.Jarriault, S., Brou, C., Logeat, F., Schroeter, E. H., Kopan, R. & Israel, A. (1995) Nature 377, 355-358. [DOI] [PubMed] [Google Scholar]
- 51.Kim, A. H., Yano, H., Cho, H., Meyer, D., Monks, B., Margolis, B., Birnbaum, M. J. & Chao, M. V. (2002) Neuron 35, 697-709. [DOI] [PubMed] [Google Scholar]