Abstract
The Arabidopsis root has a unique cellular pattern in its single-layered epidermis. Cells residing over the intercellular spaces between underlying cortical cells (H position) differentiate into hair cells, whereas those directly over cortical cells (N position) differentiate into non-hair cells. Recent studies have revealed that this cellular pattern is determined by interactions of six patterning genes CPC, ETC, GL2, GL3/EGL3, TTG, and WER, and that the position-dependent expression of the CPC, GL2, and WER genes is essential for their appropriate interactions. However, little is known about how the expressions of the pattern genes are determined. Here we show that trichostatin A (TSA) treatment of germinating Arabidopsis seedlings alters the cellular pattern of the root epidermis to induce hair cell development at nonhair positions. The effects of TSA treatment are rapid, reversible, concentration-dependent, and position-independent. TSA inhibition of histone deacetylase activity results in hyperacetylation of the core histones H3 and H4, and alters the expression levels and cell specific expression of the patterning genes CPC, GL2 and WER. Analysis of histone deacetylase mutant cellular patterning further verified the participation of histone acetylation in cellular patterning, and revealed that HDA18 is a key component in the regulatory machinery of the Arabidopsis root epidermis. We propose a working model to suggest that histone acetylation may function in mediating a positional cue to direct expression of the patterning genes in the root epidermal cells.
Keywords: histone as a signaling mediator, trichostatin A, histone deacetylase, positional cue, chromatin immunoprecipitation
Pattern formation is the process by which cells form ordered spatial arrangements, and is one of the most crucial features in the development of multicellular organisms. In Arabidopsis, single-layered root epidermal cells differentiate into hair and non-hair cells in a position-dependent manner. Epidermal cells positioned over the intercellular spaces between underlying cortical cells (H position) differentiate into hair cells, and those directly over cortical cells (N position) differentiate into non-hair cells (1). Extensive studies of mutants with various root hair phenotypes have made the Arabidopsis root epidermis a simple and experimentally accessible model for studying the regulatory mechanisms of cellular pattern formation in plants (2-4). Previous studies have shown that the position-dependent cellular pattern of root epidermal cells is determined by interactions of six patterning genes: CPC, ETC, GL2, GL3/EGL3, TTG, and WER (5-16). CPC, GL2, and WER are preferentially expressed in N position cells upon seed germination (6, 9-12), and regulate the preferential expression of GL3/EGL2 at H position cells (15). Therefore, their appropriate expression is essential for accurate regulation of root epidermal patterning. However, little is known about the mechanisms whereby the position-dependent expressions of these genes are regulated, although a “positional cue” derived from the cortical cells has been hypothesized to orchestrate the cellular patterning of the root epidermis (10). If the cellular patterning is indeed dependent on a positional cue or cues, two features are required simultaneously to mediate the preferential expression of CPC, GL2, and WER at the N cell positions. These features are: (i) sensitivity to informational cues arising from cortical cells and (ii) direct participation in the regulation of CPC, GL2, and WER expression. Recently, a receptor-like kinase, encoded by a gene SCRAMBLED (SCM), has been reported to mediate the positional signaling and the expression patterns of the CPC, GL2, and WER genes (17). However, information is still lacking about whether the SCM gene interacts with the known patterning genes or how this process might occur.
In a series of experiments, we obtained evidence that histone acetylation is involved in the cellular patterning of the root epidermis. When we applied trichostatin A (TSA), a specific inhibitor of histone deacetylase (HDAC) (18), to germinating Arabidopsis seeds, we found a striking increase in the density of root hairs in the seedlings. Because histone modification has been well established as an important regulatory mechanism of gene expression (19-21), and histone acetylation is reversible, we hypothesized that histone acetylation could provide a sensor switch to temporally and spatially modulate patterning signals during root hair differentiation. Here we report that the histone hyperacetylation induced by application of TSA alters the expression levels and cellular patterns of the CPC, GL2, and WER patterning genes, and that HDA18, a gene encoding histone deacetylase, is a key component required for appropriate cellular patterning. These findings demonstrate that histone acetylation has a mechanistic role in regulating the position-dependent expression of patterning genes in the Arabidopsis root epidermis, and most likely functions to mediate the “positional cue.”
Materials and Methods
Supporting Information. For further details, see Supporting Text, Tables 1-6, and Figs. 7-11, which are published as supporting information on the PNAS web site.
Plant Materials and Treatments. All wild-type plants and the HDAC mutants in the Salk series (see Table 5) used in the experiments are in the Columbia background. Except for the cpc mutant, all other lines were purchased from the Arabidopsis Biological Resource Center. The cpc mutant and the HDAC mutants in the CS series (Table 5) are in the WS ecotype. To rule out possible differences in ecotype background, we carried out control experiments with the WS wild type and found no noticeable differences between the Columbia and WS ecotypes in the traits investigated in this study. Seedlings were grown on Petri dishes with Murashige and Skoog (MS) agar medium, vertically oriented at 22°C under a 16-h light/8-h dark photo-period regime. TSA (Sigma T8552) treatment was carried out by (i) directly adding TSA into the agar media to final concentrations of 10, 100, 500 and 1,000 ng/ml and then germinating seeds on the agar, or (ii) transferring 8-day-old seedlings grown on MS media into liquid MS media containing corresponding concentrations of TSA for various times. Root tips (0.5 cm) were excised from treated and control seedlings for use in biochemical and molecular analyses.
Paraffin Sections. Seedlings were fixed with 3% glutaraldehyde in PBS at pH 7.2 for 1 h, preembedded, dehydrated, cleared in xylene, and embedded in paraffin as described (22). Roots were cross-sectioned in 7-μm sections from the tip to the root hair region. Sections were stained as described with Safrain O/Fast Green to obtain an optimal contrast between hair and non-hair cells, or Toluidine blue (23). Sections taken at the end of the root cap were used to show the radial cellular patterns of each treatment. Photographs were taken with an Olympus BX51 microscope and a Spot 2 digital camera (Diagnostic Instruments, Sterling Heights, MI) and images were processed with photoshop software (Adobe Systems, San Jose, CA).
HDAC Activity Assay. To evaluate the effects of TSA on HDAC activity, control samples grown in MS-grown seedling were compared with three types of TSA applications: (i) seedlings grown on MS agar containing 100 ng/ml TSA for 8 days; (ii) seedlings grown on MS agar containing 1,000 ng/ml TSA for 8 days; or (iii) seedlings grown on MS agar for 8 days and then transferred to liquid media containing 1,000 ng/ml TSA. Root tips were collected after three brief washes with PBS (pH 7.0), and enzyme extractions and fluorescence detections were carried out according to the manufacturer's instructions (AK-500, BioMol, Plymouth Meeting, PA). Fluorescence was measured with a Luminescence Spectrometer (LS55, PerkinElmer) with an excitation of 360 nm and emission of 460 nm in an arbitrary fluorescence unit.
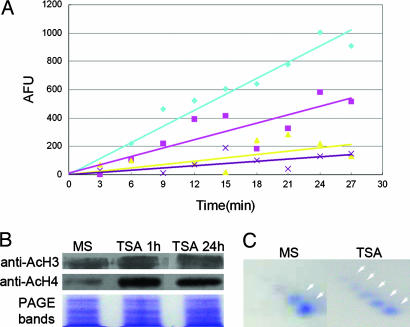
Analysis of Histone Acetylation. To assess the levels of histone acetylation, histones were isolated, separated, and immunobloted as described in Table 1. Antibodies were purchased from Upstate Biotechnology (anti-Ac H3, catalog no. 06-599; anti-Ac-H4, catalog no. 06-866; anti-rabbit IgG horseradish peroxidase-conjugated, catalog no. 12-348). Antibody signals were detected by using enhanced chemiluminscenece (SuperSignal West Pico Chemiluminescent Substracte, Pierce). To further evaluate levels of histone acetylation, AUT/AUC 2D gel analyses were performed according to Rogakou et al. (24). According to Waterborg, the spots in the framed region in Fig. 2C were H4, and there was a positive correlation between the number of spots and the number of amino acid residues that were acetylated at the N-terminal tail of H4 (25).
Fig. 2.
TSA inhibits HDAC activity and alters histone acetylation patterns. (A) Inhibition of HDAC is indicated by the decreased slope of enzyme activity vs. reaction time. Blue line (diamond), control; pink line (square), root tissue treated with100 ng/ml TSA for 8 days; yellow line (triangle), root tissue treated with 1,000 ng/ml TSA for 8 days; purple line (cross), root tissue treated with 1,000 ng/ml TSA for 15 min. (B) Immunoblot detection of H3 and H4 hyperacetylation after TSA treatment as indicated by increases in the intensity of antibody signals at the corresponding protein bands. (C) H4 hyperacetylation shown by 2D gel separation of rapidly isolated histone extracts. Arrowheads highlight the increased number of protein spots in the control (2) vs. the TSA treatment (5).
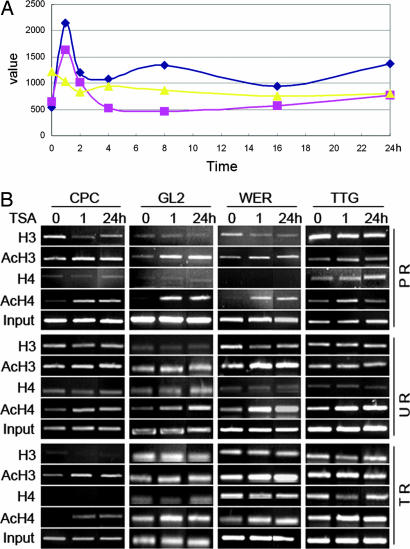
Reverse Transcription and Real-Time PCR Analyses. Total RNA was isolated from the root tips by using the RNeasy Plant Mini kit (Qiagen, Valencia, CA). The RNA was reversed-transcribed by using the SuperScript II (Invitrogen). Real-time PCR was conducted with the Amplifuor universal amplification and detection system (Intergen catalog no. S7901). The sequences used for real-time PCR are shown in Table 2. The expression values presented in Fig. 3A were adjusted by comparing values for each gene with GAPDH or glyceraldehyde-3-phosphate dehydrogenase C (GAPC) internal gene standards to correct for systematic inconsistencies generated in different reactions.
Fig. 3.
TSA directly alters expression levels of the patterning genes. (A) The time course of expression levels of CPC (blue line with diamond), GL2 (pink line with square), and WER (yellow line with triangle) after 1,000 ng/ml TSA treatment (detected by real-time RT-PCR). (B) ChIP analysis showing a correlation between the histone acetylation status and the expression of CPC, GL2, and WER. Note the increased band intensities from the control (TSA, 0 h) to the 1,000 ng/ml TSA treatments at 1 h and 24 h in the CPC, GL2, and WER genes, but not the TTG gene control. PCR primers used for the ChIP assays were designed from the sequences of promoter region (PR), upstream region (UR), and transcribed region (TR).
Chromatin Immunoprecipitations (ChIPs). ChIPs were performed according to the protocol in the kit manual (Upstate Biotechnology 17-295) and described methods (26) with minor modifications (Table 3). Primers used in the ChIP assays are shown in Table 2. All PCRs were conducted in 25-μl volumes with 32-38 cycles to optimize the PCR products of each gene. The PCR products were detected with the gel spot system (Vilber Lourmat) after electrophoresis in a 2% agarose gel.
In Situ Hybridization. An in situ hybridization protocol was adopted from the Helariutta's group (www.biocenter.helsinki.fi/bi/PLANT/Helariutta) with slight modifications. DNA sequences used for preparing probes were obtained by PCR using the primers described in Table 2. The PCR fragments were then cloned into the pGEM T-Easy vector (Promega) and linearized with NcoI or SpeI. Sense or antisense RNA probes were generated by using the DIG RNA Labeling kit (SP6/T7, Roche Applied Science catalog no. 1175025), and the probes were hydrolyzed to ≈250 bp in alkaline carbonate buffer. Tissue was embedded in Paraplast plus (Sigma), and the 7-μm sections were mounted on slides pretreated with poly(l-lysine) (Sigma). The RNA probe was added to a final concentration of 0.1-0.2 ng/μl and incubated overnight at 45°C. Hybridizations were visualized with an anti-digoxigenin-alkaline-phosphatase coupled antibody (Roche Applied Science, catalog no. 1175041), and photographed under an Olympus BX51 microscope. The images were then adjusted with photoshop to enhance the differential signals detected in epidermal cells.
Results
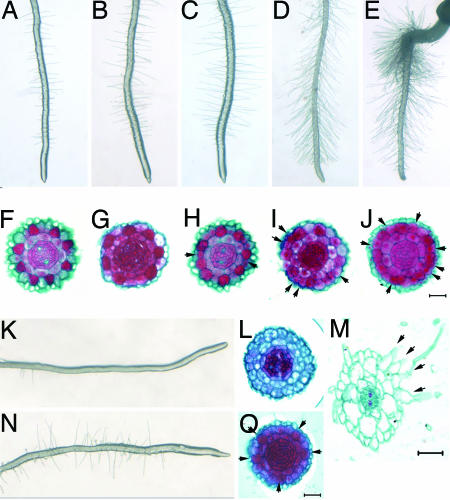
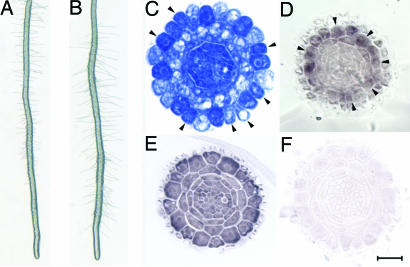
TSA Treatment Alters Cellular Pattern of Arabidopsis Root Epidermis. To determine whether developmental events could be affected by histone modification, we germinated Arabidopsis seeds on MS agar media containing various concentrations of TSA. The most striking morphological change in early seeding development was a TSA concentration-dependent increase in root hair density (Fig. 1 A-E). Morphological analyses indicated that the TSA-dependent enhancement of root hairs results from an increased proportion of N position cells that switched to a hair cell fate, and that this switch was concentration dependent (Fig. 1 F-J and M, arrowheads, and Table 4). Thus, TSA treatment promotes epidermal cells to commit to a hair cell fate independently of their spatial relationship to cortical cells.
Fig. 1.
TSA alters root hair patterning. (A-E) Concentration-dependent increases in the numbers of root hairs after growth on agar containing 0, 10, 100, 500, and 1,000 ng/ml TSA, respectively. (F-J) Cross sections of the root tip. Hair cells in the epidermis are stained in red. Extra hair cells in the roots after TSA treatment are indicated by arrowheads. (K and L) Reduced root hair phenotype of the cpc mutant. (M) A cross section at the root hair region shows side by side differentiation of hair cells after TSA treatment. (N and O) Hair cells differentiated predominately at the H positions of the cpc mutant by TSA treatment are indicated by arrowheads (O). (Bar, 20 μm.)
As the root continues to elongate due to the activity of the root apical meristem, epidermal cell patterning continues to generate new root tissue. To determine whether TSA is constantly required for the maintenance of cell pattern changes, we transferred 8-day-old seedlings grown in MS or TSA media to TSA or MS respectively. In cross sections, the MS grown seedlings developed an altered epidermal cellular pattern after 8-h treatment at 1,000 ng/ml TSA (Fig. 7). In contrast, TSA grown seedlings reverted to the normal root epidermal cellular pattern after being moved back to MS (Fig. 8). These results demonstrate that (i) TSA is required continuously for maintenance of aberrant cell patterning during root growth, and (ii) the effects of TSA treatment on the cellular patterning of newly initiated epidermal cells are rapid, reversible, and concentration-dependent, but are position-independent.
TSA Treatment Alters Cellular Pattern by Changing the Expression of Patterning Genes. According to current data and models, the cellular pattern of the Arabidopsis root epidermis is determined by the interactions of six patterning genes designated CPC, ETC, GL2, GL3/EGL3, TTG, and WER (5-16). Histone modifications have also been implicated as important players in regulating gene expression (19-21). Because TSA is a specific inhibitor of HDAC enzyme activity (18), we sought to determine whether TSA treatment results in alterations in histone acetylation that correlate with changes in expression of the root epidermal patterning genes.
We first determined whether TSA treatment results in rapid histone hyperacetylation in the root cells. A histone deacetalyase assay with root tips revealed that TSA application reduced HDAC activity in a concentration-dependent manner (Fig. 2A). Separate Western blot assays using antibodies against acetylated core histones H3 and H4 revealed that, after 1 h of treatment with TSA, hyperacetylated H3 and H4 increased substantially (Fig. 2B). Previous studies have shown that the acetylation status of H4 can be identified by separation of histone extracts on 2D gels (25). By applying this technique to roots tips, we found that a higher proportion of acetylated H4 derivatives were present in TSA-treated roots than in MS-grown roots (Fig. 2C). These results suggest that the reductions in HDAC activity are correlated with hyperacetylation of H4 amino acid residues. Taken together, these data provide persuasive evidence that the levels of histone acetylation are increased in root cells as a consequence of the inhibition of HDAC during TSA treatment.
Next we asked whether TSA treatment altered the expression of the patterning genes. To obtain an overview of the mechanisms whereby TSA treatment affects gene expression, microarray analyses were conducted with an Affymetrix Arabidopsis ATH1 GeneChip. The results revealed that of the six patterning genes (CPC, ETC, GL2, EGL3, TTG, and WER), only the expression levels of CPC, GL2 and WER were altered significantly (C.-R.X., G. Gao, L.-C.L., J.-C. Luo, Z.-H.X., and S.-N.B., unpublished data). Based on these results, we performed a real-time PCR assay to more precisely analyze the expression of the three genes. The PCR data were consistent with the microarray data in that the expression levels of CPC and GL2 were up-regulated by 3- to 4-fold within the first hour of the highest levels of the TSA treatments, and then decreased in the successive periods (Fig. 3A). In contrast, WER expression was down-regulated rapidly after application of TSA and remained low throughout the remaining time periods (Fig. 3A).
To determine whether the changes in expression levels of the three genes are affected by histone hyperacetylation, we performed ChIP experiments (27, 28). Using validated primers (Fig. 9), we found that the expression levels of CPC, GL2, and WER were highly correlated with the levels of acetylation of H3 and H4. The increased levels of acetylation were not only present in the promoter regions (CPC, GL2, WER; H3, H4), but also occurred in the upstream (WER; H3, H4; CPC, GL2, H4) and transcribed (CPC; H3, H4) regions (Fig. 3B). Therefore, the most straightforward interpretation of these results is that the elevated expression levels of the CPC and GL2 genes are directly affected by histone hyperacetylation. The down-regulation of WER gene expression hyperacetylation does not fit this pattern, but there are a number of cases where histone hyperacetylation decreases gene expression (29-31). As expected based on the microarray and semiquantitative RT-PCR results, no discernable changes in the PCR products of the TTG promoters were observed after TSA treatment, although a slight increase in the upstream region of acetylated H4 was detected (Fig. 3B). This finding suggests that the expression levels of TTG are not affected substantially by histone acetylation. Recent studies have shown that ETC, GL3, and EGL3 participate in an intercellular regulatory circuit by preferential expression in developing hair cells (15, 16), but our data revealed no obvious changes in the expression levels of ETC, GL3, and EGL3 in response to TSA treatment (data not shown).
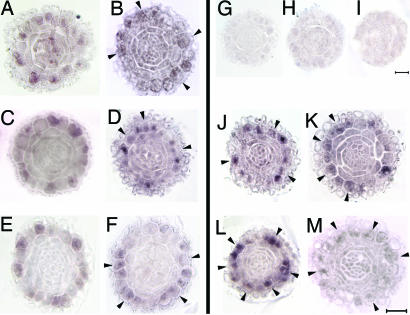
Because CPC, GL2, and WER are preferentially expressed in N position cells (6, 9, 10), in situ hybridization experiments were conducted to determine whether TSA treatment alters their patterns of expression. The results show that expression of all three genes was detected not only in the N position cells within 1 h after TSA application, as they were in the wild-type roots (Fig. 4 A, C, and E), but also in some H position cells (Fig. 4 B, D, and F, arrowheads). Current notions about the functions of the three patterning genes provide a model whereby WER induces CPC and GL2 expression in N position cells. The CPC protein moves to neighboring H position cells and suppresses both WER and CPC expression, which causes GL2 down-regulation events that release GL2-repression of root hair differentiation (2, 3). Our results add to this hypothesis by showing that, when root cells are exposed to TSA, HDAC activity is reduced, and the original patterning of CPC and GL2 expression is deregulated as a consequence of histone hyperacetylation. The in situ hybridization experiments reveal that the deregulation results in abnormal increases in expression of CPC and GL2 at the H position. Because CPC is a negative regulator of GL2, WER, and CPC itself, the increases resulting from TSA treatment were transient and declined within 2 h after initiation of treatment. This characteristic explains the decrease of the CPC expression level after the first hour of TSA treatment (Fig. 3A), as well as substantial decreases in signal intensity of the gene at 24 h revealed by in situ hybridization experiments (data not shown). Because GL2 is positively regulated by WER (11) and WER is directly down-regulated by TSA treatment (Fig. 3A), the decrease of GL2 after the first hour of TSA treatment might be a consequence of self-inhibition of expression. However, it is difficult to explain the ectopic expression of WER in the H position because the real-time PCR results indicate that WER expression is down-regulated, and we did not expect its presence in the H position after TSA treatment. Irrespective of this anomaly, the data from the in situ hybridizations demonstrate that histone hyperacetylation is correlated with the overall levels and position effects of patterning gene expression.
Fig. 4.
In situ hybridization reveals that TSA alters the patterns of CPC, GL2, and WER gene expression. (A and B) CPC expression patterns in the wild-type (WT) control (A) and TSA treatment (B). (C and D) GL2 expression patterns in the WT control (C) and TSA treatment (D). (E and F) WER expression patterns in the WT control (E) and TSA treatment (F). (G-I) Negative controls with CPC (G), GL2 (H), and WER (I) sense probes. (J and K) GL2 expression patterns in the cpc mutant control (J) and TSA treatment (K). (L and M) WER expression patterns in the cpc mutant control (L) and TSA treatment (M). The concentrations of TSA treatment used for in situ hybridization were 1,000 ng/ml. Cells abnormally expressing corresponding genes are indicated by arrows. (Bar, 20 μm.)
It is worth noting that others have previously suggested that the preferential expression of CPC and GL2 in N position cells is determined by WER (2, 11). However, our study shows that the expression of CPC, GL2, and WER are also directly affected by the histone acetylation status. To further understand how histone acetylation affects the interaction of these three patterning genes, we applied TSA to a cpc mutant because it is the only mutant among the three patterning genes with few root hairs, and therefore provides a model to evaluate the enhancing effects of TSA on root hairs. In the cpc mutant, TSA treatment induced root hairs to levels that approximated those of wild-type root hairs (Fig. 1, compare K and N), and a cell fate transition from the N to H status (Fig. 1 L and O). In agreement with other studies, we found that GL2 and WER also had increased expression levels in the cpc mutant (11) (Fig. 4 J and L). Additionally, we found that TSA treatment down-regulated the expression levels of WER (Fig. 4 L and M), which could have resulted in the increased GL2 expression in the N position (Fig. 4 J and K). Thus, these findings suggest that WER and GL2 are directly involved in the transition from the N to H cell fate depending on their relative positions to cortical cells, whereas CPC is involved in determining the sensitivity of the cells at different positions in the root epidermis to changes in histone acetylation.
Taken together, the data presented above strongly indicate that TSA treatment affects the cellular patterning of the Arabidopsis root epidermis by changing histone acetylation, which alters the expression of the CPC, GL2, and WER patterning genes. Therefore, our results clearly show that histone acetylation is a component of the mechanism that regulates the position-dependent expression of the patterning genes.
HDA18 Is a Key Component Required for Cellular Patterning of Arabidopsis Root Epidermis. To further evaluate the involvement of histone acetylation in regulation of the position-dependent expression of the patterning genes, we carried out a phenotypic screening of HDAC mutants. The rationale is that if histone acetylation is indeed involved in the regulation, the cellular patterns of the root epidermis should be altered in the HDAC mutants, and the expression patterns of the patterning genes should be altered as well. Thus far, 16 genes encoding TSA-sensitive HDAC have been identified (32). Our screening of all 23 available mutants covering these 16 genes reveals that five mutants elicit phenotypes with increased root hair densities similar to those found in seedlings treated with TSA (Fig. 5 A and B and Table 5). Cross sections of the developing roots of homozygous mutants with increased root hair densities revealed that alterations of the cellular patterns of the root epidermis were present in the hda18 mutant (At5g61070) (Fig. 5C and Table 4), but not in hda5, hda14, and hdt4 (Table 5). Using the WER gene as a CPC and GL2 model, we found that the increased expression in TSA treated roots (Fig. 4F) correlated strongly with the expression patterns of the hda18 mutant (Fig. 5D). These results provide further evidence that (i) histone acetylation is involved in regulation of the cellular patterning in the root epidermis; (ii) only some members of the HDAC gene family are required for the cellular patterning; and (iii) because not all of the epidermal cells are committed to the hair fate in the hda18 mutant, other components in addition to HDA18 are expected to be involved in the histone-mediated regulation of cellular patterning.
Fig. 5.
HDA18 is required for hair cell cellular patterning. (A) Root hair density in wild-type plants. (B) Slightly increased root hair density in the hda18 mutant. (C) The cellular pattern of root epidermis is altered in the hda18 mutant. Arrowheads indicate the differentiation of hair cells at the N position. (D) Expression pattern of WER is altered in the hda18 mutant, similar to that of wild-type roots treated with TSA (compare to Fig. 4F). (E) HDA18 RNA is present in all root cells, indicating a position-independent expression. (F) Negative control for in situ hybridization using a sense HDA18 a probe. (Bar, 20 μm.)
To determine whether HDA18 participates in the regulation of cellular patterning by decreases in enzyme activity, rather than position-dependent expression of RNA, we carried out in situ hybridizations with a HDA18-specific probe (Fig. 10) to examine whether the gene is differentially expressed in the root cells. Fig. 5E shows that the position-dependent expression of HDA18 RNA was not detected in root epidermal cells, suggesting that HDA18 participates in the regulation of cellular patterning not through gene differential expression at RNA level.
Discussion
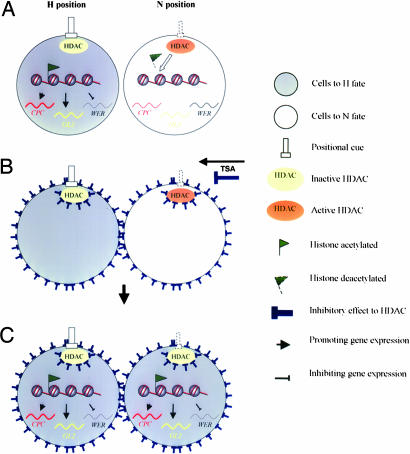
Our results provide insight into the mechanisms regulating position-dependent expression of patterning genes in the Arabidopsis root epidermis. We first observed that TSA treatment can rapidly and reversibly alter the patterns of root hair development. A series of biochemical experiments revealed that histone hyperacetylation induced by TSA treatment affects the levels and patterns of expression the CPC, GL2, and WER patterning genes. Mutant analyses also provided complementary support for the involvement of histone acetylation in the cellular patterning, and revealed that HDA18 is a key component required for the regulatory mechanism. Here, we propose a working model based on currently available data to explain the role of histone acetylation in the cellular patterning of the Arabidopsis root epidermis (Fig. 6).
Fig. 6.
Working model to explain the role of histone acetylation in the cellular patterning of the Arabidopsis root epidermis. (A) A hypothesized “positional cue” inhibits the enzyme activity of HDAC in cells at the H position but not those at the N position. This results in a differential histone hyperacetylation that is correlated with the known position-dependent CPC, GL2 and WER gene interactions (2, 3). (B) When TSA is applied, HDAC activity is unselectively inhibited independently of the positions of the H and N cells, and the differential effect of “positional cue” is interrupted. (C) When the TSA concentration is sufficient to inhibit HDAC, the expression of the CPC, GL2, and WER patterning genes of the N cells are shifted to the H position status that alters cellular patterning and results in increased differentiation of hair cells.
Our working model builds on the concept of a “positional cue” that orchestrates differential effects on N and H position according to their spatial relationship with cortical cells (10). We further hypothesize that one of the features of the yet unidentified “positional cue” involves inhibitory effects on specific member(s) of the HDAC family. Among the members of the family, HDA18 mediates differential histone acetylation of the patterning genes between N and H position cells (Fig. 6A). When TSA is applied, the small molecule rapidly spreads over the root cells and inhibits HDAC enzyme activity unselectively in both N and H cells independently of their position (Fig. 6B). As a consequence, the differential effects of the “positional cue” mediating N and H cell differentiation are interrupted. When the inhibitory effect from TSA is sufficiently high, the HDAC enzyme activity of cells at the N or H positions will be inhibited equally; this results in a substantial shift of the regulatory machinery of N position cells to an H cell status and a new histone acetylation balance leading to the stimulation of hair cell differentiation. Thus, the hyperacetylation status of the epidermal cells results in a commitment to the H cell fate (Fig. 6C). When TSA treatment is withdrawn, the position-dependent HDAC activity is restored, and the expression of the patterning genes along with root tip growth again becomes position-dependent. Thus, in our working model, histone acetylation functions as a mediator between the known network of the patterning genes and the yet unidentified positional cue.
To extend the model, additional research is needed to verify the differential histone acetylation status of N and H position cells, and to determine how histone acetylation in cells in each state are regulated by the proposed “positional cue”. Recently Kwak et al. (17) have reported that a receptor-like-kinase, SCM, has a role of in mediating the positional signaling that affects the cellular patterning of the Arabidopsis root epidermis. It is very likely that there are connections between the SCM function and the histone acetylation effects on the differential of the status of the patterning gene in the root epidermal cells.
Other interesting questions about the specificity of functions of the HDAC genes also arise based on our study. According to the tissue-specific gene expression profiling, all 16 genes encoding HDAC in the RPD3 and HD2 families that are sensitive to TSA inhibition (32, 33) have been detected in the root epidermis, except for HDA5 (At5G61060) (34). Our microarray data also detected expression of 14 of the 16 HDAC genes with various responding patterns to TSA treatment (www.cbi.pku.edu.cn/database/ATHChip/page/index.htm). However, so far, only HDA18 has been confirmed to be required for the cellular patterning in the root epidermis, whereas HDA5, HDA14, and HDT4 are not, even though HDA5 is arranged in tandem with and considered to be a duplication of HDA18 (32). These findings provide opportunities to ask how different members of the HDAC family function in position dependent patterning. Because our findings reveals that HDA18 is involved in the regulation of expression of specific patterning genes such as WER, we can investigate how a particular HDAC can selectively acetylate core histones that are involved in regulating specific target genes. In addition to revealing the involvement of histone acetylation in the regulation of cellular pattering in the Arabidopsis root epidermis, our findings clearly show that the Arabidopsis root epidermis is an attractive experimental model that can provide new understanding of the mechanisms whereby gene expression is regulated by changes in histone acetylation.
Supplementary Material
Acknowledgments
We thank K. Okada (Kyoto University, Kyoto) for graciously providing cpc mutant seeds; L.-Y. Xu, J.-S. Yuwen, Z.-P. Lin, and Y.-X. Zhu (College of Life Sciences, Peking University) and D.-P. Liu (Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences) for help with experiments; J.-Y. Li (Institute of Genetics, Chinese Academy of Sciences); X-W. Deng (Yale University, New Haven, CT); W. Gruissem (Institute of Plant Sciences, Eidgenössiche Technische Hochschule); and L. Wolpert (University College, London) and B. Scheres (Utrecht University, Utrecht, The Netherlands) for their critical readings and comments on the work. We especially thank A. Jackson (Department of Plant and Microbial Biology, University of California, Berkeley) for his invaluable contributions to final edits of the manuscript. This work was supported by grants from National Natural Science Foundation Grant 30393114 and Ministry of Science and Technology of PRC Grant 2003CB715906 (to S.-N.B.).
Author contributions: C.-R.X. and S.-N.B. designed research; C.-R.X., C.L., Y.-L.W., L.-C.L., and W.-Q.C. performed research; C.-R.X., Y.-L.W., Z.-H.X., and S.-N.B. analyzed data; and S.-N.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TSA, trichostatin A; HDAC, histone deacetylase; MS, Murashige and Skoog; ChIP, chromatin immunoprecipitation.
References
- 1.Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K. & Scheres, B. (1993) Development (Cambridge, U.K.) 119, 71-84. [DOI] [PubMed] [Google Scholar]
- 2.Schiefelbein, J. (2003) Curr. Opin. Plant Biol. 6, 74-78. [DOI] [PubMed] [Google Scholar]
- 3.Pesch, M. & Hulskamp, M. (2004) Curr. Opin. Genet. Dev. 14, 422-427. [DOI] [PubMed] [Google Scholar]
- 4.Montiel, G., Gantet, P., Jay-Allemand, C. & Breton, C. (2004) Plant Physiol. 136, 3478-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galway, M. E., Masucci, J. D., Lloyd, A. M., Walbot, V., Davis, R. W. & Schiefelbein, J. W. (1994) Dev. Biol. 166, 740-754. [DOI] [PubMed] [Google Scholar]
- 6.Masucci, J. D., Rerie, W. G., Foreman, D. R., Zhang, M., Galway, M. E., Marks, M. D. & Schiefelbein, J. W. (1996) Development (Cambridge, U.K.) 122, 1253-1260. [DOI] [PubMed] [Google Scholar]
- 7.Wada, T., Tachibana, T., Shimura, Y. & Okada, K. (1997) Science 277, 1113-1116. [DOI] [PubMed] [Google Scholar]
- 8.Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., Thumfahrt, J., Jurgens, G. & Hulskamp, M. (2002) EMBO J. 21, 5036-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wada, T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., Marks, M. D., Shimura, Y. & Okada, K. (2002) Development (Cambridge, U.K.) 129, 5409-5419. [DOI] [PubMed] [Google Scholar]
- 10.Lee, M. M. & Schiefelbein, J. (1999) Cell 99, 473-483. [DOI] [PubMed] [Google Scholar]
- 11.Lee, M. M. & Schiefelbein, J. (2002) Plant Cell 14, 611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa, S. & Dolan, L. (2003) Development (Cambridge, U.K.) 130, 2893-2901. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T. & Lloyd, A. (2003) Development (Cambridge, U.K.) 130, 4859-4869. [DOI] [PubMed] [Google Scholar]
- 14.Bernhardt, C., Lee, M. M., Gonzalez, A., Zhang, F., Lloyd, A. & Schiefelbein, J. (2003) Development (Cambridge, U.K.) 130, 6431-6439. [DOI] [PubMed] [Google Scholar]
- 15.Bernhardt, C., Zhao M., Gonzalez, A., Lloyd, A. & Schiefelbein, J. (2005) Development (Cambridge, U.K.) 132, 279-289. [DOI] [PubMed] [Google Scholar]
- 16.Kirik, V., Simon, M., Huelskamp, M. & Schiefelbein, J. (2004) Dev. Biol. 268, 506-513. [DOI] [PubMed] [Google Scholar]
- 17.Kwak, S. H., Shen, R. & Schiefelbein, J. (2005) Science 307, 1111-1113. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida, M., Kijima, M., Akita, M. & Beppu, T. (1990) J. Biol. Chem. 265, 17174-17179. [PubMed] [Google Scholar]
- 19.Brownell, J. E., Zhou, J., Ranalli, T., Kobayashi, R., Edmondson, D. G., Roth, S. Y. & Allis, C. D. (1996) Cell 84, 843-851. [DOI] [PubMed] [Google Scholar]
- 20.Robyr, D., Suka, Y., Xenarios, I., Kurdistani, S. K., Wang, A., Suka, N. & Grunstein, M. (2002) Cell 109, 437-446. [DOI] [PubMed] [Google Scholar]
- 21.Berger, S. L. (2002) Curr. Opin. Genet. Dev. 12, 142-148. [DOI] [PubMed] [Google Scholar]
- 22.Di Laurenzio, L., Wysocka-Diller, J., Malamy, J. E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M. G., Feldmann, K. A. & Benfey, P. N. (1996) Cell 86, 423-433. [DOI] [PubMed] [Google Scholar]
- 23.Masucci, J. D. & Schiefelbein, J. W. (1996) Plant Cell 8, 1505-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogakou, E. P., Redon, C., Boon, C., Johnson, K. & Bonner, W. M. (2000) BioTechniques 28, 38-40, 42, 46. [DOI] [PubMed] [Google Scholar]
- 25.Waterborg, J. H. (1998) J. Biol. Chem. 273, 27602-27609. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, L., Cao, X. & Jacobsen, S. (2002) Curr. Biol. 12, 1360-1367. [DOI] [PubMed] [Google Scholar]
- 27.Hebbes, T. R., Thorne, A. W. & Crane-Robinson, C. (1988) EMBO J. 7, 1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo, M. H. & Allis, C. D. (1999) Methods 19, 425-433. [DOI] [PubMed] [Google Scholar]
- 29.Kurdistani, S. K. & Grunstein, M. (2003) Nat. Rev. Mol. Cell. Biol. 4, 276-284. [DOI] [PubMed] [Google Scholar]
- 30.Wu, K., Tian, L., Zhou, C., Brown, D. & Miki, B. (2003) Plant J. 34, 241-247. [DOI] [PubMed] [Google Scholar]
- 31.Tian, L., Fong, M. P., Wang, J. J., Wei, N. E., Jiang, H., Doerge, R. W. & Chen, Z. J. (2005) Genetics 169, 337-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandey, R., Muller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., Bender, J., Mount, D. W. & Jorgensen, R. A. (2002) Nucleic Acids Res. 30, 5036-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marks, P. A., Miller, T. & Richon, V. M. (2003) Curr. Opin. Pharmacol. 3, 344-351. [DOI] [PubMed] [Google Scholar]
- 34.Birnbaum, K., Shasha, D. E., Wang, J. Y., Jung, J. W., Lambert, G. M., Galbraith, D. W. & Benfey, P. N. (2003) Science 302, 1956-1960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.