Abstract
There is current debate whether genetically modified (GM) plants might contain unexpected, potentially undesirable changes in overall metabolite composition. However, appropriate analytical technology and acceptable metrics of compositional similarity require development. We describe a comprehensive comparison of total metabolites in field-grown GM and conventional potato tubers using a hierarchical approach initiating with rapid metabolome “fingerprinting” to guide more detailed profiling of metabolites where significant differences are suspected. Central to this strategy are data analysis procedures able to generate validated, reproducible metrics of comparison from complex metabolome data. We show that, apart from targeted changes, these GM potatoes in this study appear substantially equivalent to traditional cultivars.
Keywords: genetically modified substantial equivalence, machine learning
There is concern that genetic engineering may allow introduction of unforeseen traits into crops, causing them to contain undesirable metabolites (1, 2). “Substantial equivalence” is used as the starting point to structure current food safety assessment and suggests comparison of intended differences between the genetically modified (GM) plant and progenitor cultivar (1, 2). We compared field-grown tubers from conventional potato cultivars and genotypes bioengineered to contain high levels of inulin-type fructans (3, 4). Inulins stimulate bifidobacteria growth in the intestine and help to boost digestive tract pathogen resistance (5). The beneficial effects of inulins as prebiotic food supplements have been well publicized; thus, this metabolic pathway provides a readily understandable scientific context. Two classes of experimental transgenic line developed in the cultivar Désirée were investigated. The first transgene coded for the enzyme sucrose:sucrose 1-fructosyltransferase (SST), which transfers a fructosyl residue from one sucrose molecule to another, producing the trisaccharide 1-kestose, and oligofructans up to 5 degrees of polymerization (DP) (3, 4). The second transgene was fructan:fructan 1-fructosyltransferase (FFT), the product of which utilizes 1-kestose (and other oligofructans) to build inulin polymers (3, 4).
In any compositional comparison it is important to develop robust metabolomics methodology allowing for, as near as possible, a global analysis of metabolite content (6-8). Established methods for metabolite analysis include gas chromatography, HPLC, or capillary electrophoresis, usually linked to mass spectrometers (9-11). Such approaches result in detailed knowledge relating to only a subset of previously characterized metabolites (6-11), and studies thus far have been restricted to single, relatively small batches of plants produced under controlled growth conditions (9, 12-14). For an initial screen of overall compositional similarity, we propose more rapid and less selective fingerprinting techniques that do not incorporate a chromatographic step (8, 15-18). Fingerprints based on MS, such as flow injection electrospray ionization (FIE)-MS, can be regarded as simplified images of total sample composition in that the measured variables (m/z) are compiled by integrating the levels of more than one metabolite (e.g., for isomers). Where compositional differences unrelated to the bioengineered trait are suggested, substantial equivalence testing can be applied to more detailed metabolome analysis involving a chromatographic step guided by the fingerprinting results.
Defining substantial equivalence does not fall neatly into a standard statistical task. Unsupervised data analysis techniques, such as principal components analysis (PCA) (19) look for regularities in unlabeled data. Supervised techniques, such as linear discriminant analysis (LDA) (19, 20) and decision tree analysis (21), build models that discriminate between labeled data (22, 23). However, for substantial equivalence we are interested in data similarity rather than the ability to discriminate classes. We reason that if an unsupervised algorithm clusters metabolome samples close together, then they can be objectively considered to be similar, and if classes cannot easily be discriminated by supervised methods then they are objectively similar.
The overall experimental approach was to initially evaluate the degree of compositional similarity between tubers of individual traditional potato cultivars. This comparison provided a context for determining whether transgenic potatoes displayed alterations in metabolite composition outside the range exhibited normally by conventional cultivars. To ensure comprehensive coverage of the metabolome, a hierarchical approach was adopted that initially involved a nonselective metabolite fingerprinting technique followed by more detailed global profiling of individual metabolites and finally a targeted analysis of any metabolites responsible for discriminating GM genotypes. Data-mining methods were used that were specifically capable of identifying metabolites responsible for differences between potato genotypes. The use of several different data analysis methods ensured that any conclusions relating to metrics of similarity were independent of specific statistical treatments.
Materials and Methods
Plant Material. The experimental transgenic genotypes derived from the progenitor cultivar Désirée are described in ref. 3. The GM plants were grown under field conditions in a block design for the 2001 and 2003 growing seasons together with the conventional cultivars Agria, Linda, Granola, Solara, and two Désirée lines [one line was propagated through tissue culture (De2), and the other was obtained from tuber propagation]. Approximately 48 tubers were selected at random from each of four randomly arranged field blocks and stored at 4°C for 4 weeks before sample preparation. Potato tuber disks (fresh weight, 200 mg each) were excised from 3 mm below the tuber peel, perpendicular to the main tuber axis. Immediately after cutting, disks were frozen in liquid nitrogen and kept frozen at -80°C before extraction.
Sample Preparation and Metabolite Analysis. Tuber slice homogenization and extraction in 1 ml of prechilled water/methanol/chloroform (2:5:2, vol/vol/vol) and GC TOF-MS analysis were carried out as described in ref. 24. FIE-MS was performed with an LCT mass spectrometer (Micromass, Manchester, U.K.) as described in detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Randomized extracts were diluted 1:50 in water/methanol (60:40, vol/vol), and aliquots of 40 μl were injected into a flow of 100 μl·min-1 water/methanol (60:40, vol/vol) with a Waters Alliance 2690 liquid chromatography (LC) system.
LC-MS-targeted analysis for glycoalkaloids and oligofructans was performed with a LCQ Quantum triple quadrupole mass spectrometer (ThermoFinnigan, San Jose, CA) running xcalibur software (version 1.3, ThermoFinnigan) as described in ref. 25.
Confirmation of 1-kestose presence besides raffinose in the Solara, Linda, SST, and SST/FFT lines was performed by hydrophilic-interaction LC (10) and triple-quadrupole MS in MRM mode on fragmentations of parent ion m/z 522 to m/z 325, 163, 145, 127, and 85. Chromatograms were processed with lcquan (xcalibur, version 1.3).
Data Analysis. FIE-MS raw data were first log-transformed and then normalized to the total ion current before analysis. All GC-TOF data were normalized to total peak area and then log-transformed. The latter data matrix contained 15.4% missing values, being either below detection limit (true low values) or missed because of failures of the automatic deconvolution and peak detection software (missing values). The 1-kestose (expected new metabolite in GM lines) region in all 2,253 chromatograms was manually checked and corrected because this molecule was found to have a retention time very close to that of raffinose. Undetected peaks were excluded in the univariate analysis.
Boundaries delimiting the relative concentration range of each metabolite observed by GC-TOF in the conventional cultivars were first determined, and the level of each metabolite in GM lines was then compared to the specific limits set for it. From frequency distributions of metabolites in cultivars regarded as “safe,” upper and lower limits of commonly detected relative metabolite levels were calculated. One-sigma deviations from cultivar mean levels were regarded as a conservative borderline of typically found food metabolite levels. For each, one standard deviation from each comparator group mean was calculated, and the overall maximum and minimum were taken as conservative estimations of the extents of acceptability. As a further test, it was determined also whether the mean of an individual GM line differed significantly from the mean of each of the cultivars. Nonparametric multiple comparisons corrected for unequal sample sizes with tied ranks (described in ref. 26) were performed with the r environment (27) and the results presented as Q values.
For multivariate analysis, each initial data matrix was split randomly into a training set and a test set (two-thirds and one-third, respectively). This method of division allows a direct comparison of the accuracies of any model using McNemar's test (28, 29). Some multivariate methods (e.g., PCA) require complete data matrices (19, 30, 31); therefore, when required, the overall mean of the peak intensity taken from the training set was applied to in-fill the missing values in the training and validation sets. PCA (30) as carried out by using matlab (version 6.5, release 13; Mathworks, Natick, MA) on the mean-centered covariance matrix of the training set. The training set only was used to build PCA models. LDA (19) [also referred to in chemometrics literature (17) as discriminant function analysis] was implemented in matlab according to the procedure described in ref. 17. Decision tree analysis was carried out on the original data matrix (without in-filling) and in the mean in-filled data matrix using an implementation of the c4.5 algorithm (21) in the rpart package in r (27). The results of the analysis on the original data are presented, but broadly similar overall classification accuracies were achieved by using both data sets.
Results
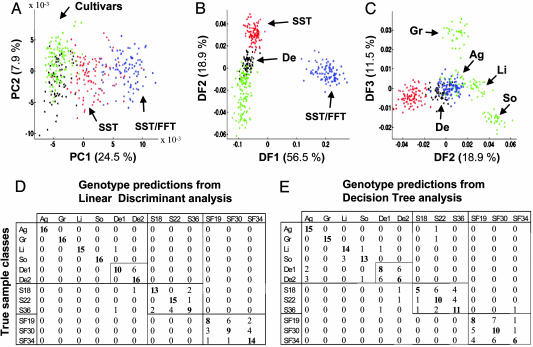
Potato Genotypes Have Distinct Metabolomes. FIE-MS fingerprints were generated for 600 samples representing all genotypes selected randomly from four field plots. PCA showed that metabolome variation was dominated by the three major genotype metaclasses (cultivars, SST, and SST/FFT) (Fig. 1A). We further applied two different supervised data analysis approaches, LDA (8, 19, 20, 23) and a decision tree method (8, 12, 21), both of which produce interpretable results. Visualizing the data with the first three discriminant functions (DFs) reveals a more comprehensive separation of the classes (Fig. 1 B and C). The class membership of unseen samples (test set) can be visualized in a confusion matrix when evaluating the predictive power of the LDA model (Fig. 1D). Misclassification was restricted largely within the three main genotype groupings with only ≈4% of SST samples misclassified as Désirée. Within the group containing conventional cultivars significant confusion occurred only between the two Désirée genotypes, suggesting that each cultivar has a distinct metabolome. Although decision trees rely on a different mechanism to develop a model from the original FIE-MS fingerprints, a pattern almost identical to that seen in the LDA was evident (Fig. 1E).
Fig. 1.
FIE-MS (+) metabolite fingerprints of tuber extracts from five conventional potato cultivars [Ag, Agria; De, Désirée (1 and 2); Gr, Granola; Li, Linda; So, Solara] and two types of transgenic lines (SST and SST/FFT) analyzed by different multivariate data analysis methods. (A) PCA with Désirée genotypes are colored black, other cultivars are colored green, SST genotypes are colored red, and SST-FFT genotypes are colored blue. The PCA scores plots for PC1 versus PC2 are presented. (B and C) LDA with color coding as in A. The LDA scores plots for DF1 versus DF2 (B) and DF2 versus DF3 (C) are presented. (D and E) Confusion matrix of the LDA class predictions and decision tree class predictions in the test set. The confusion matrices are read in rows, with the numbers indicating the frequency with which samples are predicted to be either of the true class or an alternative genotype. Correct classifications are highlighted in bold.
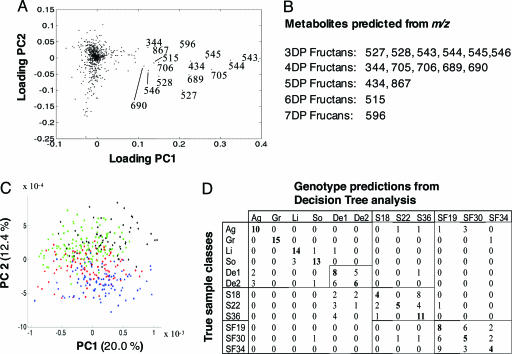
The Most Discriminatory Ions Are Derived from Fructans. The GM lines had been engineered to synthesize novel metabolites; therefore, the virtually complete separation of GM and non-GM lines in PCA space was not unexpected. Investigation of the relative contribution (loadings) of individual variables in the PC1 dimension highlighted 15 ions with a significant impact on genotype separation (Fig. 2A). All of these top-ranked variables were predicted to represent fructan molecules of increasing DP (Fig. 2B and Table 2, which is published as supporting information on the PNAS web site). Reanalysis of representative extracts by hydrophilic interaction LC-MS confirmed this proposition (see Fig. 3A for an example chromatogram). When the analysis was repeated with top-ranking ions (>0.05 in PC1) omitted from the data, although separation of GM and non-GM genotypes on the vector of major variance (PC1) was no longer achieved, some general grouping of samples in the three metaclasses was still evident in PC2 (Fig. 2C). This observation was corroborated by decision tree analysis of the reduced data, which showed that classification of individual cultivars and discrimination between SST/FFT lines and other genotypes was still excellent; however, there was a significant increase (McNemar's test (28, 29) = 7.2; P = 0.007) in confusion between SST genotypes and Désirée (Fig. 2D). The lack of total collapse in the classification models when these ions were removed from the data suggested that further metabolic differences could exist that might be revealed only by a more comprehensive profiling method.
Fig. 2.
Identification of top-ranking ions for genotype separation in PCA and effect on multivariate models when omitted from data. From a PCA, it is possible to investigate the contribution of each variable to each of the principal components, a metric referred to as the loadings score. (A) Loadings plot of PC1 versus PC2 of FIE-MS fingerprint data representing GM and non-GM potatoes used to derive Fig. 1 A. (B) The m/z of 15 ions with high-loading scores (>0.1) in the PC1 dimension are labeled, and all were found to be masses typical of fructans with varying DP (Table 2). (C) PCA using FIE-MS data with top-ranked ions (>0.05 in PC1) omitted. (D) Decision tree analysis using FIE-MS data with top-ranked ions (>0.05 in PC1) omitted.
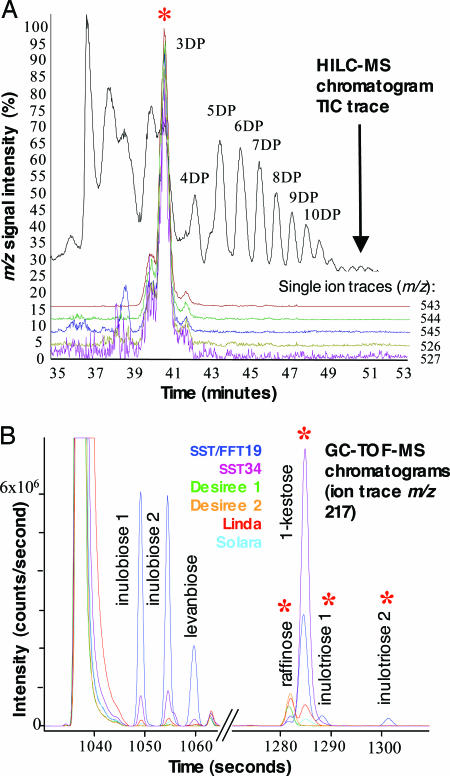
Fig. 3.
Identification of discriminatory metabolites in GM potato tubers by LC-MS and GC-MS. (A) Overlaid single-ion chromatograms of top-ranked variables predicted to represent ions derived from a fructan with 3 DP in an example SF30 extract analyzed by hydrophilic interaction LC-MS. The major peak with coincident signals from extracted ion chromatograms of m/z 543, 544, 545, 526, and 527 at the retention time of 3-DP fructans in the total ion current (TIC) trace is indicated with a red asterisk. The position of peaks representing fructans of increasing DP are indicated. (B) Exemplary GC-TOF-extracted ion chromatogram m/z 217 for GM and non-GM potato tubers, enlarged for discriminatory disaccharide and trisaccharide regions. Separation of the discriminatory peaks of inulobiose 1, inulobiose 2, and levanbiose from the major disaccharide sucrose and separation of the discriminatory trisaccharide peaks of inulotriose 1 and inulotriose 2 from 1-kestose and raffinose (red asterisk) are indicated. The increase in discriminatory abundances of 2- and 3-DP fructans in GM lines and the presence of 1-kestose in Linda and Solara cultivars is shown, whereas 1-kestose is absent in the direct GM comparator Désirée.
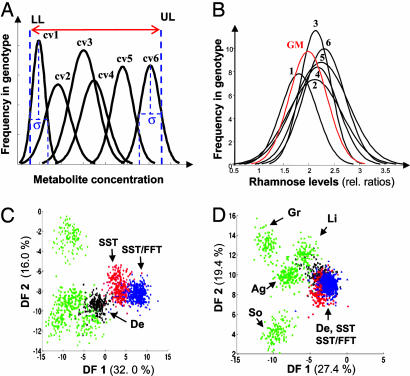
Only Anticipated Metabolites Were Found in GM Lines. Analysis was extended in scope and depth in the next layer of data acquisition and testing for which 2,182 tubers were analyzed from the 12 genotypes, again randomized over all field plots. GC-TOF-MS (24) recorded 252 metabolite peaks in an automated manner (90 positively identified, 89 assigned to a specific metabolite class, and 73 classified as unknowns). The chromatographic region associated with the retention time of major disaccharides and trisaccharides of several chromatograms representative of the major genotype groups is shown in Fig. 3B. Because each of the conventional cultivars can on the basis of consumption be regarded as safe, single metabolites were sought initially that were “out of range” in a GM line (Fig. 4 A and B). Two metabolites present in GM lines were not detected in cultivars, and a further four metabolites had means above the upper limit of the range set for cultivars (Table 1 and see Fig. 3B). By further targeted analysis, these six peaks were characterized from authentic standards (or isolated fractions from chicory), corresponding mass spectra and chromatographic retention indices as fructose-containing trisaccharides (1-kestose and inulotriose) as predicted by the FIE-MS analysis and in addition the disaccharide fructans levanbiose and inulobiose (Fig. 5, which is published as supporting information on the PNAS web site). In a test comparing metabolite mean concentrations in GM lines to each cultivar mean (26) only the same six metabolite peaks were identified as significantly different (Q = > 3.72; P = <0.001) (Table 3, which is published as supporting information on the PNAS web site).
Fig. 4.
GC-TOF profiling to detect and assess the impact of out-of-range metabolites in comparison to GM genotypes with conventional potato cultivars. (A) Visualization of the concept of metabolite concentration out-of-range assessment in substantial equivalence analysis. Determination of frequency distributions of metabolites in six cultivars (cv) regarded as a safe result in an upper limit (UL) and lower limit (LL) of concentration for each commonly detected metabolite. (B) Illustration of the out-of-range assessment concept using rhamnose levels (relative ratio of metabolite peak area in data normalized to total peak area in each chromatogram). Frequency distributions of ≈150 tubers per potato line have been curve-fitted. 1, Linda; 2, Désirée1; 3, Désirée2; 4, Solara; 5, Granola; 6, Agria; GM, single transformant of line S22. Average rhamnose levels in S22 are found significantly different from the Désirée parental lines in univariate statistics but fall well within the overall range typical of potato cultivars. (C) LDA scores plot of GC-TOF data. (D) Scores plot of LDA performed on the same data but with the omission of the six discriminatory fructan peaks representing levanbiose, 1-kestose, inulobiose, and inulotriose (see Fig. 3B).
Table 1. Metabolites that are out of range in at least one GM group compared with non-GM cultivars.
| Fructans | S18 | S22 | S36 | SF19 | SF30 | SF34 | LL | UL |
|---|---|---|---|---|---|---|---|---|
| Levanbiose | 2.29 | 2.79 | 2.57 | 3.60 | 3.76 | 3.57 | 1.54 | 2.20 |
| Inulobiose 1 | 2.75 | 3.7 | 2.96 | 3.90 | 4.02 | 3.84 | 1.39 | 2.49 |
| Inulobiose 2 | 2.83 | 3.30 | 3.10 | 3.95 | 4.05 | 3.87 | 1.71 | 2.77 |
| Inulotriose 1 | nd | 3.05 | 2.79 | 2.63 | 2.84 | 2.48 | nd | nd |
| Inulotriose 2 | 1.60 | 2.85 | 1.60 | 2.63 | 2.85 | 2.49 | nd | nd |
| 1-Kestose | 3.91 | 3.89 | 3.68 | 3.48 | 3.67 | 3.72 | 2.07 | 3.58 |
LL, lower limit; UL, upper limit; nd, not detectable.
Analysis of GC-TOF data by PCA, LDA, and decision trees revealed a similar pattern of genotype clustering/discrimination to that observed in the fingerprinting analysis (Fig. 4C and Figs. 6A, 7A, and 8A, which are published as supporting information on the PNAS web site). The same fructose-derived oligosaccharides highlighted in univariate analyses were important in multivariate models (Fig. 6B). When these oligosaccharides were omitted from the data, PCA failed to separate any classes (Fig. 6C). With this reduced data set, LDA resulted in distinct genotype clustering in which it was difficult to satisfactorily discriminate the GM lines from the two cultivar Désirée background groups, whereas the other cultivars remained isolated (Figs. 4D and 7B). Genotype classification accuracy was similarly compromised in decision tree analysis (Fig. 8B).
Glycoalkaloid Levels Are Normal in GM Potatoes. We have concluded from a metabolomics study incorporating a range of different data analysis techniques that only six important fructosyl peaks resulted from the genetic modifications in potato. Disregarding this finding of only minor changes in oligosaccharide metabolism, the possibility of changes in possibly toxic, low level, secondary metabolites could not be excluded apriori. Further targeted analysis (Fig. 9, which is published as supporting information on the PNAS web site) revealed no changes in the levels of glycosidic steroidal alkaloids (α-chaconine and α-solanine), which usually comprise up to 95% of the total glycoalkaloid content of tubers from domesticated Solanum tuberosum cultivars (32).
Discussion
The nature of food in terms of safety cannot be assessed in an absolute manner. As a first pass in any compositional comparison, we suggest that a rapid but sensitive comprehensive and comparably inexpensive first screen can be provided by mass spectrometric fingerprinting, which may be complemented by more detailed analyses using GC-TOF or LC-MS, depending on the level of similarity to other cultivars as determined by statistical analysis.
A major finding from the present study was the large variation in metabolite profile between the conventional cultivars. These significant differences were never sought as desired traits in traditional breeding programs, and overall composition has not given cause for public safety concerns in conventionally bred cultivars. In the context of substantial equivalence, we show that the metabolite composition of field-grown inulin-producing potatoes were within the natural metabolite range of classical cultivars and were, in fact, very similar to the progenitor line Désirée, with the exception of the introduced genes and, therefore, the predictable up-regulation of fructans and their expected derivatives. In the comparative assessment framework, such metabolic side products might eventually be subjected to more detailed investigations if deemed necessary with respect to toxicity, abundance, and chemical structure.
The cultivar-based compositional heterogeneity we describe emphasizes the importance of comparison with a range of equivalent cultivars and not solely the parental line. For example, although 1-kestose was not found in the genetic background line of the GM plants, Désirée, a trisaccharide indistinguishable from 1-kestose was found in Solara and Linda tubers (see Fig. 3B). According to the GC-TOF data, supervised multivariate statistics demonstrated continuing cultivar distinction despite omitting the fructosyl-oligosaccharides found in GM tubers. This result indicated that metabolic changes caused through conventional breeding techniques were, in these cases, at least of a comparable magnitude to those resulting as an unintended effect of genetic engineering techniques.
Supplementary Material
Acknowledgments
We thank Bernd Hommel, Pia Roppel, and colleagues for designing and undertaking the field trials under Bundesanstalt für Land und Forstwirtschaft Project 0312632; Karin Koehl for study design; Arnd Heyer (Max Planck Institute for Molecular Plant Physiology) for generation and supply of transgenic material and helpful discussion; André van Laere and Wim van den Ende (Katholieke University, Leuven, Belgium) and Jerry Chatterton and Phil Harrison (Utah State University, Logan) for providing 2- to 4-DP fructan reference compounds; Jim Heald and Robert Darby for supporting LCT analysis; and Roy Goodacre and David Broadhurst for advice on data analysis. The metabolite analysis and statistical work was funded by the Food Standards Agency (London) as part of its G02006 project.
Author contributions: N.H., A.S., R.D.K., D.B.K., O.F., and J.D. designed research; G.S.C., M.B., M.M., B.Z., and O.F. performed research; G.S.C., M.B., D.P.E., J.T., N.H., R.D.K., and J.D. analyzed data; G.S.C., M.B., D.P.E., R.D.K., O.F., and J.D. wrote the paper; and J.D. coordinated the project consortium.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GM, genetically modified; SST, sucrose:sucrose 1-fructosyltransferase; FFT, fructan:fructan 1-fructosyltransferase; DP, degree(s) of polymerization; PCA, principal components analysis; FIE, flow injection electrospray ionization; LDA, linear discriminant analysis; DF, discriminant functions.
References
- 1.Organisation for Economic Cooperation and Development (2001) Report of the OECD Workshop on the Nutritional Assessment of Novel Foods and Feeds (Org. Econ. Cooperation Dev., Ottawa).
- 2.Kok, E. J. & Kuiper, H. A. (2003) Trends in Biotechnol. 21, 439-444. [DOI] [PubMed] [Google Scholar]
- 3.Hellwege, E. M., Czapla, S., Jahnke, A., Willmitzer, L. & Heyer, A. G. (2000) Proc. Natl. Acad. Sci. USA 97, 8699-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edelman, J. & Jefford, T. G. (1968) New Phytol. 67, 517-531. [Google Scholar]
- 5.Gibson, G. R., Beatty, E. R., Wang, X. & Cummings, J. H. (1995) Gastroenterology 108, 968-975. [DOI] [PubMed] [Google Scholar]
- 6.Fiehn, O. (2002) Plant Mol. Biol. 48, 155-171. [PubMed] [Google Scholar]
- 7.Sumner, L. W., Mendes, P. & Dixon, R. A. (2003) Phytochemistry 62, 817-836. [DOI] [PubMed] [Google Scholar]
- 8.Goodacre, R., Vaidyanathan, S., Dunn, W. B., Harrigan, G. G. & Kell, D. B. (2004) Trends Biotechnol. 22, 439-444. [DOI] [PubMed] [Google Scholar]
- 9.Roessner, U., Wagner, C., Kopka, J., Trethewey, R. N. & Willmitzer, L. (2000) Plant J. 23, 131-142. [DOI] [PubMed] [Google Scholar]
- 10.Tolstikov, V. V. & Fiehn, O. (2003) Anal. Biochem. 301, 298-307. [DOI] [PubMed] [Google Scholar]
- 11.Sato, S., Soga, T., Nishioka, T. & Tomita, M. (2004) Plant. J. 40, 151-163. [DOI] [PubMed] [Google Scholar]
- 12.Taylor, J., King, R. D., Altmann, T. & Fiehn, O. (2002) Bioinformatics 18, S241-S248. [DOI] [PubMed] [Google Scholar]
- 13.Fiehn, O., Kopka, J., Altmann, T., Trethewey, R. & Willmitzer, L. (2000) Nat. Biotechnol. 18, 1157-1161. [DOI] [PubMed] [Google Scholar]
- 14.Roessner, U., Willmitzer, L. & Fernie, A. R. (2001) Plant Physiol. 127, 749-764. [PMC free article] [PubMed] [Google Scholar]
- 15.Ward, J. L., Harris, C., Lewis, J. & Beale, M. H. (2003) Phytochemistry 62, 949-957. [DOI] [PubMed] [Google Scholar]
- 16.Aharoni, A., De Vos, C. H. R., Verhoeven, H. A., Maliepaard, C. A., Kruppa, G., Bino, R. & Goodenowe, D. B. (2002) OMICS 6, 217-234. [DOI] [PubMed] [Google Scholar]
- 17.Allen, J., Davey, H. M., Broadhurst, D., Heald, J. K., Rowland, J. J., Oliver, S. G. & Kell, D. B. (2003) Nat. Biotechnol. 21, 692-696. [DOI] [PubMed] [Google Scholar]
- 18.Scholz, M., Gatzek, S., Sterling, A., Fiehn, O. & Selbig, J. (2004) Bioinformatics 20, 2447-2454. [DOI] [PubMed] [Google Scholar]
- 19.Manley, B. F. J. (1994) Multivariate Statistical Methods: A Primer (Chapman & Hall, London).
- 20.Goodacre, R., Timmins, E. M., Burton, R., Kaderbhai, N., Woodward, A. M., Kell, D. B. & Rooney, P. J. (1998) Microbiology 144, 1157-1170. [DOI] [PubMed] [Google Scholar]
- 21.Quinlan, J. R. (1993) c4.5: Programs for Machine Learning (Morgan Kaufmann, San Mateo, CA).
- 22.Wu, B., Abbott, T., Fishman, D., McMurray, W., Mor, G., Stone, K., Ward, D., Williams, K. & Zhao, H. (2004) Bioinformatics 19, 1636-1643. [DOI] [PubMed] [Google Scholar]
- 23.Kell, D. B., Darby, R. M. & Draper, J. (2001) Plant Physiol. 126, 943-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weckwerth, W., Loureiro, M. E., Wenzel, K. & Fiehn, O. (2004) Proc. Natl. Acad. Sci. USA 101, 7809-7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zywicki, B., Catchpole, G., Draper, J. & Fiehn, O. (2004) Anal. Biochem. 336, 178-186. [DOI] [PubMed] [Google Scholar]
- 26.Zar, J. H. (1984) in Biostatistical Analysis (Prentice-Hall, Englewood Cliffs, NJ), 2nd Ed, p. 201.
- 27.Gentleman, R. & Ihaka, R. (2004) r: A Language and Environment for Statistical Computing and Graphics (Univ. Auckland, Auckland, Australia).
- 28.McNemar, Q. (1947) Psychometrika 12, 153-157. [DOI] [PubMed] [Google Scholar]
- 29.Dietterich, T. G. (1998) Neural Comput. 10, 1895-1923. [DOI] [PubMed] [Google Scholar]
- 30.Jolliffe, I. T. (1986) Principal Component Analysis (Springer, New York).
- 31.Little, R. J. A. & Rubin, D. B. (1987) Statistical Analysis with Missing Data (Wiley, New York).
- 32.Van Gelder, W. M. J. (1991) in Poisonous Plant Contamination of Edible Plant, ed. Abdel-Fattah, M. (CRC Press, Boca Raton, FL).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.