Abstract
The extent to which lateral genetic transfer has shaped microbial genomes has major implications for the emergence of community structures. We have performed a rigorous phylogenetic analysis of >220,000 proteins from genomes of 144 prokaryotes to determine the contribution of gene sharing to current prokaryotic diversity, and to identify “highways” of sharing between lineages. The inferred relationships suggest a pattern of inheritance that is largely vertical, but with notable exceptions among closely related taxa, and among distantly related organisms that live in similar environments.
Keywords: lateral genetic transfer, microbial genomes, molecular phylogeny
Beginning in the 1980s, recognition of the 16S ribosomal RNA gene (rDNA) as a molecular chronometer (1), the development of automated sequencing technology and PCR, and improved phylogenetic methods (2) converged to yield a universal phylogenetic tree (1, 3) that was often interpreted as the “tree of life.” However, trees inferred from protein-coding genes or proteins are not always topologically congruent with the rDNA tree (4, 5) or with one another (6, 7). Instances of incongruence are often attributed to historical transfers of genetic information from one genealogical lineage to another (8). Mechanisms for lateral genetic transfer (LGT) are well characterized, and in a laboratory context underpin much of the biotechnology industry. At issue, however, is the extent to which LGT has contributed to the natural diversity of prokaryotes. If LGT has been rampant and consequential, there may in fact be no universal tree of life, and attempts to construct a phylogenetic classification of prokaryotes based on molecular sequence information will ultimately be futile (9).
Least controversial among proposed LGT events are transfers that span relatively short evolutionary distances, where the donor and recipient organisms are members of the same species. DNA exchanges between close relatives are likely to be successful because of compatible methods of genetic exchange such as conjugation and the increased likelihood of homologous recombination between the donated DNA and the recipient genome (10). Linkage disequilibrium analysis of environmental samples has revealed extensive homologous recombination within many species of prokaryotes (11). Transfers between distantly related taxa are much less likely to succeed, because conjugation or viral transduction of genes between different species is less common (although not impossible), and foreign DNA must be integrated into the genome via illegitimate rather than homologous recombination (10). However, if organisms in the environment are subjected to a constant “rain” of DNA (12), then these rare processes will occur in evolutionary time, and will be fixed in a lineage especially if they confer a selective advantage on the recipient organism. LGT has tremendous implications for the genesis and evolution of microbial communities: if extensive LGT can occur among distantly related organisms, then processes such as niche invasion may be greatly accelerated, and genes that confer advantages against host defense mechanisms may be shared widely among pathogens.
There is considerable evidence for long-distance LGT events. To date, this evidence has typically been based on either the broad application of nonphylogenetic “surrogate” methods across a wide range of taxa (13) or the use of phylogenetic methods on a subset of available taxa with restricted phylogenetic (14) or environmental (15) distributions. However, the set of sequenced genomes represents taxa that are phylogenetically and ecologically diverse, and can be used to test broad hypotheses about the sharing of genes. Here we apply rigorous phylogenetic methods to annotated proteins from 144 completely sequenced genomes sampled across 15 phyla of prokaryotes. We derive a reference supertree from 22,432 orthologous protein families, and by comparing individual protein trees with this reference tree, we infer for this data set the frequency and phyletic extent of LGT, the taxa implicated as partners in LGT events, and the tendency of different cellular functions to be subjected to transfer.
Methods
Additional details are provided in Supporting Text, Tables 2-7, and Figs. 4-20, which are published as supporting information on the PNAS web site. The 422,971 conceptually translated protein sequences recognized for all 144 prokaryotic genomes publicly available as of November 15, 2003, were downloaded from the National Center for Biotechnology Information and clustered by using a Markov algorithm (16), yielding 5,864 Markov clusters of size n ≥ 4 containing a total of 382,991 proteins. Sequences within each cluster were hierarchically clustered, and maximal subsets in which no genome is represented more than once (maximally representative clusters, MRCs; ref. 16) were identified, yielding 22,437 MRCs containing 220,240 sequences. Sequences in each MRC were aligned by using several different algorithms, and the alignment yielding the highest score according to the word-oriented objective function (WOOF) (17) was chosen for subsequent analysis. Ambiguously aligned regions were removed (18) to yield 22,432 alignment sets. Bayesian phylogenetic analysis (19, 20) was used to associate posterior probability (PP) values with all possible groupings of taxa (bipartitions) for each alignment set. Models and parameters were selected after extensive calibration (supporting information). Bipartitions (internal edges) having PP ≥0.95 were assessed for topological consistency with a reference supertree generated by the MRP method (21) from all strongly supported (PP ≥ 0.95) bipartitions among the 22,432 protein trees. In the absence of eukaryotic nuclear genomes from our analysis, the supertree was arbitrarily rooted on the edge connecting the bacterial and archaeal subtrees. This rooting does not imply that prokaryotes constitute a monophyletic group.
We developed an algorithm to identify the minimal set of subtree prune-and-regraft operations (22), here simply termed edits, required to make our supertree topologically consistent with a given protein tree (Fig. 1). Of the 19,672 protein trees fully or partially resolved at PP ≥0.95, we computed the minimal edit path exactly for 19,351 (13,849 completely congruent with the supertree, and 5,502 with a nonzero edit distance) and used ratchet-based heuristics (see supporting information) to recover a result for 237 of the remaining 321. The minimum number of proteins in any of these 321 data sets was 14; in simulations with smaller data sets, where our heuristics returned a result it was minimal in >95% of cases. Edit distances ranged from 1 (3,694 trees) to 22 (1 tree).
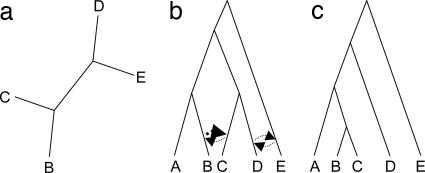
Fig. 1.
Reconciliation of an unrooted protein tree with a rooted reference tree. Successive subtree prune-and-regraft (edits) operations (edits) are applied to the reference tree (b), until (ideally) all topologies, or for complex comparisons at least one topology, consistent with the protein tree (a), is obtained. In this example, only a single operation is needed to generate a tree (c) with which the tree in a is topologically congruent. Four alternative edits, indicated with arrows representing the direction of gene flow, can reconcile the reference and protein trees in a single step (thus with an edit path of length 1). The tree in c is the result of the edit operation implied by the boldface arrow in the tree in b: an ancestor of taxon B donates genetic material to an ancestor of taxon C. This is implemented algorithmically by breaking the terminal edge subtending C and reannealing it along the terminal edge subtending B. The resulting rooted tree (c) contains no bipartitions that are discordant with protein tree (see supporting information for details). In this simple example there is no obligate edit path; (B,C) and (D,E) are possible (but mutually exclusive) partner pairs in the implied LGT event.
Results
A Supertree of Prokaryotic Life. From 422,971 proteins in 144 genomes (Table 2) we generated and aligned 22,432 orthologous sets (families) of size 4 or greater, covering 220,240 proteins (52.1%) in total. For each family, we inferred a Bayesian phylogenetic tree. Of the 152,808 bipartitions in these trees, 95,950 have rounded PP ≥0.95 and were used to compute a supertree by the method of matrix representation with parsimony (21). This supertree (Fig. 4), our reference hypothesis about relationships among these 144 prokaryotes, is remarkably congruent with taxonomy based on 16S rDNA. Of the nine phyla represented by more than one genome, our supertree reconstructs eight as monophyletic, with only Euryarchaeota paraphyletic.
Individual protein trees that strongly (PP strictly ≥0.95) support the bipartition of taxa implied by a given internal edge or node in the supertree are concordant with that node, and support a regime of vertical inheritance at that node. Protein trees that are strongly incongruent with a supertree node are discordant, and provide prima facie evidence of LGT. Of the 95,194 strongly supported bipartitions among our 22,432 protein trees, 82,473 (86.6%) are concordant with the supertree. Discordance is highly variable across the supertree: for many nodes that subtend a single genus or species, <5% of the corresponding protein tree bipartitions with PP ≥0.95 are discordant, whereas for “backbone” nodes that define the branching order of phyla, frequently >40% are discordant (see ref. 23). The implied relationships among members of a single genus are sometimes strongly supported (e.g., Bordetella and Staphylococcus) but are more often contradicted by many protein trees (e.g., Clostridium, Prochlorococcus, and some relationships within Streptococcus and Escherichia). Only 22 of 110 protein trees strongly support the basal Aquifex + Thermotoga clade seen in our supertree and in many previous studies (24-26). The phylogenetic approach strongly supports alternative partners for these two genomes.
To examine whether methodological artefacts could be responsible for this level of discordance, we carried out extensive statistical analyses (see supporting information) to test whether inferred discordance was more prevalent among data sets that are most prone to artefacts of clustering, alignment or phylogenetic inference. For some tests of protein clustering and G+C content biases, increasing threshold stringency eliminates more discordant than concordant conclusions. We also performed a bootstrapped parsimony analysis of insertion and deletion states in the aligned protein sequences. Over all cases where strongly supported bipartitions (PP ≥ 0.95) are paired with strong parsimony conclusions (bootstrap ≥ 70%), the level of agreement for discordant bipartitions (92%) is only slightly lower than for concordant ones (94%). These tests imply that, at the stringent PP thresholds we employ here, erroneous conclusions are only slightly biased toward discordance, i.e., toward LGT.
Genome Partners and the Phylogenetic Network. Proteins with discordant histories can be identified by simple comparison against a reference tree. It is much more difficult to identify the partners implicated in a transfer, or the shortest transfer path. The edit path between the supertree and a discordant protein tree represents a hypothesis about the set of historical LGT events responsible for the observed discordance. We developed an algorithm (supporting information) to search recursively for the shortest edit path(s) between the supertree and each discordant protein tree. We define a transfer as obligate if it is implied by every path in the set of most-parsimonious edit paths resolving the discordance of a given MRC, and as possible if it appears in some, but not necessarily all, of the most-parsimonious edit paths. Implied LGT events found in the edit paths of many discordant protein trees define “highways” of LGT between taxa.
We observe that many common obligate edit operations (putative LGT events) affect taxa that are topologically close to each other and relatively terminal in the supertree. One such event, inferred for no fewer than 175 protein trees, implies transfer between an ancestor of Yersinia pestis and a common ancestor of Escherichia coli plus Salmonella. More than 250 LGT events are implicated within the Synechococcus-Prochlorococcus clade, consistent with the low support values seen in the reference supertree. Because LGT between immediate sister taxa cannot be inferred by using topological comparisons, our inferred counts of “short-distance” edits likely underestimate the true extent of sharing between closely related genomes.
“Long-distance” edits imply LGT between taxa from different phyla or divisions, typically crossing basal or “backbone” nodes in the supertree. β-Proteobacteria are implicated in a particularly large number of obligate long-distance LGT events, >150 with the γ-proteobacterial genus Pseudomonas alone. The two best-represented phyla, Proteobacteria and Low-G+C Gram-positives, are implicated in many long-distance edits; this is not wholly due to sampling frequency, as the four proteobacterial divisions (α, β, γ, and ε) preferentially exchange genes with different partners (Fig. 2) to an extent not simply proportional to the number of genomes represented in each division. Aquifex shares a substantial number of transfer events only with ε-proteobacteria, whereas clostridia (here including Fusobacterium nucleatum and Thermoanaerobacter tengcongensis) show diverse transfer relationships including with euryarchaeotes, Thermotoga, and ε-proteobacteria. The β-proteobacteria exhibit more obligate transfers with other proteobacterial groups than among themselves, whereas pseudomonads and xanthomonads are frequently intermingled with β-proteobacteria, and sometimes with α-proteobacteria, in the protein trees.
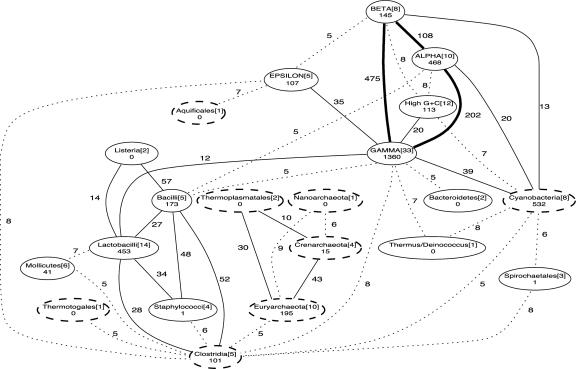
Fig. 2.
Highways of obligate gene transfer within and among phyla and divisions of prokaryotes, based on analysis of the 22,348 protein trees for which a minimal edit path could be resolved. Each oval represents a prokaryotic group, with the name and number of taxa in that group indicated on the first line. Numbers below taxon names report inferred transfers within that taxon, whereas numbers on the linking edges report inferred transfers between taxa. Ovals representing groups with one or more thermophilic organisms are drawn with dashed lines. The type of line shown between each pair of taxa indicates the number of obligate edits in this analysis: >100 with thick solid lines, 10-99 with thin solid lines, and 5-9 with dashed lines. Relationships between taxonomic groups with fewer than five obligate edits are not shown. Note that transfers cannot be identified within phyla with one (e.g., Nanoarchaeota) or two (e.g., Bacteroidetes) genomes in our data set.
In analyzing LGT involving individual taxa, it is often useful to consider (as above for higher-order taxa) not only obligate transfers, but also the more-numerous possible transfers. Among the five clostridia, T. tengcongensis shows the strongest affinity for T. maritima (34 possible transfers), with lesser affinities for F. nucleatum (23 possible transfers) and the three species of Clostridium (fewer than 10 in each case). T. tengcongensis also has the largest number of possible transfers (40 and 33) with the Archaea in general and the Euryarchaeotes in particular, whereas no other member of the clostridia has >19 possible transfers with the Archaea. Within the Proteobacteria, there is extensive evidence for transfers within genera such as Escherichia, Vibrio, and Xanthomonas. The most ecologically versatile organisms tend to be implicated in the largest number of transfers between major proteobacterial divisions: Pseudomonas aeruginosa, a soil- and water-borne bacterium, and a prominent pathogen in plants and animals, is implicated in possible transfers with organisms such as Ralstonia solanacearum and Caulobacter crescentus, which live in soil and water, as well as animal pathogens including Pasteurella multocida and Photorhabdus luminescens. The generalist plant pathogen R. solanacearum in turn has shared many genes with plant pathogens and symbionts including Pseudomonas syringae, Bradyrhizobium japonicum, and Mesorhizobium loti.
The genome partnerships identified by edit path analyses are substantially supported by phylogenetic profiling of orthologous (Table 1) and homologous (Table 3) groups of proteins, after correcting for the relative representation of different taxonomic groups in our data set. Phylogenetic profiles (27, 28) identify which taxonomic groups are jointly represented (or co-occur) in homologous or orthologous sets of proteins. Clostridial proteins, for example, occur both exclusively and nonexclusively with those of several thermophilic phyla, ε-proteobacteria, and other Gram-positive divisions. Proteins of Aquifex and Thermotoga frequently co-occur, often with euryarchaeal proteins as well. Proteins of F. nucleatum and T. tengcongensis also co-occur, supporting their arrangement in the supertree, but share many orthologs and paralogs with representatives of other Gram-positive divisions and with T. maritima. Profiles do not support monophyly of all Gram-positive devisions, as the high-G+C Gram-positive divisions show a much stronger affinity for the γ- and α-proteobacteria than for the low-G+C Gram-positive divisions, even when size corrections are applied. Pseudomonads and xanthomonads often show stronger affinities for β- and α-proteobacteria than for each other, or for other subdivisions of the γ-proteobacteria such as Enterobacteraceae.
Table 1. Preferred genome partners based on maximally representative clusters.
| Shared nonexclusively
|
Shared exclusively
|
|||
|---|---|---|---|---|
| Phylum/division | Uncorrected | Corrected | Uncorrected | Corrected |
| Aquificales | γ-Proteobacteria | Thermotogales | Euryarchaeota | ε-Proteobacteria |
| Bacteroidetes | γ-Proteobacteria | Listeria | γ-Proteobacteria | Chlorobi |
| Chlamydiales | γ-Proteobacteria | Aquificales | Lactobacillales | ε-Proteobacteria |
| Chlamydiales | γ-Proteobacteria | Aquificales | γ-Proteobacteria | Bacteroidetes |
| Crenarchaeota | Euryarchaeota | Clostridia | Euryarchaeota | Euryarchaeota |
| Cyanobacteria | γ-Proteobacteria | Clostridia | α-Proteobacteria | α-Proteobacteria |
| Euryarchaeota | γ-Proteobacteria | Clostridia | Crenarchaeota | Crenarchaeota |
| High-G + C Firmicutes | γ-Proteobacteria | Clostridia | γ-Proteobacteria | α-Proteobacteria |
| Nanoarchaeota | Euryarchaeota | Aquificales | γ-Proteobacteria | Chlamydiales |
| Planctomycetes | γ-Proteobacteria | Clostridia | γ-Proteobacteria | γ-Proteobacteria |
| Spirochaetales | γ-Proteobacteria | Clostridia | γ-Proteobacteria | Cyanobacteria |
| Thermotogales | Clostridia | Aquificales | Euryarchaeota | Clostridia |
| Thermus/Deinococcus | γ-Proteobacteria | Aquificales | γ-Proteobacteria | Cyanobacteria |
| α-Proteobacteria | γ-Proteobacteria | β-Proteobacteria | γ-Proteobacteria | γ-Proteobacteria |
| β-Proteobacteria | γ-Proteobacteria | Clostridia | γ-Proteobacteria | γ-Proteobacteria |
| γ-Proteobacteria | β-Proteobacteria | β-Proteobacteria | β-Proteobacteria | β-Proteobacteria |
| ε-Proteobacteria | γ-Proteobacteria | Aquificales | γ-Proteobacteria | Aquificales |
| Bacilli | γ-Proteobacteria | Clostridia | Clostridia | Clostridia |
| Clostridia | γ-Proteobacteria | Listeria | Bacilli | Bacilli |
| Lactobacilli | γ-Proteobacteria | Listeria | Listeria | Listeria |
| Listeria | Bacilli | Staphylococci | Lactobacilli | Lactobacilli |
| Mollicutes | γ-Proteobacteria | Listeria | γ-Proteobacteria | Staphylococci |
| Staphylococci | Bacilli | Listeria | Bacilli | Bacilli |
Genomes are grouped according to the National Center for Biotechnology Information level 4 taxonomy, except for proteobacteria and low-G + C Gram-positive divisions, which we subdivide into four and six divisions, respectively, as implied by the MRP supertree (see Fig. 6). For each defined taxonomic group G, we determine the group G′ ≠ G that is represented most frequently in MRCs that also contain proteins from group G. The requirement for co-occurrence can be either nonexclusive, in which case all MRCs containing proteins from G and G′ are counted, or exclusive, with counts derived from only those MRCs that contain proteins from G and G′ but no other taxonomic group. Counts are presented based on the raw count of shared MRCs, and after normalization (dividing each raw count by the total number of MRCs that contain a protein from G and/or G′). For instance (first line), MRCs in which proteins from Aquificales (Aquifex aeolicus) and γ-Proteobacteria co-occur nonexclusively are more numerous than those containing proteins from Aquificales and any other single phylum represented in our data set (column 2). However, normalization for the large number of MRCs that contain γ-proteobacterial proteins (column 3) reveals that proteins from Aquificales co-occur preferentially with those from Thermotogales (Thermotoga maritima). The preferred exclusive partner based on raw count is Euryarchaeota (column 4), but after normalization is ε-Proteobacteria (column 5).
Functions of Putatively Transferred Genes. We performed χ2 tests to examine functional correlates of the concordant versus discordant bipartitions among our 22,432 protein family trees. The National Center for Biotechnology Information clusters of orthologous groups (COG) database defines four major groupings: metabolism, cellular processes, information storage and processing, and poorly characterized or hypothetical genes (29). These groupings are further subdivided into 25 categories. The overall χ2 for the four major groupings (Table 4) was 128.45 (3 df, P = 1.17 × 10-27), with “metabolism” and “cellular processes” overrepresented among the set of discordant bipartitions relative to their frequency among the concordant ones. A test of distribution across the 25 functional categories (Table 5) yields a χ2 value of 414.29 (24 df, P = 8.70 × 10-73). Among proteins with annotated function, the only category with a distributional bias substantially different from its parent grouping is “inorganic ion transport and metabolism,” which is underrepresented among discordant bipartitions, although its parent (“cellular processes”) is overrepresented.
We also assigned functions to our orthologous families by using The Institute for Genomic Research role categories database (30) in which 19 major groupings relevant to our analysis are further subdivided into 120 categories. χ2 tests as above yielded P values of <1 × 10-56 for both sets of functional categories. Among the 19 major groupings (Table 6), the functions most strongly overrepresented in the set of discordant bipartitions are “energy metabolism” and “mobile and extrachromosomal element functions,” whereas “DNA metabolism,” “protein synthesis,” “protein fate,” and “regulatory functions” were strongly underrepresented (i.e., highly concordant with the reference topology). Fig. 3 shows the ratio of observed to expected discordance for major role category groupings: proteins with metabolic functions tend to be more discordant than informational proteins. Among the 120 subsidiary categories (Table 7), several relating to sugar metabolism are among those most strongly overrepresented among our discordant bipartitions, as are proteins involved in amino acid metabolism and detoxification. Ribosomal proteins are less prone to discordance, as are proteins involved in DNA replication and repair, cell wall synthesis, and cell division.
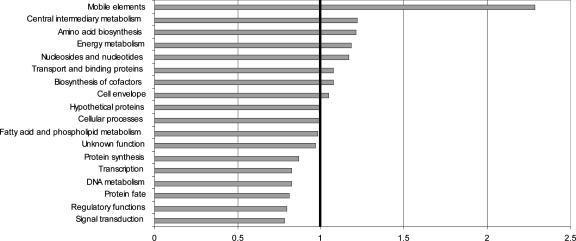
Fig. 3.
Ratio of observed to expected discordant bipartitions among proteins in major TIGR role category groupings. The expected number of discordant bipartitions in each category is equal to the total number of strongly supported bipartitions (concordant and discordant) within that category, multiplied by 0.134, the proportion of bipartitions that are discordant across all categories at PP ≥0.95. Observed numbers of discordant bipartitions range from 78 for “mobile elements” to 3,015 for “hypothetical proteins.”
Discussion
Phylogenetic analysis of 220,240 proteins from 144 sequenced genomes yielded a supertree of prokaryotic life that is remarkably congruent with 16S rDNA and other molecular-sequence trees, and supports many recognized systematic groupings. However, some features of this supertree are more often incongruent than congruent with strongly supported nodes of the constituent protein trees. For example, the thermophiles Aquifex and Thermotoga group together at the base of the bacterial supertree, but protein trees and phylogenetic profiles associate the Aquifex genome more closely with the ε-proteobacteria, and Thermotoga with the low-G+C Gram-positive divisions, probably in the branch that includes T. tengcongensis and F. nucleatum. These alternative affiliations were identified by Cavalier-Smith (31) on the basis of cell wall and other characters. The particularly strong nature of these alternative affiliations suggests that the arrangement of the Aquifex and Thermotoga genomes in the supertree may reflect strong lateral signal, rather than the shared history that is strongly supported in most of the tree. Given the extremely weak support (22 of 110 constituent trees at PP ≥0.95) for the grouping of Aquifex and Thermotoga in the supertree, it may be surprising that this arrangement is proposed at all. However, this arrangement appears to be a compromise among conflicting alternative affiliations for these taxa with various bacterial and archaeal partners, none of which is individually supported more strongly than is their association with each other. The true phylogenetic position of these taxa, and which proteins have vertical or lateral histories, may become clearer as more genomes from their respective phyla are sequenced. Extensive LGT has occurred among certain mesophiles as well. Some proteins from the Xanthomonas group of plant pathogens, and from soil bacteria of genus Pseudomonas, have closest relatives among the α- and β-proteobacteria. There is also evidence of extensive transfer within Cyanobacteria, particularly among strains of Prochlorococcus and Synechococcus.
The picture of protein functions that are relatively susceptible, or resistant, to LGT that has been developed by using surrogate methods is largely confirmed by the rigorous phylogenetic approach described here, which is likely to be more robust to the effects of amelioration (32). In particular, informational genes including 16S rDNA (see Fig. 7) are particularly resistant to LGT (5, 33), and cell wall and cell division proteins tend to be inherited vertically, and are therefore informative for high-level systematics of prokaryotes (31). However, high-level functional groupings of proteins encompass multiple biochemical processes that may have different degrees of susceptibility to LGT. For instance, among proteins implicated in protein synthesis, integral components of the ribosome show a strong tendency toward vertical descent, whereas aminoacyl-tRNA synthetases, which interact at a single common site on the exterior of the ribosome, frequently show evidence of LGT (6). Strong vertical tendencies observed in many classes of proteins contradict the idea that phylogenetic classification of organisms is impossible in the face of massive LGT (9), and supports the proposal that prokaryotes do in fact have unique, characteristic histories (1). However, we do find extensive evidence for the preferential transfer of metabolic genes: acquisition of such genes would allow organisms to gain access to new energy and nutrient sources, thereby increasing their ability to colonize or compete in the environment.
Our results clearly show that genetic modification of organisms by lateral transfer is a widespread natural phenomenon. It will likely be impossible to assess exactly the footprint of LGT on prokaryotic genomes, because the percentage of genes or proteins that yield discordant tree topologies depends on the taxa sampled. This is particularly true for “orphan” proteins (≈6.5% in our data set), which lack recognizable homologs. However, for the diverse prokaryotes in our sample, we find a pervasive coherent vertical genetic signal with significant modulation by LGT, particularly among thermophiles, pathogens, and cyanobacteria. Coupled with rigorous phylogenetic methodology such as we employ here, the growth of community genomics (34-36) will lead to an increasingly precise delineation of the genomic, functional, and environmental determinants of vertical and lateral genetic transfer in nature.
Supplementary Material
Acknowledgments
We acknowledge support of the Australian Research Council, the Institute for Molecular Bioscience, and the Australian Partnership in Advanced Computing, and valuable input from Cheong Xin Chan, Nicholas Hamilton, and Phil Hugenholtz.
Author contributions: R.G.B. and M.A.R. designed research; R.G.B. and T.J.H. performed research; R.G.B. and T.J.H. contributed new reagents/analytic tools; R.G.B., T.J.H., and M.A.R. analyzed data; and R.G.B. and M.A.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: rDNA, rRNA gene; LGT, lateral genetic transfer; MRC, maximally representative cluster; PP, posterior probability.
References
- 1.Woese, C. R. (1987) Microbiol. Rev. 51, 221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felsenstein, J. (2004) Inferring Phylogenies (Sinauer, Sunderland, MA).
- 3.Woese, C. R. (2000) Proc. Natl. Acad. Sci. USA 97, 8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, J. R., Masuchi, Y., Robb, F. T. & Doolittle, W. F. (1994) J. Mol. Evol. 38, 566-576. [DOI] [PubMed] [Google Scholar]
- 5.Jain, R., Rivera, M. C. & Lake, J. A. (1999) Proc. Natl. Acad. Sci. USA 96, 3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woese, C. R., Olsen, G. J., Ibba, M. & Soll, D. (2000) Microbiol. Mol. Biol. Rev. 64, 202-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nesbø, C. L., Boucher, Y. & Doolittle, W. F. (2001) J. Mol. Evol. 53, 340-350. [DOI] [PubMed] [Google Scholar]
- 8.Syvanen, M. & Kado, C. I. (2002) Horizontal Gene Transfer (Academic, London).
- 9.Doolittle, W. F. (1999) Science 284, 2124-2129. [DOI] [PubMed] [Google Scholar]
- 10.Ochman, H., Lawrence, J. G. & Groisman, E. A. (2000) Nature 405, 299-304. [DOI] [PubMed] [Google Scholar]
- 11.Papke, R. T., Koenig, J. E., Rodriguez-Valera, F. & Doolittle, W. F. (2004) Science 306, 1928-1929. [DOI] [PubMed] [Google Scholar]
- 12.Doolittle, W. F. (1998) Trends Genet. 14, 307-311. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura, Y., Itoh, T., Matsuda, H. & Gojobori, T. (2004) Nat. Genet. 36, 760-766. [DOI] [PubMed] [Google Scholar]
- 14.Lerat, E., Daubin, V., Ochman, H. & Moran, N. A. (2005) PloS Biol. 3, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raymond, J., Zhaxybayeva, O., Gogarten, J. P., Gerdes, S. Y. & Blankenship, R. E. (2002) Science 298, 1616-1620. [DOI] [PubMed] [Google Scholar]
- 16.Harlow, T. J., Gogarten, J. P. & Ragan, M. A. (2004) BMC Bioinformatics 5, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beiko, R. G., Chan, C. X. & Ragan, M. A. (2005) Bioinformatics 21, 2230-2239. [DOI] [PubMed] [Google Scholar]
- 18.Castresana, J. (2000) Mol. Biol. Evol. 17, 540-552. [DOI] [PubMed] [Google Scholar]
- 19.Huelsenbeck, J. P., Ronquist, F., Nielsen, R. & Bollback, J. P. (2001) Science 294, 2310-2314. [DOI] [PubMed] [Google Scholar]
- 20.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- 21.Ragan, M. A. (1992) Mol. Phylogenet. Evol. 1, 53-58. [DOI] [PubMed] [Google Scholar]
- 22.Allen, B. L. & Steel, M. (2001) Ann. Combinatorics 5, 1-15. [Google Scholar]
- 23.Creevey, C. J., Fitzpatrick, D. A., Philip, G. K., Kinsella, R. J., O'Connell, M. J., Pentony, M. M., Travers, S. A., Wilkinson, M. & McInerney, J. O. (2004) Proc. R. Soc. London Ser. B 271, 2551-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravind, L., Tatusov, R. L., Wolf, Y. I., Walker, D. R. & Koonin, E. V. (1998) Trends Genet. 14, 442-444. [DOI] [PubMed] [Google Scholar]
- 25.Nelson, K. E., Clayton, R. A., Gill, S. R., Gwinn, M. L., Dodson, R. J., Haft, D. H., Hickey, E. K., Peterson, J. D., Nelson, W. C., Ketchum, K. A., et al. (1999) Nature 399, 323-329. [DOI] [PubMed] [Google Scholar]
- 26.Nesbø, C. L., L'Haridon, S., Stetter, K. O. & Doolittle, W. F. (2001) Mol. Biol. Evol. 18, 362-375. [DOI] [PubMed] [Google Scholar]
- 27.Gaasterland, T. & Ragan, M. A. (1998) Microb. Comp. Genomics 3, 177-192. [DOI] [PubMed] [Google Scholar]
- 28.Pellegrini, M., Marcotte, E. M., Thompson, M. J., Eisenberg, D. & Yeates, T. O. (1999) Proc. Natl. Acad. Sci. USA 96, 4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., Krylov, D. M., Mazumder, R., Mekhedov, S. L., Nikolskaya, A. N., et al. (2003) BMC Bioinformatics 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson, J. D., Umayam, L. A., Dickinson, T., Hickey, E. K. & White, O. (2001) Nucleic Acids Res. 29, 123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavalier-Smith, T. (2002) Int. J. Syst. Evol. Microbiol. 52, 7-76. [DOI] [PubMed] [Google Scholar]
- 32.Ragan, M. A. (2001) FEMS Microbiol. Lett. 201, 187-191. [DOI] [PubMed] [Google Scholar]
- 33.Fox, G. E., Pechman, K. R. & Woese, C. R. (1977) Int. J. Syst. Bacteriol. 27, 44-57. [Google Scholar]
- 34.Stahl, D. A., Lane, D. J., Olsen, G. J. & Pace, N. R. (1985) Appl. Environ. Microbiol. 49, 1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, T. M., DeLong, E. F. & Pace, N. R. (1991) J. Bacteriol. 173, 4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tringe, S. G., von Mering, C., Kobayashi, A., Salamov, A. A., Chen, K., Chang, H. W., Podar, M., Short, J. M., Mathur, E. J., Detter, J. C., et al. (2005) Science 308, 554-557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.