Abstract
Misfolded proteins are eliminated from the endoplasmic reticulum (ER) by retrotranslocation into the cytosol, a pathway hijacked by certain viruses to destroy MHC class I heavy chains. The translocation of polypeptides across the ER membrane requires their polyubiquitination and subsequent extraction from the membrane by the p97 ATPase [also called valosin-containing protein (VCP) or, in yeast, Cdc48]. In higher eukaryotes, p97 is bound to the ER membrane by a membrane protein complex containing Derlin-1 and VCP-interacting membrane protein (VIMP). How the ubiquitination machinery is recruited to the p97/Derlin/VIMP complex is unclear. Here, we report that p97 interacts directly with several ubiquitin ligases and facilitates their recruitment to Derlin-1. During retrotranslocation, a substrate first interacts with Derlin-1 before p97 and other factors join the complex. These data, together with the fact that Derlin-1 is a multispanning membrane protein forming homo-oligomers, support the idea that Derlin-1 is part of a retrotranslocation channel that is associated with both the polyubiquitination and p97-ATPase machineries.
Keywords: AAA ATPase, ER-associated degradation, dislocation, Derlin, Hrd1
Retrotranslocation is a cellular quality control mechanism that removes aberrantly folded proteins from the endoplasmic reticulum (ER) (for review, see ref. 1). The pathway is co-opted by certain viruses, for example, the human cytomegalovirus (HCMV), to eliminate MHC class I molecules, allowing infected cells to evade destruction by cytotoxic T cells (for review, see ref. 2). HCMV encodes two proteins, US11 and US2, both of which induce retrotranslocation of newly synthesized class I heavy chains (3, 4). A crucial step for all substrates undergoing retrotranslocation is their transfer across the ER membrane. For many substrates, the conserved multispanning membrane protein Derlin-1 could play a central role, perhaps as a component of a retrotranslocation channel (5, 6). Derlin-1 is a mammalian homolog of Der1p in Saccharomyces cerevisiae, which is involved in the degradation of a class of misfolded proteins (7, 8). During retrotranslocation induced by the HCMV protein US11, Derlin-1 receives its substrate, MHC class I heavy chains, from US11, which is involved in substrate recognition in the ER lumen (5, 6). In the cellular pathway, Derlin-1 may receive misfolded protein substrates from ER chaperones (for review, see ref. 1). Derlin-1 also associates with a cytosolic ATPase complex, consisting of p97 and its cofactors Ufd1 and Npl4 (6, 9). This interaction is mediated at least in part by VCP-interacting membrane protein (VIMP), a single-spanning membrane protein with a cytosolic domain (6). The p97 ATPase complex seems to receive substrates from Derlin-1 by binding to both unmodified, presumably unfolded, segments of the polypeptide chain, and to polyubiquitin chains attached to it (10). It subsequently uses ATP hydrolysis to “pull” the polypeptide chain into the cytosol, where it is degraded by the proteasome (11–15).
Because polyubiquitination of the substrate is a prerequisite for the function of the p97 ATPase (16), we asked how the ubiquitination machinery is recruited to the site of retrotranslocation containing the Derlin-1/p97 ATPase complex. Here, we demonstrate that ubiquitin ligases dedicated to retrotranslocation associate with the Derlin-1/p97 complex. The assembly of a retrotranslocation complex seems to occur in at least two steps. An early step involves the targeting of a retrotranslocation polypeptide to Derlin-1 that is not associated with the p97 ATPase. At a later stage, the ATPase joins Derlin-1 to form a larger retrotranslocation complex, which may also contain specific ubiquitin ligases. Derlin-1 simultaneously associates with both the pulling ATPase and ubiquitination ligases, bringing together two crucial components required for retrotranslocation. Together with our observation that Derlin-1 forms homooligomers, these data support our previous hypothesis that Derlin-1 might be part of a retrotranslocation channel.
Methods
Constructs. The plasmids pcDNA3.1-Hrd1-Myc/His, pcDNA3.1-Hrd1-C291A-Myc/his, and pcDNA3.1-KIAA1810-Myc/His have been described (17). The plasmid pcDNA3.1-Hrd1-GFP was constructed by cloning the human Hrd1 ORF into the pcDNA3.1-CT-GFP-topo vector (Invitrogen), thereby creating a fusion of GFP at the C terminus of Hrd1. For the pcDNA3.1-gp78-FLAG construct, the gp78 was amplified from pCIneo-gp78 (a kind gift from A. Weissman, National Cancer Institute, Frederick, MD) by using primers containing the FLAG tag-encoding sequence. The PCR product was cloned into pcDNA3.1(-) by using NotI and HindIII restriction sites. Plasmids encoding FLAG-Parkin or Derlin-1-GFP were kindly provided by R. Takahashi (RIKEN Brain Science Institute, Tokyo) and B. Lilley (Harvard Medical School, Boston), respectively. GST-Hrd1c and His-Derlin-1c constructs were generous gifts from M. Seeger (Humboldt University, Berlin).
Antibodies and Proteins. Antibodies to Derlin-1 and VIMP have been described (6). Antibodies to p97 were raised against a mixture of the peptides MASGADSKGDDLSTAILKQKNR(C) and (C)GSVYTEDNDDDLYG. For immunostaining and coimmunoprecipitation, VIMP and p97 antibodies were affinity-purified by using the corresponding antigenic peptides. Affinity-purified Derlin-1 antibodies used in fluorescent studies were kindly provided by B. Lilley. Hrd1 antibodies have been described in ref. 17.
The purification of His-tagged p97 variants and of GST-gp78c have been described in ref. 10. The purification of GST-VIMPc has been described in ref. 6. GST-Hrd1c was purified according to standard procedure by using glutathione beads. His-Derlin-1c was purified under denaturing condition (8 M urea), following the instructions provided by Qiagen (Valencia, CA). Purified His-Derlin-1c was refolded through sequential dialysis, first in buffer R (20 mM Tris·HCl, pH 8.0/150 mM sodium chloride/10% glycerol/0.1 mM DTT) in the presence of 3 M urea, then in the same buffer in the absence of urea. Refolded proteins were subjected to centrifugation at 20,000 × g for 10 min, and the supernatant fraction was used in binding experiments.
Mammalian Cell Culture, Transient Gene Expression, and Coimmunoprecipitation. HeLa cells and 293T cells were maintained according to standard procedures. Transfections were done either with polyfect (Qiagen) for HeLa cells, or by using Trans IT (Mirus, Madison, WI) for 293T cells. Immunofluorescence experiments were done as described (6). For coimmunoprecipitation experiments, tissue culture cells or dog pancreatic microsomes were extracted with buffer N (30 mM Tris·HCl, pH 7.4/150 mM potassium acetate/4 mM magnesium acetate/1% DeoxyBigC-HAP/a protease inhibitor mixture). The extract was cleared by centrifugation (20,000 × g) and subjected to immunoprecipitation with various antibodies.
In Vitro Binding Experiments. GST-pull down experiments were performed as described in ref. 10. Briefly, GST-fusion proteins (5 μg) bound to glutathione beads were incubated with various purified recombinant proteins in 50 mM Hepes (pH 7.3), 150 mM potassium chloride, 2.5 mM magnesium chloride, 5% glycerol, 0.1% Triton X-100, and 1 mg/ml BSA. Bound material was analyzed by SDS/PAGE and stained with Coomassie blue. To test binding of His-tagged p97 to His-Derlin-1c, purified p97 variants (5 μg) were immobilized on protein A beads containing 3 μg of affinity-purified p97 antibodies.
Association of the Derlin-1 Complex with MHC Class I Heavy Chains. US11-expressing astrocytoma cells were treated with 20 μM MG132 and labeled with [35S]methionine as described (12). Cellular extracts obtained with buffer N were subjected to immunoprecipitation with various antibodies, followed by reprecipitation with heavy chain antibodies.
Sucrose Gradient Centrifugation. US11-expressing cells were labeled with [35S]methionine, permeabilized, and incubated with cytosol in the presence of a p97 mutant (AK) as described (10). Cells were solubilized in buffer N. The detergent extract were layered on top of a 10–25% sucrose gradient, and centrifuged at 150,000 × g for 15 h. Fractions were collected and analyzed by either direct immunoprecipitation with heavy chain antibodies, or immunoblotting with Derlin-1 and p97 antibodies.
Results
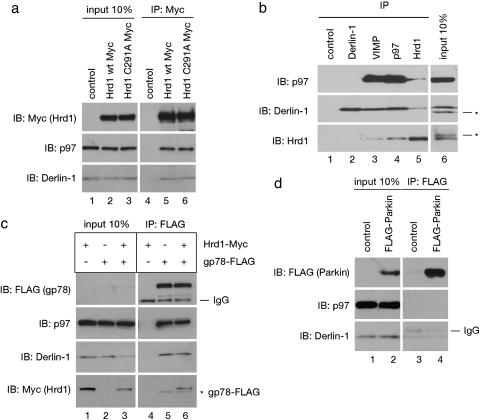
The Derlin-1/p97 Complex Contains Ubiquitin Ligases. We first tested whether the Derlin-1 complex associates with Hrd1, an ER-anchored ubiquitin ligase (E3 ligase) that is required for the retrotranslocation of some misfolded substrates in both S. cerevisiae and mammals (17–20). We expressed Myc-tagged human Hrd1 (Hrd1-Myc) in 293T cells, and subjected a detergent extract of these cells to immunoprecipitation with Myc antibodies. Immunoblotting with specific antibodies showed that Derlin-1 and p97 were coprecipitated with Hrd1-Myc (Fig. 1a, lane 5). VIMP, a previously identified membrane partner of Derlin-1, was also coprecipitated (data not shown). Hrd1 consists of an N-terminal domain with six predicted transmembrane segments, and a C-terminal cytosolic domain containing a ring finger fold with enzymatic activity (17). A Myc-tagged Hrd1 isoform (KIAA1810) that lacks a transmembrane segment also interacted with Derlin-1 and p97 (Fig. 7, which is published as supporting information on the PNAS web site). The E3 ligase activity of Hrd1 is not required for the interaction because an inactive Hrd1 variant containing a cysteine to alanine substitution in its ring finger motif (C291A) still bound Derlin-1 and p97 (Fig. 1a, lane 6).
Fig. 1.
Interaction of E3 ligases with components of the Derlin-1 complex. (a) Extracts from 293T cells, transfected with an empty vector (control) or with constructs encoding Myc-tagged wild-type Hrd1 (Hrd1 wt Myc), or Myc-tagged Hrd1 mutant containing a mutation in its ring finger domain (Hrd1 C291A Myc), were analyzed by immunoprecipitation (IP) with Myc antibodies, followed by immunoblotting (IB) with the indicated antibodies. Ten percent of the extracts were analyzed directly by immunoblotting (input). (b) A detergent extract of dog pancreatic microsomes was analyzed by immunoprecipitation (IP) with the indicated antibodies, followed by immunoblotting (IB) with various antibodies. The asterisks indicate proteins crossreacting with the antibodies. (c) As in a, except that cells expressed Hrd1-Myc, FLAG-tagged gp78 (gp78-FLAG), or both. The asterisk indicates residual FLAG-gp78 left after incomplete stripping of the FLAG immunoblot (Top). (d) As in a, except that cells expressed FLAG-tagged Parkin (FLAG-Parkin). IgG, immunoglobins detected by the secondary antibodies.
To test whether endogenous Hrd1 also interacts with Derlin-1, VIMP, and p97, dog pancreatic ER membranes were solubilized, and subjected to immunoprecipitation with different antibodies. We found that antibodies to VIMP or p97 precipitated Derlin-1 (11% and 14%, respectively) in complex with the p97 ATPase (12% and 18%, respectively) and the Hrd1 ubiquitin ligase (4.5% and 12%, respectively) (Fig. 1b, lanes 3 and 4). Considering that, under our experimental conditions, the antibodies precipitated <20% of their respective antigens, the partners seem to be coprecipitated efficiently. Hrd1 antibodies precipitated ≈3% Derlin-1 and very little p97 (lane 5), probably because the antibodies sterically prevented efficient binding of p97 and Derlin-1 to Hrd1. Antibodies to Derlin-1 precipitated ≈14% of this protein alone (lane 2), likely because they do not recognize Derlin-1 when in complex with the other partners (see below). These data confirm an interaction between the ubiquitin ligase Hrd1 and the previously identified components of the Derlin-1 complex, but they also suggest that a large fraction of Derlin-1 may not be present in the complex.
We next tested other E3 ligases for their interaction with Derlin-1 and p97. FLAG-tagged gp78/AMFR, another membrane-anchored ubiquitin ligase implicated in retrotranslocation (21–23), also associated with Derlin-1 and p97 (Fig. 1c, lane 5), whereas FLAG-tagged Parkin, an E3 ligase involved in the degradation of misfolded Pael receptor (24), coprecipitated <5% of Derlin-1 (Fig. 1d, lane 4). Because similar amounts of Derlin-1 were precipitated from extracts that do not contain FLAG-Parkin (lane 3), the coprecipitation of Derlin-1 with Parkin seems to be nonspecific. Interestingly, when Hrd1-Myc and gp78-FLAG were coexpressed, FLAG antibodies precipitated both proteins, as well as Derlin-1 and p97 (Fig. 1c, lane 6). Myc antibodies also precipitated all these proteins (data not shown). These data suggest that the two E3 enzymes are associated with one another, a finding that is consistent with the reported involvement of both Hrd1 and gp78 in the retrotranslocation of CD3δ (17, 23). Thus, our data indicate the existence of membrane protein complexes containing Derlin-1, p97, and ubiquitin ligases.
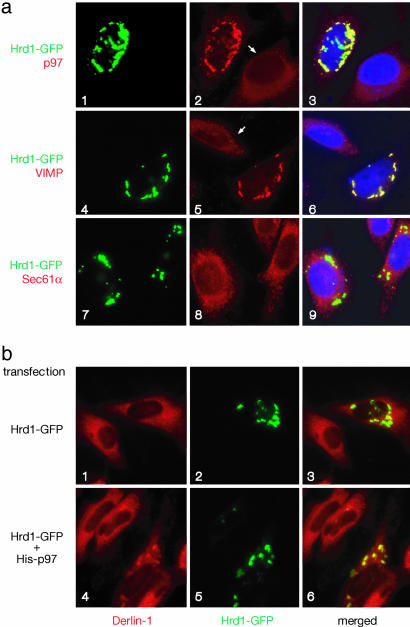
p97 Promotes the Interaction Between Derlin-1 and Hrd1. To further study how Hrd1 binds to Derlin-1, we used immunofluorescence microscopy. We have shown previously that Derlin-1 and VIMP are distributed throughout the ER network, whereas the p97 ATPase is localized mainly to the cytosol (6). When a fusion protein, consisting of Hrd1 and GFP at its C terminus (Hrd1-GFP), was expressed in HeLa cells, it formed aggregates around the nucleus, reminiscent of organized smooth ER structures (OSER) (25) (Fig. 2a). In cells expressing Hrd1-GFP, the staining pattern of endogenous p97 was altered; it formed dotted structures that colocalized with the Hrd1-GFP-containing aggregates (Fig. 2 a1–a3). The localization of endogenous VIMP was also changed to a similar punctate pattern (Fig. 2 a4–a6). In contrast, Hrd1-GFP overexpression did not change the localization of Sec61α, the major component of the ER translocation channel required for moving polypeptides into the ER lumen (Fig. 2 a7–a9). The ER staining pattern of endogenous Derlin-1 also remained largely unchanged (Fig. 2 b1–b3), consistent with the observation that a large part of Derlin-1 is not in a complex with Hrd1 and the p97 ATPase (see Fig. 1b). In addition, these data suggest that Derlin-1 may not have a strong direct interaction with Hrd1; rather, other factors may cause the binding seen in immunoprecipitation experiments (Fig. 1).
Fig. 2.
Colocalization of Hrd1 and components of the Derlin-1 complex. (a) HeLa cells expressing GFP-tagged Hrd1 (Hrd1-GFP) (a1, a4, and a7, green) were also stained with antibodies to p97, VIMP, or Sec61α (a2, a5, and a8, red). (a3, a6, and a9) Merged images, in which the nuclei were also stained with DAPI (in blue). Note that punctae are seen only in Hrd1-Myc-expressing cells (for comparison, see nonexpressing cells indicated by arrows). (b) HeLa cells expressing Hrd1-GFP on its own (b1–b3) or together with His-tagged p97 (b4–b6, His-p97) were visualized for GFP expression (in green) and stained with Derlin-1 antibodies (in red). (b3 and b6) Merged images.
We therefore tested whether expression of VIMP or p97 could enhance the association of Derlin-1 with Hrd1-GFP. Expression of VIMP together with Hrd1-GFP had no effect on the localization of endogenous Derlin-1 (data not shown). In contrast, in cells cotransfected with p97 and Hrd1-GFP, Derlin-1 was relocalized to the Hrd1-GFP-containing punctae in 78% of Hrd1-GFP-expressing cells (Fig. 2 b4–b6), presumably because these cells also express p97. Because we cannot visualize all three proteins at the same time, we verified the expression of p97 in Hrd1-GFP-expressing cells in a parallel experiment, in which the localization of overexpressed p97 was determined by anti-p97 antibodies. This experiment showed that p97 was indeed localized to the GFP-Hrd1-containing punctae in a similar fraction of cells (data not shown). Taking into account that the transfection efficiency is not 100%, these data argue that the punctae contain all three proteins. The data therefore suggest that p97 facilitates the recruitment of Hrd1 to Derlin-1, possibly by interacting directly with these proteins.
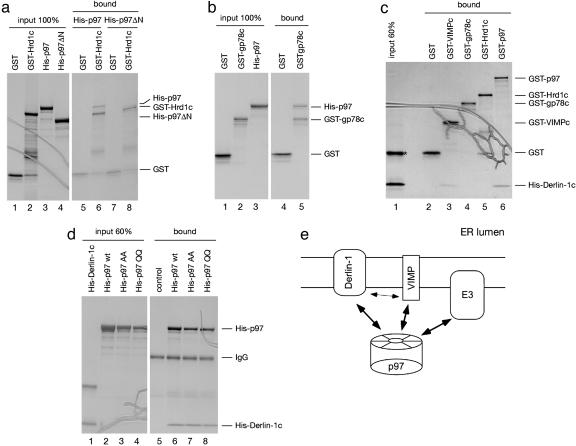
Mapping the Interactions in Vitro. We next examined whether p97 can bind directly to the cytosolic domain of Hrd1. Indeed, a recombinant protein containing the GST fused to the cytosolic domain of Hrd1 (GST-Hrd1c) bound purified His-tagged wild-type p97 (His-p97), but not a mutant lacking its N domain (Fig. 3a, lane 6 vs. 8). A GST-fusion to the cytosolic domain of the ubiquitin ligase gp78 (GST-gp78c) also interacted with p97 (Fig. 3b, lane 5). Our data extend the previously observed coprecipitation of gp78 and p97 from cell extracts (23) by demonstrating a direct interaction. It should be noted that, in the GST-pull-down experiments, stringent washing conditions were used to detect only the strongest interactions.
Fig. 3.
Mapping interactions among components of the Derlin-1 complex. (a) A GST fusion protein containing the cytosolic domain of Hrd1 (GST-Hrd1c) was immobilized on glutathione beads and incubated with either purified His-tagged p97 (His-p97) or a His-tagged p97 mutant lacking its N domain (His-p97ΔN). Bound proteins were separated by SDS/PAGE and stained with Coomassie blue. GST was used as a control. (b)Asin a, except that a GST fusion to the cytosolic domain of gp78 (GST-gp78c) was used and binding to His-p97 was tested. (c) GST fusions to either p97 (GST-p97) or the cytosolic domain of VIMP (GST-VIMPc) were tested together with GST-Hrd1c and GST-gp78c for binding to the purified His-tagged C-terminal, cytosolic domain of Derlin-1 (His-Derlin-1c). The asterisk indicates a contaminated protein, copurified with His-Derlin-1c. (d) His-p97 or His-tagged p97 mutants, defective in either ATP-binding (His-p97 AA) or -hydrolysis (His-p97 QQ) were immobilized with p97-specific antibodies on protein A beads, and tested for binding of His-Derlin-1c. As a control, His-p97 was omitted from the reaction. (e) Scheme of protein–protein interactions within the Derlin-1 complex. The thin arrow indicates a weak interaction.
Next, we tested whether p97 can also bind to Derlin-1. The largest cytosolic domain of Derlin-1 is at its C terminus (Derlin-1c), and we therefore tested whether it can bind p97. GST-p97 indeed bound a His-tagged version of this domain (His-Derlin-1c) (Fig. 3c, lane 6). We also found that a GST-fusion to the cytosolic domain of VIMP (GST-VIMPc) interacts weakly with His-Derlin-1c (Fig. 3c lane 3), indicating that the interaction between the full-length proteins is at least in part mediated by their cytosolic domains. In contrast, we did not observe binding between GST fusions of the cytosolic domains of the ubiquitin ligases Hrd1 or gp78 and His-Derlin-1c, in agreement with the observation that Derlin-1 and Hrd1 do not associate strongly in immunofluorescence experiments (Fig. 2). The interaction between p97 and Derlin-1c is not dependent on the nucleotide state of the ATPase, because mutants defective in either ATP binding (AA) or hydrolysis (QQ) have the same affinity for Derlin-1c as wild-type p97 (Fig. 3d, lanes 6–8). Together, these data indicate that p97 could mediate the association between ubiquitin ligases and Derlin-1 by directly interacting with both partners. In the final complex, p97 may be bound simultaneously to the cytosolic domains of the ubiquitin ligases, Derlin-1, and VIMP (see scheme in Fig. 3e).
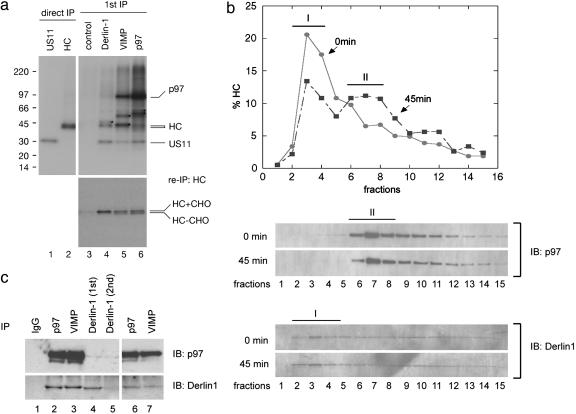
Assembly of a Retrotranslocation Complex. To further investigate the assembly of the Derlin-1 complex during retrotranslocation, we examined its association with a retrotranslocation substrate. In US11-expressing cells treated with a proteasome inhibitor, glycosylated MHC class I heavy chains undergo retrotranslocation and are converted into deglycosylated species when they arrive in the cytosol (3). To follow this process, the cells were incubated with [35S]methionine and solubilized. A detergent extract was subjected to immunoprecipitation with antibodies to Derlin-1, VIMP, p97, or Hrd1. The precipitated proteins were subjected to a second round of immunoprecipitation with antibodies to MHC class I heavy chains. Derlin-1 antibodies precipitated only glycosylated heavy chains (Fig. 4a Lower, lane 4), i.e., substrate molecules at an early stage of retrotranslocation. In contrast, antibodies to either VIMP or p97 precipitated both glycosylated and deglycosylated heavy chains (Fig. 4a Lower, lanes 5 and 6); the latter represent substrate molecules that have already emerged into the cytosol. These differences correlate with the fact that antibodies to Derlin-1 precipitated a complex that is distinct from the one isolated with VIMP or p97 antibodies; Derlin-1 antibodies precipitated US11, an unknown protein of ≈46 kDa, and Derlin-1 itself (Fig. 4a Upper, lane 4 and Fig. 1b, lane 2), whereas antibodies to VIMP or p97 precipitated US11, p97, and some other unidentified proteins (Fig. 4a Upper, lanes 5 and 6). They also precipitated Derlin-1 and VIMP [Derlin-1 and VIMP were not efficiently radiolabeled, and could be detected only after longer exposure of the film (data not shown)], or by immunoblotting (Fig. 8, which is published as supporting information on the PNAS web site, and Fig. 1b, lanes 3 and 4). Hrd1 does not seem to interact with retrotranslocating heavy chains in US11 cells (data not shown). Because overexpression of a dominant-negative Hrd1 mutant had no effect on the retrotranslocation of class I heavy chain in US11 cells (M.K., unpublished results), we assume that a different E3 ligase is required for the retrotranslocation of class I molecules.
Fig. 4.
Assembly of the Derlin-1 complex. (a) US11-expressing astrocytoma cells were treated with a proteasome inhibitor and labeled with [35S]methionine. A detergent extract of these cells was subjected to immunoprecipitation under native condition (1st IP) with specific antibodies, as indicated (Upper). A fraction of the precipitated material was analyzed by a second immunoprecipitation (re-IP) with antibodies to MHC class I heavy chains (HC) (Lower). A portion (10%) of the extract was directly precipitated (direct IP) with either US11 or heavy chain (HC) antibodies under denaturing conditions (lanes 1 and 2). The samples were analyzed by SDS/PAGE and autoradiography. The asterisks (Upper) designate several unknown proteins that were coprecipitated with the indicated antibodies. HC + CHO and HC - CHO indicate glycosylated and deglycosylated HC, respectively. (b) US11-expressing cells were labeled with [35S]methionine, permeabilized, and incubated with cytosol containing a p97 mutant defective in ATP hydrolysis for 45 min before they were solubilized in detergent (45 min). An aliquot of cells was solubilized immediately after permeabilization (0 min). The detergent extracts were fractionated on 10–25% sucrose gradient. The amount of MHC class I heavy chains in each fraction was determined by immunoprecipitation and autoradiography, and was plotted (Upper). A portion of each fraction was analyzed by immunoblotting (IB) with Derlin-1 and p97 antibodies. (c) Detergent extracts of US11-expressing cells were subjected to immunoprecipitation with either control IgG or indicated antibodies (lanes 1–4). The supernatant from the Derlin-1 precipitation (1st) was subjected to a second (2nd) immunoprecipitation with Derlin-1 antibodies (lane 5). The resulting supernatant was analyzed by immunoprecipitation with either p97 (lane 6) or VIMP (lane 7) antibodies.
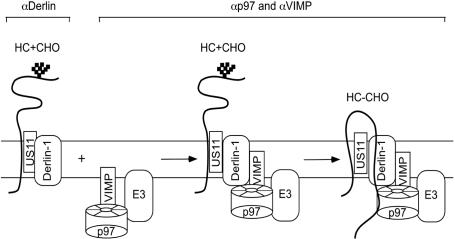
Our data suggest that two distinct complexes exist, one isolated with Derlin-1 antibodies corresponding to an early stage of retrotranslocation, and one isolated by VIMP and p97 antibodies corresponding to a later stage in the process. To further test this idea, US11-expressing cells labeled with [35S]methionine were permeabilized, and incubated with cytosol in the presence of a p97 mutant defective in ATP hydrolysis (KA) to accumulate retrotranslocating intermediates in complex with p97 (10). Aliquots of cells taken before (0 min) or after (45 min) the incubation were solubilized. The detergent extracts were fractionated by sucrose gradient centrifugation, and the distribution of MHC class I heavy chains was analyzed by immunoprecipitation and that of Derlin-1 and p97 by immunoblotting. Before incubation, heavy chains were mainly present in low molecular weight fractions (Fig. 4b Upper, peak I), which contained Derlin-1, but not p97 (Fig. 4b Lower). A small fraction of heavy chains was in high molecular weight fractions that contained both p97 and Derlin-1, suggesting that they had entered a distinct complex even after the short labeling period. After incubation, more heavy chains were found in this high molecular weight, p97-containing complex (Fig. 4b Upper, peak II). As expected, the distribution of total Derlin-1 or p97 remained unchanged during the incubation period (Fig. 4b Lower). To exclude the possibility that Derlin-1 dissociates from p97 during sample preparation or centrifugation, a detergent extract of US11-expressing astrocytoma cells was subjected to two rounds of immunoprecipitation with Derlin-1 antibodies (Fig. 4c, lanes 4 and 5). Very little Derlin-1 was found in the second immunoprecipitation (Fig. 4c, lane 5), whereas significant amounts of Derlin-1 could still be precipitated from the depleted extract with p97 or VIMP antibodies (Fig. 4c, lanes 6 and 7). These data indicate that there are at least two distinct populations of Derlin-1: one that does not bind to p97 and VIMP and preferentially interacts with an early retrotranslocation intermediate, and another one that forms a complex with p97 and VIMP and associates with both early and late retrotranslocation intermediates. The Derlin-1 antibodies can deplete only the former species, presumably because they do not recognize Derlin-1 when in complex with the p97 ATPase. These data also suggest that, during retrotranslocation, heavy chains are initially targeted to a complex containing Derlin-1 and US11, which is joined by p97, VIMP, and other factors only at a later stage (see scheme in Fig. 6).
Fig. 6.
A model for the assembly of the Derlin-1 complex during the retrotranslocation of MHC class I heavy chains. US11 initially targets glycosylated heavy chain (HC + CHO) to Derlin-1, resulting in a complex containing Derlin-1, US11, and HC plus CHO, which can be precipitated with antibodies to Derlin-1 (αDerlin-1). Substrate-bound Derlin-1 is then joined by a membrane complex containing VIMP, p97, and an E3 ligase (E3). The unknown ligase would be recruited to the complex in a similar way as Hrd1 and gp78. The final complex can be immunoprecipitated with antibodies to either VIMP or p97. A fraction of heavy chains (HC - CHO) associated with this complex has emerged into the cytosol because these lack carbohydrate chains, removed by a cytosolic glycanase.
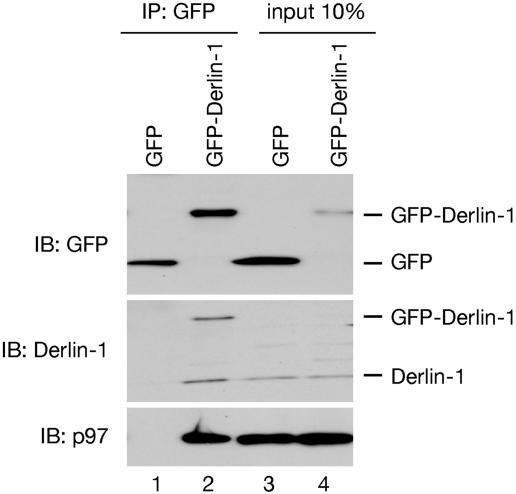
Derlin-1 Forms Homo-Oligomer. The fact that Derlin-1 is associated with class I heavy chains throughout their retrotranslocation from the ER lumen to the cytosol supports our previous proposal that Derlin-1 may be part of a retrotranslocation channel (6). However, Derlin-1 has only four transmembrane segments, which are likely not sufficient to form a protein-conducting channel. We therefore tested whether Derlin-1 can form homooligomers. Indeed, when GFP-tagged Derlin-1 was expressed in 293T cells, it was found associated with endogenous Derlin-1 at an ≈1:1 molar ratio (Fig. 5, lane 2). The complex also contained p97. These data are thus consistent with the assumption that Derlin-1 can form a retrotranslocation channel. It should be noted that GFP-Derlin-1 was not detected with Derlin-1 antibodies in the 10% input sample (Fig. 5 Middle, lane 4), but it was detected with GFP antibodies (Fig. 5 Top, lane 4). These data suggest that GFP-Derlin-1 was expressed at a significantly lower level than endogenous Derlin-1.
Fig. 5.
Derlin-1 forms homo-oligomer. A detergent extract of 293T cells, transfected with constructs encoding either GFP or a GFP-fusion to Derlin-1 (Derlin-1-GFP), was subjected to immunoprecipitation (IP) with GFP antibodies, followed by immunoblotting (IB) with the indicated antibodies. Ten percent of the extract was directly analyzed by immunoblotting (input).
Discussion
Our results provide a link between Derlin-1, a central membrane component, and two important machineries that act on retrotranslocating substrates at the cytosolic side of the ER membrane: the p97 ATPase and ubiquitin ligases. Although the precise order of events in the assembly of this retrotranslocation complex remains to be clarified, the process seems to start with the targeting of a substrate molecule to Derlin-1 (Fig. 6). MHC class I heavy chains are targeted by US11, and unfolded proteins by unknown targeting factors, likely including chaperones. In the next step, the Derlin-1/substrate complex is joined by VIMP, p97, and a ubiquitin ligase, which might all be preassembled (Fig. 6). Although we have identified two ubiquitin ligases (Hrd1 and gp78) that associated with Derlin-1, the ligase responsible for the polyubiquitination of MHC class I heavy chains during their US11-mediated retrotranslocation remains to be identified.
How the assembly of the final retrotranslocation complex is regulated in cells is unclear. The fact that formation of such complex seems to follow the targeting of MHC class I heavy chains to Derlin-1 raises the possibility that assembly of this complex may depend on initial substrate binding by Derlin-1. The p97 ATPase seems to play an important role in this process: it binds to unmodified segments of the substrate, and to cytosolic domains of Derlin-1, VIMP, and ubiquitin ligases. At a later stage, it also binds to polyubiquitin chains attached to the substrate. These multiple interactions might be facilitated by the hexameric state of the p97 ATPase. The direct association of the p97 ATPase with ubiquitin ligases is reminiscent of the interaction between the Hsp70 ATPase and the ubiquitin ligase carboxyl terminus of Hsc70-interacting protein (CHIP), which has been implicated in the degradation of cytosolic and ER membrane proteins (26, 27). Many other ubiquitin ligases also interact with chaperone ATPases (28, 29), suggesting that the activity of the two enzyme systems may generally be coupled. The recruitment of ubiquitin ligases to Derlin-1 by the p97 ATPase would ensure that polypeptide modification is restricted to substrates undergoing retrotranslocation, conferring another layer of specificity to ubiquitin ligases. The simultaneous recruitment of an ubiquitin ligase and the p97 ATPase to Derlin-1 also suggests that Derlin-1 represents the site from which retrotranslocation substrates exit the ER into the cytosol. Because Derlin-1 is also associated with early translocation intermediates, it might guide substrates all of the way through the membrane. The oligomeric nature of Derlin-1 is consistent with its being part of a retrotranslocation channel. However, because the ubiquitin ligases Hrd1 and gp78 are also multispanning membrane proteins, it is possible that they contribute to the formation of a channel. In addition, from experiments in yeast, it is clear that there are other classes of substrates that do not require Der1p, the yeast homolog of Derlin-1, for their retrotranslocation (7, 8). These substrates could either use a different retrotranslocation channel or perhaps be extracted from the membrane without a channel.
We thank M. Seeger, A. Weissman, R. Takahashi, B. Lilley, and H. Ploegh (Harvard Medical School) for reagents, and the Nikon Imaging Center (Harvard Medical School) for assistance in microscopy. We thank E. Buerger (Humboldt University, Berlin) for making the pGEX-Hrd1c and pQE30-Derlin-1c constructs, M. Seeger for friendly exchange of ideas, and M. Rape and B. Burton for critical reading of the manuscript. The work was supported by National Institutes of Health Grant RO1 GM05586-10 (to T.A.R.). M.K. is supported by the Dutch Arthritis Foundation. T.A.R. is a Howard Hughes Medical Institute Investigator.
Supplementary Material
Author contributions: Y.Y., E.W., and T.A.R. designed research; Y.Y., Y.S., M.K., and S.v.V. performed research; Y.Y., M.K., E.W., and T.A.R. analyzed data; and Y.Y. and T.A.R. wrote the paper.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 3, 2005.
Abbreviations: ER, endoplasmic reticulum; VIMP, VCP-interacting membrane protein.
See accompanying Profile on page 14129.
References
- 1.Tsai, B., Ye, Y. & Rapoport, T. A. (2002) Nat. Rev. Mol. Cell. Biol. 3, 246-255. [DOI] [PubMed] [Google Scholar]
- 2.Tortorella, D., Gewurz, B. E., Furman, M. H., Schust, D. J. & Ploegh, H. L. (2000) Annu. Rev. Immunol. 18, 861-926. [DOI] [PubMed] [Google Scholar]
- 3.Wiertz, E. J. H. J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T. R., Rapoport, T. A. & Ploegh, H. L. (1996) Nature 384, 432-438. [DOI] [PubMed] [Google Scholar]
- 4.Wiertz, E. J. H. J., Jones, T. R., Sun, L., Bogyo, M., Geuze, H. J. & Ploegh, H. L. (1996) Cell 84, 769-779. [DOI] [PubMed] [Google Scholar]
- 5.Lilley, B. N. & Ploegh, H. L. (2004) Nature 429, 834-840. [DOI] [PubMed] [Google Scholar]
- 6.Ye, Y., Shibata, Y., Yun, C., Ron, D. & Rapoport, T. A. (2004) Nature 429, 841-847. [DOI] [PubMed] [Google Scholar]
- 7.Knop, M., Finger, A., Braun, T., Hellmuth, K. & Wolf, D. H. (1996) EMBO J. 15, 753-763. [PMC free article] [PubMed] [Google Scholar]
- 8.Vashist, S. & Ng, D. T. (2004) J. Cell Biol. 165, 41-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer, H. H., Shorter, J. G., Seemann, J., Pappin, D. & Warren, G. (2000) EMBO J. 19, 2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye, Y., Meyer, H. H. & Rapoport, T. A. (2003) J. Cell Biol. 162, 71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bays, N. W., Wilhovsky, S. K., Goradia, A., Hodgkiss-Harlow, K. & Hampton, R. Y. (2001) Mol. Biol. Cell 12, 4114-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye, Y., Meyer, H. H. & Rapoport, T. A. (2001) Nature 414, 652-656. [DOI] [PubMed] [Google Scholar]
- 13.Braun, S., Matuschewski, K., Rape, M., Thoms, S. & Jentsch, S. (2002) EMBO J. 21, 615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D. H. & Sommer, T. (2002) Nat. Cell Biol. 4, 134-139. [DOI] [PubMed] [Google Scholar]
- 15.Rabinovich, E., Kerem, A., Frohlich, K. U., Diamant, N. & Bar-Nun, S. (2002) Mol. Cell. Biol. 22, 626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flierman, D., Ye, Y., Dai, M., Chau, V. & Rapoport, T. A. (2003) J. Biol. Chem. 278, 34774-34782. [DOI] [PubMed] [Google Scholar]
- 17.Kikkert, M., Doolman, R., Dai, M., Avner, R., Hassink, G., van Voorden, S., Thanedar, S., Roitelman, J., Chau, V. & Wiertz, E. (2004) J. Biol. Chem. 279, 3525-3534. [DOI] [PubMed] [Google Scholar]
- 18.Bordallo, J., Plemper, R. K., Finger, A. & Wolf, D. H. (1998) Mol. Biol. Cell 9, 209-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilhovsky, S., Gardner, R. & Hampton, R. (2000) Mol. Biol. Cell 11, 1697-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bays, N. W., Gardner, R. G., Seelig, L. P., Joazeiro, C. A. & Hampton, R. Y. (2001) Nat. Cell Biol. 3, 24-29. [DOI] [PubMed] [Google Scholar]
- 21.Fang, S., Ferrone, M., Yang, C., Jensen, J. P., Tiwari, S. & Weissman, A. M. (2001) Proc. Natl. Acad. Sci. USA 98, 14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, J. S., Kim, T., Fang, S., Yamaguchi, J., Weissman, A. M., Fisher, E. A. & Ginsberg, H. N. (2003) J. Biol. Chem. 278, 23984-23988. [DOI] [PubMed] [Google Scholar]
- 23.Zhong, X., Shen, Y., Ballar, P., Apostolou, A., Agami, R. & Fang, S. (2004) J. Biol. Chem. 279, 45676-45684. [DOI] [PubMed] [Google Scholar]
- 24.Imai, Y., Soda, M., Inoue, H., Hattori, N., Mizuno, Y. & Takahashi, R. (2001) Cell 105, 891-902. [DOI] [PubMed] [Google Scholar]
- 25.Snapp, E. L., Hegde, R. S., Francolini, M., Lombardo, F., Colombo, S., Pedrazzini, E., Borgese, N. & Lippincott-Schwartz, J. (2003) J. Cell Biol. 163, 257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meacham, G. C., Patterson, C., Zhang, W., Younger, J. M. & Cyr, D. M. (2001) Nat. Cell Biol. 3, 100-105. [DOI] [PubMed] [Google Scholar]
- 27.Connell, P., Ballinger, C. A., Jiang, J., Wu, Y., Thompson, L. J., Hohfeld, J. & Patterson, C. (2001) Nat. Cell Biol. 3, 93-96. [DOI] [PubMed] [Google Scholar]
- 28.Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U. & Jentsch, S. (1999) Cell 96, 635-644. [DOI] [PubMed] [Google Scholar]
- 29.Hatakeyama, S., Matsumoto, M., Yada, M. & Nakayama, K. I. (2004) Genes Cells 9, 533-548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.