Abstract
Polypeptides that fail to pass quality control in the endoplasmic reticulum (ER) are dislocated from the ER membrane to the cytosol where they are degraded by the proteasome. Derlin-1, a member of a family of proteins that bears homology to yeast Der1p, was identified as a factor that is required for the human cytomegalovirus US11-mediated dislocation of class I MHC heavy chains from the ER membrane to the cytosol. Derlin-1 acts in concert with the AAA ATPase p97 to remove dislocation substrate proteins from the ER membrane, but it is unknown whether other factors aid Derlin-1 in its function. Mammalian genomes encode two additional, related proteins (Derlin-2 and Derlin-3). The similarity of the mammalian Derlin-2 and Derlin-3 proteins to yeast Der1p suggested that these as-yet-uncharacterized Derlins also may play a role in ER protein degradation. We demonstrate here that Derlin-2 is an ER-resident protein that, similar to Derlin-1, participates in the degradation of proteins from the ER. Furthermore, we show that Derlin-2 forms a robust multiprotein complex with the p97 AAA ATPase as well as the mammalian orthologs of the yeast Hrd1p/Hrd3p ubiquitin-ligase complex. The data presented here define a set of interactions between proteins involved in dislocation of misfolded polypeptides from the ER.
Keywords: Derlin, HRD1, SEL1L, p97, VIMP
Elimination of misfolded proteins from both cytoplasmic and membrane-delimited compartments is critical for cellular homeostasis. Protein folding in the secretory pathway is an imperfect process that generates by-products, which are disposed of largely by the cytosolic 26S proteasome (1). The definition of the molecular machinery in protein quality control and protein degradation has relied mostly on biochemical analyses and, in yeast, a genetic approach (2-4). The extent to which the known factors physically interact and function in a coordinated manner to clear the endoplasmic reticulum (ER) of misfit proteins is not known.
High-throughput approaches have examined physical interactions between proteins and resulted in the construction of interaction maps for the Saccharomyces cerevisiae and Caenorhabditis elegans proteomes (5, 6). However, the interactome maps reported thus far for S. cerevisiae or C. elegans do not include the interactions revealed by a more refined and targeted genetic analysis of protein degradation from the ER. For example, Der1p is important for the degradation of a restricted set of protein substrates (7, 8) and acts in concert with multiple factors to degrade these substrates (2, 3, 9-12). Data regarding both the mechanism of Der1p action and physical interactions formed by Der1p are lacking.
A human homolog of Der1p, Derlin-1, is required (in concert with p97) for the dislocation of class I MHC heavy chains mediated by human cytomegalovirus US11 but not by US2 (13, 14). It is likely that still other factors assist Derlin-1 and p97 during substrate dislocation to the cytosol (13). It also is unknown whether the two additional Derlin proteins that are present in mammalian genomes (13) play any role in the degradation of proteins that misfold in the ER. Here we explore physical interactions formed by two human Der1-like proteins (Derlin-1 and -2) and identify a set of contacts that suggest a mechanism for the dislocation of proteins from the ER to the cytosol and directly link a ubiquitin ligase to this process.
Materials and Methods
Antibodies (Abs), DNA Constructs, and Cell Lines. The following Abs have been described: anti-Derlin-1, anti-Derlin-2, anti-GRP94 (13), anti-VIMP [ref. 14; generously provided by Yihong Ye and Tom Rapoport (both of Harvard Medical School, Boston)], anti-HRD1 (ref. 15; generously provided by Emmanuel Wiertz, Leiden University, Leiden, The Netherlands), and anti-calnexin mAb AF8 (16). The anti-Derlin-2 Ab was affinity-purified as described (13) by using a synthetic peptide with the sequence (C)EERPGGFAWGEGQRLGG. Human SEL1L (National Center for Biotechnology Information gene ID: 6400) was cloned by RT-PCR from a human pancreas cDNA pool (Stratagene). The anti-p97 and anti-SEL1L Abs were generated as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Anti-GST and anti-GFP Abs were from Abcam (Cambridge, MA), and the anti-CHOP (CCAAT/enhancer-binding protein homologous protein) Ab was from Santa Cruz Biotechnology.
The Derlin-3GFP construct was made by cloning a PCR product from IMAGE clone 30338943 (encoding Derlin-3 isoform b cDNA; GenBank accession no. BC057830) into pEGFP-N1 as described for the Derlin-1GFP and Derlin-2GFP constructs (13). All three DerlinGFP constructs were subcloned into the pMSCV-Puro retroviral vector (CLONTECH). The pRETRO-SUPER vector (17) was used to deliver constructs for stable short hairpin RNA (shRNA) expression. Oligonucleotides targeting 19- or 20-bp regions of the indicated genes were designed as described (17). The sequences used, with the numbers in parentheses indicating the nucleotide coordinates of the respective cDNAs (with 1 representing the adenosine of the start codon) were TGGATATGCAGTTGCTGAT (347-365) (Derlin-1); ATGAGGATCCAAATTACAAT (638-657) (Derlin-2); and ACGGAAATCGGACAGAAAG (450-468) (VIMP). The sequence targeting GFP has been described (18).
HeLa cells, 293T cells, the US11WT and US11Q192L cell lines (19), WT, and Xbp-1-deficient mouse embryonic fibroblasts (ref. 20; generously provided by Laurie Glimcher, Harvard School of Public Health, Boston) were cultured as reported (19). Retroviral stocks were generated in 293T cells essentially as described (21), and infections were performed as described (19). Infected U373 and HeLa cells were selected in DMEM containing 0.375 and 1.0 μg/ml puromycin, respectively.
Metabolic Labeling, Immunoprecipitations, SDS/PAGE, and Immunoblotting. Methods for pulse labeling of cells, preparation of digitonin lysates, SDS/PAGE, and fluorography have been described (19). Steady-state metabolic labeling of cells, preparation of lysates, immunoprecipitations, and SDS/PAGE analysis were performed as described in Supporting Materials and Methods. Preparation of mouse tissue extracts and immunoblotting experiments were performed as described (13). Cells were treated with tunicamycin (Calbiochem) at concentrations and for durations of time as indicated in the figure legends. Lysates for immunoblotting experiments were prepared as described in Supporting Materials and Methods. Examination of Derlin-associated poly-GST-ubiquitinconjugated material was performed essentially as described (14) except that lysates of the membrane pellets were prepared as described in Supporting Materials and Methods. GST-ubiquitin was purchased from Boston Biochem (Cambridge, MA). Immunohistochemistry and confocal microscopy were performed as described in Supporting Materials and Methods.
Results
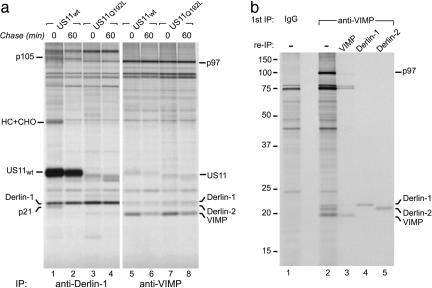
In our study of the role of Derlin-1 in US11-mediated dislocation of class I MHC heavy chains, we observed that proteins with molecular masses of 105 and 21 kDa associated with Derlin-1 in US11WT-expressing cells in a pulse-chase experiment (13) (Fig. 1a). We hypothesized that these species, which we refer to as p105 and p21, form a complex with Derlin-1 that catalyzes dislocation, because they associate with Derlin-1 only in the presence of active US11WT but not with the inactive US11Q192L (Fig. 1a, compare lanes 1 and 2 with 3 and 4). One candidate for the 21-kDa species is the VIMP protein (14) [also known as selenoprotein S (22) and Tanis (23)]. VIMP is believed to be an ER membrane receptor for the p97 ATPase complex, but its significance in the dislocation reaction is unclear (14). US11WT, the class I MHC heavy chain, p21, and p105 all coimmunoprecipitated with Derlin-1 in US11WT cells but not in US11Q192L cells (Fig. 1a, lanes 1 and 3). Denaturation of the Derlin-1 immunoprecipitate followed by reimmunoprecipitation with the anti-VIMP antiserum failed to recover p21. Examination of VIMP immunoprecipitates from identical amounts of the same lysates (Fig. 1a, lanes 5-8) revealed the presence of three polypeptides with molecular masses between 18 and 22 kDa; the smallest of the three was shown to be VIMP by reimmunoprecipitation (data not shown). Thus, the observed molecular weight of VIMP is inconsistent with that of p21.
Fig. 1.
Derlins interact with VIMP. (a) Interactions of US11WT, class I HC, Derlin-1, and VIMP. Immunoprecipitations from digitonin lysates of US11WT or US11Q192L cell lines pulse-labeled for 30 min and chased for the indicated times were performed with anti-Derlin-1 (lanes 1-4) or anti-VIMP (lanes 5-8) Abs. The positions of the relevant polypeptides are indicated. (b) VIMP associates with Derlin proteins. U373 cells were labeled to steady state, and immunoprecipitations from digitonin lysates were performed with control rabbit IgG (lane 1) or anti-VIMP Abs (lanes 2-5). The anti-VIMP immunoprecipitate (IP) was either analyzed directly (lane 2) or subjected to reimmunoprecipitation (re-IP) in sequential fashion by using the Abs indicated.
Introduction of an ATPase-deficient p97 protein into semipermeabilized US11 cells allowed visualization of the association between VIMP and class I MHC heavy chains (14). In intact cells, we did not detect the association of class I MHC heavy chain with VIMP, although anti-Derlin-1 immunoprecipitates from the same cell lysates yielded a readily detectable amount of the class I MHC heavy chain (Fig. 1a, compare lanes 1 and 5). Arresting the dislocation process with p97 mutants may be required to observe what might be a very transient interaction between VIMP and the class I MHC heavy chain. VIMP interacts with US11, although the stoichiometry of the complex is not known (14). In our experiments, the anti-VIMP Ab retrieved small and equivalent amounts of both US11WT and US11Q192L (Fig. 1a, lanes 5-8), demonstrating that VIMP does not associate selectively with the active US11WT as does Derlin-1 (Fig. 1a, compare lanes 1 and 2 with 3 and 4). The transient nature of the interaction between VIMP and the Derlin-1-US11-class I MHC heavy chain complex may account for our inability to observe such interactions.
We confirmed coimmunoprecipitation of Derlin-1 and p97 with VIMP (Fig. 1 a, lanes 5-8, and b, lane 2). In VIMP immunoprecipitates from steady-state-labeled cells, we observed an additional polypeptide that migrated slightly faster than Derlin-1 (Fig. 1b, lane 2), suggestive of its identity as the additional Derlin family member, Derlin-2 (13). Use of a Derlin-2-specific Ab confirmed that Derlin-2 associates with VIMP (Fig. 1b, lane 5) and demonstrated that Derlin-2 was not p21 despite nearly identical migration in SDS/PAGE (data not shown).
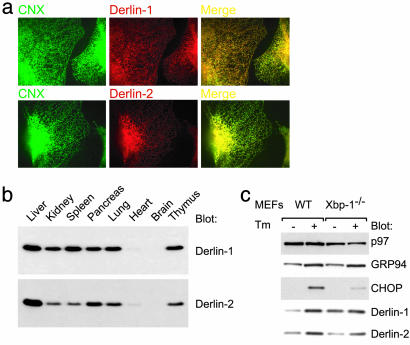
Derlin-2 [also known as F-LANa (24)] is a 239-aa protein that shares ≈30% sequence identity with Derlin-1 and possesses four transmembrane domains of similar topology to Derlin-1. The similarity of Derlin-2 to yeast Derlp, as well as to Derlin-1, suggested that it, too, may play a role in the dislocation of misfolded membrane proteins (13). Both Derlin-1 and Derlin-2 colocalize with the ER chaperone calnexin (Fig. 2a), indicating that both proteins are present in the ER. The pattern of Derlin-2 expression in mouse tissues is largely similar to that of Derlin-1; expression of Derlin-2 is highest in the liver (Fig. 2b; see also ref. 24).
Fig. 2.
Characterization of Derlin-2. (a) Derlin-2 is an ER-resident protein. U373 cells were subjected to immunohistochemistry by using Abs against calnexin (CNX; green) and Derlin-1 or -2 (red) followed by confocal microscopy. The merged images are shown, and areas of colocalization are indicated in yellow. (b) Analysis of Derlin protein expression in mouse tissues by immunoblotting. Longer exposures of the immunoblot shown reveal low levels of Derlin-1 and -2 expression in heart and brain. (c) Induction of Derlin protein expression by the unfolded protein response. Shown is immunoblot analysis of WT or Xbp-1-deficient mouse embryonic fibroblasts (MEFs) either mock-treated or treated with 100 ng/ml tunicamycin (Tm) for 24 h. CHOP, CCAAT/enhancer-binding protein homologous protein.
Yeast Der1p is up-regulated strongly by conditions that induce the accumulation of misfolded proteins in the ER lumen, presumably to assist clearance of these misfits (25, 26). In mouse embryonic fibroblasts treated with tunicamycin, both mDerlin-1 and mDerlin-2 were induced to a degree similar to that of the unfolded protein response target GRP94 (20) (Fig. 2c, lanes 1 and 2). The full induction of mDerlin-1 expression in response to ER stress required Xbp-1, a transcription factor that regulates expression of genes involved in the unfolded protein response (20). Induction of mDerlin-2 was less dependent on Xbp-1 (Fig. 2c, compare lanes 1 and 2 with 3 and 4).
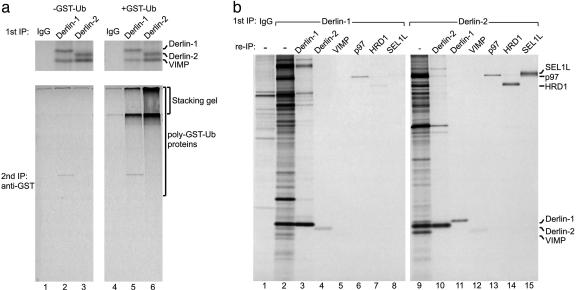
Does Derlin-2 also play a role in the dislocation of membrane proteins? To examine whether Derlin-2 associates with dislocation substrates as they exit the ER, we used a permeabilized cell system in which the addition of GST-ubiquitin prevents the release of newly synthesized dislocation substrates from the membrane (14). Immunoprecipitations with anti-Derlin-1 or anti-Derlin-2 Abs were then examined directly or reimmunoprecipitated with anti-GST Abs to reveal poly-GST-ubiquitinated proteins associated with the specific Derlin protein. In the pulse-labeling experiment shown in Fig. 3a, a significant amount of newly synthesized VIMP coimmunoprecipitates with both Derlin-1 and -2, although in experiments in which cells were labeled to steady state, only a weak signal for VIMP was observed in Derlin-1 and -2 immunoprecipitates. This discrepancy is likely caused by the difference in labeling methods. In the presence of GST-ubiquitin, high-molecular-weight poly-GST-ubiquitinated material (much of which failed to enter the resolving gel) was recovered with both the Derlin-1 and -2 Abs (Fig. 3a). More poly-GST-ubiquitinated material associated with Derlin-2 relative to Derlin-1 (Fig. 3a, compare lanes 6 and 5). Thus, Derlin-2 also associates with dislocation substrates on their way out of the ER, as was proposed for Derlin-1 (13, 14).
Fig. 3.
Association of Derlin proteins with dislocation substrates and proteins involved in ER degradation. (a) U373 cells were pulse-labeled and then incubated without (-GST-Ub; lanes 1-3) or with (+GST-Ub; lanes 4-6) GST-ubiquitin. Immunoprecipitations (IPs) using the indicated Abs were either analyzed directly (Upper) or subjected to reimmunoprecipitation using anti-GST antibodies (Lower). The position of the stacking gel is indicated to demonstrate the presence of high-molecular-weight poly-GST-ubiquitinated material. (b) Immunoprecipitations from lysates of steady-state-labeled U373 cells were performed with control rabbit IgG (lane 1), anti-Derlin-1 Abs (lanes 2-8), or anti-Derlin-2 Abs (lanes 9-15). The respective immunoprecipitates were either analyzed directly (lanes 1, 2, and 9) or reimmunoprecipitated (re-IP) in sequential fashion using the indicated Abs.
Additional factors may help Derlin proteins to extract misfolded proteins from the ER membrane. Comparison of the Derlin-1 and -2 immunoprecipitates shows a set of shared polypeptides, the patterns of migration of which are immediately recognizable as p97 and VIMP (Figs. 3b and 4). We used specific Abs and reimmunoprecipitation to identify these proteins unambiguously (Fig. 3b). The p97 and VIMP signals are consistently weak in the reimmunoprecipitation experiments (relative to the amount retrieved in the first immunoprecipitation) because of the fact that the anti-p97 and anti-VIMP Abs inefficiently retrieve their cognate antigen (Fig. 3b). Both p97 and VIMP coprecipitate with Derlin-1 and, to an even greater extent, Derlin-2. We consistently observed greater recovery of both p97 and VIMP with the anti-Derlin-2 Ab despite the fact that, based on the intensity of the radiolabeled band and similar methionine and cysteine content for Derlin-1 and -2, more Derlin-1 than Derlin-2 was recovered (Figs. 3b and 4). A small but significant amount of Derlin-1 coimmunoprecipitated with Derlin-2 (Fig. 3b, lane 11). The reciprocal immunoprecipitation demonstrated that some Derlin-2 coimmunoprecipitated with Derlin-1 (Fig. 3b, lane 4). The observed interactions between Derlin-1 and -2 are not caused by cross-reactivity of the Abs (data not shown). These results suggest that the Derlin proteins are capable of forming heterooligomers in the ER membrane, an issue addressed in more detail below. Several distinct polypeptides associate with both Derlin-1 and -2. Two strongly radiolabeled proteins, migrating at ≈82 and 102 kDa (a diffuse band migrating immediately above p97), coimmunoprecipitate with Derlin-2 (Figs. 3b and 4). Digestion of these immunoprecipitates with PNGaseF demonstrates that the 102-kDa protein possesses multiple N-linked glycans, whereas the 82-kDa polypeptide is not glycosylated (data not shown).
Fig. 4.
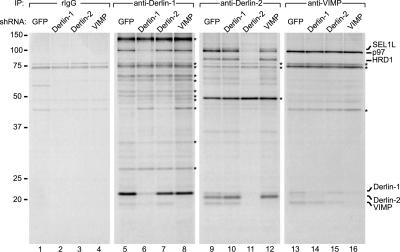
Knockdown of Derlin-1, Derlin-2, and VIMP reveals specificity of interactions. U373 cells expressing the indicated shRNA construct were labeled to steady state, and immunoprecipitations (IPs) were performed by using control rabbit IgG (rIgG; lanes 1-4), anti-Derlin-1 (lanes 5-8), anti-Derlin-2 (lanes 9-12), or anti-VIMP (lanes 13-16) Abs. The asterisks indicate nonspecifically bound polypeptides.
Although there are no biochemical data that indicate associations between Der1p and other proteins, genetic data in yeast suggest that Der1p acts in concert with other factors to degrade misfolded proteins (2, 3, 9-12). We hypothesized that the Derlin-2-associated proteins may represent mammalian orthologs of such yeast proteins. The mammalian orthologs of Hrd1p/Der3p and Hrd3p, the subunits of a ubiquitin-ligase complex implicated in degradation of misfolded proteins from the ER (10, 27, 28), represent possible candidates for the Derlin-2-associated proteins. The mammalian ortholog of Hrd1p/Der3p, HRD1 (15, 29, 30) or synoviolin (31), is a nonglycosylated ER membrane protein of ≈80 kDa containing a RING domain that acts as an active E3 ubiquitin ligase in vitro (15, 30). HRD1 participates in the degradation of ER proteins and protects cells from ER stress-induced apoptosis (15, 29, 30). The mammalian homolog of Hrd3p, SEL1L (32), is predicted to be a type I membrane protein with five N-linked glycans and a molecular mass of ≈100 kDa. SEL1L is highly expressed in the pancreas and is implicated in cell growth in a variety of tissue culture systems (32-34). Sel-1 RNA interference in C. elegans induces ER stress, as measured by using a strain that is transgenic for an unfolded protein response reporter construct (35), but a role for SEL1L in mammalian ER degradation as well as a physical association of SEL1L with HRD1 have yet to be shown.
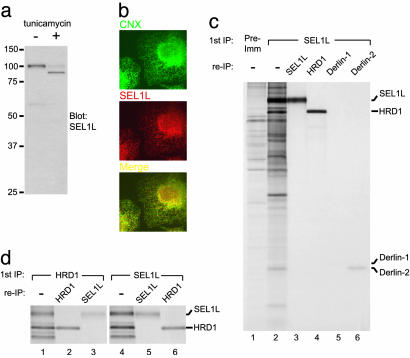
We used Abs against HRD1 (15) and SEL1L to identify unambiguously the 82- and 102-kDa species that associate with Derlin-2 as mammalian HRD1 and SEL1L, respectively (Fig. 3c, lanes 14 and 15). In immunoblotting experiments, the anti-SEL1L Ab generated here recognized a single glycosylated protein that corresponded to endogenous SEL1L (Fig. 5a). Immunohistochemistry experiments with affinity-purified anti-SEL1L Ab indicated that SEL1L largely colocalizes with calnexin (Fig. 5b). In Derlin-1 immunoprecipitates, a modest amount of HRD1 and SEL1L was recovered together with Derlin-1 when compared with Derlin-2 immunoprecipitates (Fig. 3c, compare lane 9 with 2 and lane 14 with 7). Using affinity-purified anti-SEL1L Abs, we observe that more Derlin-2 than Derlin-1 associates with SEL1L (Fig. 5c). A similar result was obtained by using anti-HRD1 Abs (data not shown). Coimmunoprecipitation of p97 with SEL1L was also observed (B.N.L. and H.L.P., unpublished results).
Fig. 5.
Analysis of SEL1L complexes. (a) Immunoblot of U373 cells either mock-treated or treated with 0.75 μg/ml tunicamycin for 14 h. (b) SEL1L largely colocalizes with calnexin (CNX) in U373 cells. (c) Immunoprecipitations (IPs) from lysates of steady-state-labeled U373 cells were performed with preimmune serum (lane 1) or anti-SEL1L Abs (lanes 2-6) and were either analyzed directly (lanes 1 and 2) or reimmunoprecipitated (re-IP) in sequential fashion using the indicated Abs. (d) As described for c except anti-HRD1 and anti-SEL1L Abs were used.
We also used the anti-HRD1 and anti-SEL1L Abs to examine whether these proteins form a complex, as do their orthologs in yeast (27). A large amount of SEL1L was coimmunoprecipitated with the anti-HRD1 Ab (Fig. 5 c, lane 4, and d Left), and significant quantities of HRD1 were coimmunoprecipitated with the anti-SEL1L Ab (Fig. 5d Right). In fact, we routinely recovered from identical amounts of lysate as much or more HRD1 by using the anti-SEL1L Ab than when using the anti-HRD1 Ab. Based on the intensities of the radiolabeled polypeptides, we estimate that HRD1 and SEL1L form a complex with 1:1 stoichiometry, as do their counterparts in yeast (27). We examined whether the HRD1/SEL1L complex associated with the class I MHC heavy chain during US11-mediated dislocation, but we failed to detect an association between SEL1L and the class I heavy chain substrate (data not shown). This result may indicate that dislocation induced by US11, although dependent on polyubiquitination (36, 37), is independent of HRD1/SEL1L ubiquitin-ligase complex.
The specificities of the interactions described were confirmed by using shRNA directed against Derlin-1, Derlin-2, or VIMP. Reduction in the levels of Derlin-2 results in a concomitant reduction in the amounts of coimmunoprecipitating SEL1L, p97, HRD1, Derlin-1, and VIMP (Fig. 4, compare lanes 9 and 11). The intensities of several other coimmunoprecipitating polypeptides remained unchanged; these may be species that cross-react with the anti-Derlin-2 Ab and should be considered nonspecific. Similar results were observed for Derlin-1 shRNA (Fig. 4, compare lanes 5 and 6).
The amount of p97 coimmunoprecipitating with Derlin-1 was not affected when VIMP levels were reduced by shRNA expression (Fig. 4, compare lanes 5 and 8). VIMP may be dispensable for the interaction of p97 with Derlin-1. When Derlin-2 levels were reduced by shRNA, the modest quantity of HRD1 that coprecipitated with Derlin-1 persisted despite a reduction in the amount of Derlin-2 recovered (Fig. 4, lane 7). Although we saw an ≈80% reduction in Derlin-1 levels when using shRNA, the amount of Derlin-1 that coimmunoprecipitated with both Derlin-2 and VIMP was hardly affected (Fig. 4, compare lanes 9 with 10 and 13 with 14). Derlin-1 therefore may be present in excess over what is required to form complexes with VIMP, Derlin-2, and perhaps other factors such as p97. Such an excess of Derlin-1 may explain why we do not observe an effect on the rate of class I MHC heavy chain degradation in US11WT cells when Derlin-1 levels are reduced significantly by the appropriate shRNA (B.N.L. and H.L.P., unpublished results). Reduction of VIMP levels by shRNA expression resulted not only in decreased amounts of Derlin-2-associated VIMP but also less p97 coimmunoprecipitated with Derlin-2 (Fig. 4, compare lanes 9 and 12). Thus, the association of p97 with Derlin-2 may depend more strictly on the presence of VIMP. The reduction of VIMP levels, however, did not impair the association of HRD1/SEL1L with Derlin-2, indicating that this complex forms independently of VIMP.
Because Derlin-1 plays a crucial role in the movement of dislocation substrates from the ER to the cytosol, it has been proposed that Derlin-1 forms a type of channel that spans the ER lipid bilayer (13, 14). Because a single Derlin-1 protein contains only four transmembrane domains, oligomerization would be required to form a channel large enough to accommodate large proteins containing N-linked glycans (13, 38). Endogenously expressed Derlin-1 and -2 are capable of forming heterooligomers (Fig. 3b). We further examined Derlin protein oligomerization by using HeLa cells that express in stable fashion GFP-tagged Derlin-1, -2, or -3 (isoform b; GenBank accession no. NP_001002862). Derlin-3 is another Derlin family member whose function and biochemical properties have not been characterized. Immunoprecipitations from radiolabeled lysates of cells expressing the different GFP-tagged Derlin proteins (Derlin-1GFP, Derlin-2GFP, or Derlin-3GFP) showed that a small amount of both Derlin-2GFP and Derlin-3GFP coimmunoprecipitated with Derlin-1 (see Fig. 7a, lanes 7 and 8, which is published as supporting information on the PNAS web site). Furthermore, Derlin-1GFP and Derlin-3GFP coimmunoprecipitated with Derlin-2 (Fig. 7a, lanes 10 and 12). Immunoprecipitations from the same cell lysates using an anti-GFP Ab showed that the endogenous Derlin-1 and -2 proteins coimmunoprecipitate with all three of the GFP-tagged Derlin proteins (Fig. 7b, lanes 7 and 8, 11 and 12, and 15 and 16). The Derlin proteins are thus capable of forming hetero- as well as homooligomers.
Retrieval of Derlin-1GFP using the anti-GFP Ab revealed a large quantity of associated p97, but we did not observe detectable amounts of HRD1 or SEL1L associating with Derlin-1GFP (Fig. 7, lane 5). p97, HRD1, and SEL1L also coimmunoprecipitated with Derlin-2GFP and Derlin-3GFP, suggesting that Derlin-3 may also participate in degradation of misfolded ER proteins.
Discussion
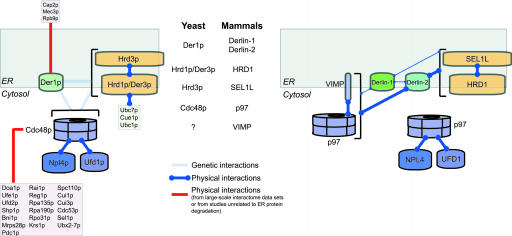
Derlin-1 is a factor that participates in the movement of certain proteins from the ER to the cytosol during dislocation (13, 14). We now demonstrate that Derlin-2, also an ER membrane protein expressed in tissues with a high secretory load, associates with dislocation substrates as they exit the ER. We used steady-state-labeled cells and coimmunoprecipitations under mild conditions to visualize, in an unbiased manner, the complement of endogenously expressed proteins that associate with two of the three human Derlin proteins. This approach revealed a set of interactions not previously observed in any model system. Data for Cdc48p from high-throughput approaches to analyzing protein-protein interactions are consistent with that protein's involvement in a number of cellular activities (39). However, the proteins that interact with Der1p in such screens (shown in Fig. 6) are not reconciled easily with current knowledge of ER protein degradation. Approaches that specifically address interactions of membrane proteins will reveal the molecular composition of the complexes that catalyze dislocation.
Fig. 6.
Map of interactions involved in the degradation of a subset of misfolded ER proteins. Previously demonstrated genetic or physical interactions observed in yeast compared with the physical interactions are shown here for the human orthologs. Interactions observed in high-throughput analyses or in analyses not pertaining to ER degradation are depicted separately from those observed in focused studies of ER protein degradation. The relative strengths of the interactions observed in this study are indicated by the thickness of the lines. The interaction maps are based on data either cited in the body of the text or from the Incyte Genomics Proteome Bioknowledge Library (39, 50) and references therein. The Der1p pathway only processes a subset of substrate proteins in yeast. Currently, the extent to which the Derlin-1 and Derlin-2 pathways are used in mammalian cells is not known.
Der1p is required for the degradation of a subset of proteins that contain luminal domains defective in folding (8). For the known substrates that require Der1p for degradation, the Hrd1p/Der3p-Hrd3p ubiquitin-ligase complex and the Cdc48p/Npl4p/Ufd1p complex are also required (8, 10, 40), although no such physical interactions between these protein complexes has been reported. We now demonstrate that the mammalian orthologs of the yeast Hrd1p/Der3p-Hrd3p complex (HRD1/SEL1L), p97, and Derlin proteins form a large, multiprotein complex (schematically depicted in Fig. 6). Additional analysis with Abs specific for p97 cofactors will be required to examine whether they, too, are present in complexes with Derlin proteins. Misfolded proteins with lesions in different topological compartments are dealt with by distinct sets of machinery associated with the ER in yeast (7, 8). Defining the types of substrates that are processed by different Derlin proteins, and those that are processed by other pathways (41), is an important goal for future studies of ER protein degradation in mammalian cells.
The requirement for the Hrd1p/Hrd3p complex for degradation of substrates that require Der1p indicates that Der1p may act as an adaptor molecule that allows Hrd1p/Hrd3p to degrade certain classes of substrates. It remains to be determined whether a similar situation applies for mammalian Derlins. We have been unable to demonstrate interactions between the HRD1/SEL1L complex and the class I MHC heavy chain in cells that express US11, which raises the question of the identity of the ubiquitin ligase involved in US11-mediated dislocation. The gp78 ubiquitin ligase is known to interact with p97 and mediate degradation of the CD3δ substrate protein (42). Could this ubiquitin ligase, or others known to associate with the ER membrane (43-45), aid the Derlin proteins in their function(s)?
The mechanism of Derlin protein action is also unknown. One hypothesis that has been put forward is that the Derlin proteins form a channel through oligomerization or association with other factors (13, 14). We observe both homo- and heterooligomerization of the Derlin proteins as well as association of Derlins with other proteins. The aggregate number of transmembrane segments in such complexes could be quite large and may have the ability to form a channel structure when properly assembled. It also remains a possibility that Derlins act as adaptor proteins to deliver the substrate molecule to the bona fide channel that allows substrate molecules to pass through the ER.
The observed complexes suggest an intimate link between movement of substrate proteins across the ER membrane, ubiquitination, and extraction. A complex that is capable of these activities would ensure directionality to dislocation by introducing the cytosolic polyubiquitin recognition tag (46) onto emerging substrates in immediate proximity to p97, the subunit of the complex that provides the energy for dislocation (47). Although Derlins, p97, and ER-associated ubiquitin ligases clearly are important for movement of substrate molecules across the ER membrane (13, 14, 36, 37, 47, 48), recognition of misfolded proteins in the lumen of the ER is an important step for initiating dislocation (49). We are in the process of identifying additional Derlin-2-associated proteins, which may represent luminal factors that target dislocation substrates to the multiprotein complexes identified here.
Note Added in Proof. In this issue of PNAS, Ye et al. (51) also report an interaction between Derlin-1 and HRD1 and homooligomerization of Derlin-1.
Supplementary Material
Acknowledgments
We thank Margo Furman for generation of the anti-p97 Ab; Jatin Vyas for assistance with confocal microscopy; Howard Hang and Chris Love for critical reading of the manuscript; Emmanuel Wiertz, Ann-Hwee Lee, Laurie Glimcher, Gustavo Mostoslavsky, and Richard Mulligan for reagents; and Yihong Ye and Tom Rapoport for reagents and communication of results before publication. B.N.L. was a Howard Hughes Medical Institute Predoctoral Fellow. This work was supported by grants from the National Institutes of Health.
Author contributions: B.N.L. designed research; B.N.L. performed research; B.N.L. and H.L.P. analyzed data; and B.N.L. and H.L.P. wrote the paper.
Abbreviations: ER, endoplasmic reticulum; shRNA, short hairpin RNA.
References
- 1.Hirsch, C., Jarosch, E., Sommer, T. & Wolf, D. H. (2004) Biochim. Biophys. Acta 1695, 215-223. [DOI] [PubMed] [Google Scholar]
- 2.Knop, M., Finger, A., Braun, T., Hellmuth, K. & Wolf, D. H. (1996) EMBO J. 15, 753-763. [PMC free article] [PubMed] [Google Scholar]
- 3.Medicherla, B., Kostova, Z., Schaefer, A. & Wolf, D. H. (2004) EMBO Rep. 5, 692-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hampton, R. Y., Gardner, R. G. & Rine, J. (1996) Mol. Biol. Cell 7, 2029-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141-147. [DOI] [PubMed] [Google Scholar]
- 6.Li, S., Armstrong, C. M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P. O., Han, J. D., Chesneau, A., Hao, T., et al. (2004) Science 303, 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taxis, C., Hitt, R., Park, S. H., Deak, P. M., Kostova, Z. & Wolf, D. H. (2003) J. Biol. Chem. 278, 35903-35913. [DOI] [PubMed] [Google Scholar]
- 8.Vashist, S. & Ng, D. T. (2004) J. Cell Biol. 165, 41-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiller, M. M., Finger, A., Schweiger, M. & Wolf, D. H. (1996) Science 273, 1725-1728. [DOI] [PubMed] [Google Scholar]
- 10.Bordallo, J., Plemper, R. K., Finger, A. & Wolf, D. H. (1998) Mol. Biol. Cell 9, 209-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hitt, R. & Wolf, D. H. (2004) FEMS Yeast Res. 4, 815-820. [DOI] [PubMed] [Google Scholar]
- 12.Buschhorn, B. A., Kostova, Z., Medicherla, B. & Wolf, D. H. (2004) FEBS Lett. 577, 422-426. [DOI] [PubMed] [Google Scholar]
- 13.Lilley, B. N. & Ploegh, H. L. (2004) Nature 429, 834-840. [DOI] [PubMed] [Google Scholar]
- 14.Ye, Y., Shibata, Y., Yun, C., Ron, D. & Rapoport, T. A. (2004) Nature 429, 841-847. [DOI] [PubMed] [Google Scholar]
- 15.Kikkert, M., Doolman, R., Dai, M., Avner, R., Hassink, G., van Voorden, S., Thanedar, S., Roitelman, J., Chau, V. & Wiertz, E. (2004) J. Biol. Chem. 279, 3525-3534. [DOI] [PubMed] [Google Scholar]
- 16.Hochstenbach, F., David, V., Watkins, S. & Brenner, M. B. (1992) Proc. Natl. Acad. Sci. USA 89, 4734-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 18.Tiscornia, G., Singer, O., Ikawa, M. & Verma, I. M. (2003) Proc. Natl. Acad. Sci. USA 100, 1844-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lilley, B. N., Tortorella, D. & Ploegh, H. L. (2003) Mol. Biol. Cell 14, 3690-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, A. H., Iwakoshi, N. N. & Glimcher, L. H. (2003) Mol. Cell. Biol. 23, 7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soneoka, Y., Cannon, P. M., Ramsdale, E. E., Griffiths, J. C., Romano, G., Kingsman, S. M. & Kingsman, A. J. (1995) Nucleic Acids Res. 23, 628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kryukov, G. V., Castellano, S., Novoselov, S. V., Lobanov, A. V., Zehtab, O., Guigo, R. & Gladyshev, V. N. (2003) Science 300, 1439-1443. [DOI] [PubMed] [Google Scholar]
- 23.Gao, Y., Walder, K., Sunderland, T., Kantham, L., Feng, H. C., Quick, M., Bishara, N., de Silva, A., Augert, G., Tenne-Brown, J., et al. (2003) Diabetes 52, 929-934. [DOI] [PubMed] [Google Scholar]
- 24.Ying, H., Yu, Y. & Xu, Y. (2001) Biochem. Biophys. Res. Commun. 286, 394-400. [DOI] [PubMed] [Google Scholar]
- 25.Casagrande, R., Stern, P., Diehn, M., Shamu, C., Osario, M., Zuniga, M., Brown, P. O. & Ploegh, H. (2000) Mol. Cell 5, 729-735. [DOI] [PubMed] [Google Scholar]
- 26.Travers, K. J., Patil, C. K., Wodicka, L., Lockhart, D. J., Weissman, J. S. & Walter, P. (2000) Cell 101, 249-258. [DOI] [PubMed] [Google Scholar]
- 27.Gardner, R. G., Swarbrick, G. M., Bays, N. W., Cronin, S. R., Wilhovsky, S., Seelig, L., Kim, C. & Hampton, R. Y. (2000) J. Cell Biol. 151, 69-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bays, N. W., Gardner, R. G., Seelig, L. P., Joazeiro, C. A. & Hampton, R. Y. (2001) Nat. Cell Biol. 3, 24-29. [DOI] [PubMed] [Google Scholar]
- 29.Kaneko, M., Ishiguro, M., Niinuma, Y., Uesugi, M. & Nomura, Y. (2002) FEBS Lett. 532, 147-152. [DOI] [PubMed] [Google Scholar]
- 30.Nadav, E., Shmueli, A., Barr, H., Gonen, H., Ciechanover, A. & Reiss, Y. (2003) Biochem. Biophys. Res. Commun. 303, 91-97. [DOI] [PubMed] [Google Scholar]
- 31.Amano, T., Yamasaki, S., Yagishita, N., Tsuchimochi, K., Shin, H., Kawahara, K., Aratani, S., Fujita, H., Zhang, L., Ikeda, R., et al. (2003) Genes Dev. 17, 2436-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biunno, I., Appierto, V., Cattaneo, M., Leone, B. E., Balzano, G., Socci, C., Saccone, S., Letizia, A., Della Valle, G. & Sgaramella, V. (1997) Genomics 46, 284-286. [DOI] [PubMed] [Google Scholar]
- 33.Cattaneo, M., Orlandini, S., Beghelli, S., Moore, P. S., Sorio, C., Bonora, A., Bassi, C., Talamini, G., Zamboni, G., Orlandi, R., et al. (2003) Oncogene 22, 6359-6368. [DOI] [PubMed] [Google Scholar]
- 34.Cattaneo, M., Canton, C., Albertini, A. & Biunno, I. (2004) Gene 326, 149-156. [DOI] [PubMed] [Google Scholar]
- 35.Urano, F., Calfon, M., Yoneda, T., Yun, C., Kiraly, M., Clark, S. G. & Ron, D. (2002) J. Cell Biol. 158, 639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shamu, C. E., Flierman, D., Ploegh, H. L., Rapoport, T. A. & Chau, V. (2001) Mol. Biol. Cell 12, 2546-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kikkert, M., Hassink, G., Barel, M., Hirsch, C., van der Wal, F. J. & Wiertz, E. (2001) Biochem. J. 358, 369-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misaghi, S., Pacold, M. E., Blom, D., Ploegh, H. L. & Korbel, G. A. (2004) Chem. Biol. 11, 1677-1687. [DOI] [PubMed] [Google Scholar]
- 39.Costanzo, M. C., Hogan, J. D., Cusick, M. E., Davis, B. P., Fancher, A. M., Hodges, P. E., Kondu, P., Lengieza, C., Lew-Smith, J. E., Lingner, C., et al. (2000) Nucleic Acids Res. 28, 73-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D. H. & Sommer, T. (2002) Nat. Cell Biol. 4, 134-139. [DOI] [PubMed] [Google Scholar]
- 41.Mancini, R., Aebi, M. & Helenius, A. (2003) J. Biol. Chem. 278, 46895-46905. [DOI] [PubMed] [Google Scholar]
- 42.Zhong, X., Shen, Y., Ballar, P., Apostolou, A., Agami, R. & Fang, S. (2004) J. Biol. Chem. 279, 45676-45684. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, Y., Chiba, T., Tokunaga, F., Kawasaki, H., Iwai, K., Suzuki, T., Ito, Y., Matsuoka, K., Yoshida, M., Tanaka, K. & Tai, T. (2002) Nature 418, 438-442. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida, Y., Tokunaga, F., Chiba, T., Iwai, K., Tanaka, K. & Tai, T. (2003) J. Biol. Chem. 278, 43877-43884. [DOI] [PubMed] [Google Scholar]
- 45.Hassink, G., Kikkert, M., van Voorden, S., Lee, S. J., Spaapen, R., van Laar, T., Coleman, C. S., Bartee, E., Fruh, K., Chau, V. & Wiertz, E. (2005) Biochem. J. 388, 647-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flierman, D., Ye, Y., Dai, M., Chau, V. & Rapoport, T. A. (2003) J. Biol. Chem. 278, 34774-34782. [DOI] [PubMed] [Google Scholar]
- 47.Ye, Y., Meyer, H. H. & Rapoport, T. A. (2003) J. Cell Biol. 162, 71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye, Y., Meyer, H. H. & Rapoport, T. A. (2001) Nature 414, 652-656. [DOI] [PubMed] [Google Scholar]
- 49.Ellgaard, L. & Helenius, A. (2003) Nat. Rev. Mol. Cell Biol. 4, 181-191. [DOI] [PubMed] [Google Scholar]
- 50.Hodges, P. E., Carrico, P. M., Hogan, J. D., O'Neill, K. E., Owen, J. J., Mangan, M., Davis, B. P., Brooks, J. E. & Garrels, J. I. (2002) Nucleic Acids Res. 30, 137-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye, Y., Shibata, Y., Kikkert, M., van Voorden, S., Wiertz, E. & Rapoport, T. A. (2005) Proc. Natl. Acad. Sci. USA 102, 14132-14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.