Abstract
The cell cycle-regulated Aurora-B kinase is a chromosomal passenger protein that is implicated in fundamental mitotic events, including chromosome alignment and segregation and spindle checkpoint function. Aurora-B phosphorylates serine 10 of histone H3, a function that has been associated with mitotic chromatin condensation. We find that activation of poly(ADP-ribose) polymerase (PARP) 1 by DNA damage results in a rapid block of H3 phosphorylation. PARP-1 is a NAD+-dependent enzyme that plays a multifunctional role in DNA damage detection and repair and maintenance of genomic stability. Here, we show that Aurora-B physically and specifically associates with the BRCT (BRCA-1 C-terminal) domain of PARP-1. Aurora-B becomes highly poly(ADP-ribosyl)ated in response to DNA damage, a modification that leads to a striking inhibition of its kinase activity. The highly similar Aurora-A kinase is not regulated by PARP-1. We propose that the specific inhibition of Aurora-B kinase activity by PARP-1 contributes to the physiological response to DNA damage.
Keywords: mitosis, poly(ADP-ribose) polymerase-1, histone H3, phosphorylation
Mitosis is a highly orchestrated process that entails a plethora of control mechanisms. Signaling events that coordinate mitosis induce a wave of protein phosphorylation governed by the cyclin-dependent kinase Cdc2. After Cdc2 activation, downstream mitotic controllers are recognized within the families of Aurora, Polo, and NIMA-related kinases (1). Mitotic phosphorylation of histone H3 at serine 10 (P-H3/Ser-10) occurs highly synchronously and is associated with the initiation of chromosome condensation (2, 3). The kinase implicated in this event is Aurora-B, one of the three members of the Aurora family of kinases (4-6).
The cell cycle-regulated Aurora-B kinase is a chromosomal passenger protein that has been shown to play essential roles in mitosis (7-9). A number of critical proteins have been shown to be substrates of Aurora-B, including the myosin II regulatory light chain, vimentin, desmin, and GFAP (glial fibrillary acidic protein) (10-12), which suggests a role for Aurora-B at the cleavage furrow during cytokinesis. Other targets of Aurora-B in cytokinetic processes are INCENP (inner-centromere protein) and ZEN-4/CeMKLP1, as reported in Caenorhabditis elegans and mammalian cells (13, 14). Interference of Aurora-B activity causes defects in chromosome congression because of its involvement in the regulation of the kinetochore-microtubule interactions (15-17).
Aurora-B and its related kinase Aurora-A (7) are overexpressed in a number of human tumors, and their ectopic overexpression in cultured cells results in cellular transformation, centrosome abnormalities, and aneuploidy (18-21). Failure of mitotic chromosome segregation leads to aneuploidy that may contribute to cancer onset (22). Therefore, identification of the pathways that control Aurora-B function is of central importance.
Genotoxic stress causes activation of cellular checkpoints (23), leading to diverse responses such as cell cycle arrest, DNA repair, and cell death. The identification of specific mediators of DNA damaging signals, such as ATM (ataxia telangiectasia mutated) and ATR (ATM- and Rad3-related) (24), and the deciphering of how they influence the progression through the cell cycle has greatly improved our understanding of the cellular response to genotoxic agents.
Poly(ADP-ribosyl)ation is an immediate cellular response to DNA strand breaks that is catalyzed by NAD+-dependent enzymes, poly(ADP-ribose) polymerases (PARPs) (25). PARPs catalyze the stepwise addition of ADP-ribose moeities to substrate proteins by using intracellular NAD+ as source of ADP-ribose. The PARP family contains several members, although >90% of cellular poly(ADP-ribosyl)ation is ascribed to PARP-1 (26), the most active and best-characterized member of the PARP family. It is a nuclear protein with DNA damage scanning activity that is implicated in the maintenance of genomic integrity (27-29), leading to the control of cellular proliferation and carcinogenesis (30).
Here, we report that mitotic P-H3/Ser-10 phosphorylation is drastically reduced upon DNA damage. This event is coupled to the inhibition of the Aurora-B kinase by PARP-1-mediated poly(ADP-ribosyl)ation. Our findings establish an intriguing link among DNA damage, chromatin modifications, and regulation of mitotic events. The NAD+ dependence of PARP-1 enzymatic activity may extend the physiological implications of these observations to the control of cellular metabolism.
Materials and Methods
Cell Cultures. NIH 3T3 mouse fibroblast cells were cultured in DMEM supplemented with 10% serum and treated with H2O2 (1 mM) for different times. After incubation, cells were collected and lysed in Laemmli buffer; equal amounts of proteins, determined by Coomassie staining, were loaded onto a SDS-acrylamide gel and processed for Western blot analysis. When the PARP-1 inhibitor 3-aminobenzamide (3-AB) (Sigma) was used, a 3-h pretreatment was performed before adding H2O2.
Mouse embryonic fibroblasts (MEFs) were isolated by micro-dissection of embryos at 13.5 days of gestation resulting from intercrosses between wild-type or mutant mice. The spermatogonial cell line GC-spg1 was purchased from American Type Culture Collection. For immunofluorescence analysis, cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.2% Triton X-100, blocked with 5% BSA, and then incubated overnight with primary antibodies, washed, and revealed with secondary FITC- or Cy3-conjugated antibodies. Treatment of cells with 0.1 μg/ml nocodazole was extended for various periods as indicated.
Immunoprecipitation. GC-spg1 cells and testis from adult mice were homogenized in a lysis buffer (50 mM Hepes, pH 7.4/100 mM KCl/0.5% Nonidet P-40/proteinase and phosphatase inhibitors) and immunoprecipitated overnight with the following antibodies: anti-rabbit Aurora-A, anti-rat or rabbit Aurora-B, anti-mouse PARP-1, and anti-rabbit IgG. The immunocomplexes were collected by protein A- or G- Sepharose beads, and bound proteins were washed several times with lysis buffer and eluted with Laemmli buffer to be processed for Western blot.
Western Blot Analysis. Protein extracts were separated by SDS-acrylamide and transferred to nitrocellulose membrane; blots were probed with the indicated antibodies and visualized by using the Pierce chemiluminescence detection system.
Poly(ADP-ribosyl)ation Assay. Recombinant Myc-tagged Aurora-B or Aurora-A was expressed in Cos-1 cells and immunoprecipitated with anti-Myc monoclonal antibody 9E10 (Santa Cruz Biotechnology). Poly(ADP-ribosyl)ation assays were performed on the immunoprecipitated proteins in 40 μl of reaction buffer (50 mM Tris·HCl, pH 8.0/4 mM MgCl2/0.2 mM DTT/2 μg/ml DNaseI-activated calf thymus DNA) containing 1 μg of purified PARP-1 and 0.3 μCi (1 Ci = 37 GBq) of [32P]-NAD+ for 10 min at 25°C. Proteins were separated by 10% SDS/PAGE, transferred to nitrocellulose membranes, and analyzed by autoradiography. Subsequently, to evaluate the kinase activity of the modified Aurora-B, cold NAD+ was used in the poly(ADP-ribosyl)ation assay.
Aurora-B Kinase Assay. Immunoprecipitated Aurora-B from treated and control cells or in vitro poly(ADP-ribosyl)ated Aurora-B was used in the presence of 5 μCi of [32P]-γ-ATP. One microgram of total histones or GST-H3 was used as a substrate as indicated. The reaction was carried out for 30 min at 30°C and stopped by the addition of Laemmli buffer. Proteins were analyzed by SDS/PAGE followed by autoradiography.
Results
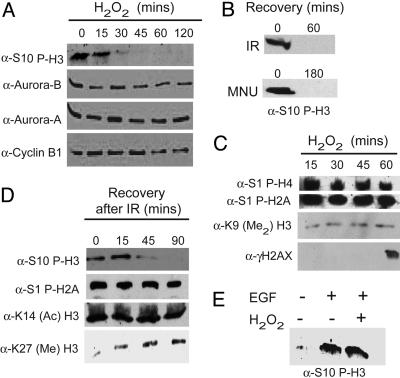
Specific Reduction of Histone H3 Ser-10 Mitotic Phosphorylation in Response to DNA Damage. Because genotoxic stress is involved in the etiology of cancer, we wanted to explore the effect of DNA damaging agents on mitotic histone H3 phosphorylation at serine 10 and possibly on Aurora-B function. To address this question, we imposed on growing cultured cells the oxidative stress elicited by H2O2 and ionizing radiation (IR); both treatments cause DNA strand breaks. There is a rapid and striking decrease in the levels of P-H3/Ser-10 mitotic phosphorylation within 45 min upon H2O2 treatment (Fig. 1A). This effect is not related to short-time changes in the cell cycle, as indicated by the unaltered levels of Aurora-A, Aurora-B, and cyclin B1 proteins (Fig. 1 A) and cell sorting analysis (Fig. 6, which is published as supporting information on the PNAS web site). H3 phosphorylation is similarly reduced in response to IR and N-nitroso-N-methylurea (Fig. 1B).
Fig. 1.
Effect of DNA damage on mitotic histone H3 phosphorylation. NIH 3T3 mouse fibroblasts were cultured in DMEM supplemented with 10% serum, and DNA damage was induced by various means. (A) Western analyses of Ser-10 H3 phosphorylation in cells treated with H2O2 (1 mM) at different times. The protein levels of Aurora-A, Aurora-B, and cyclin B1 were assayed as controls. (B) Cells exposed to IR (6 Gy) or treated with N-nitroso-N-methylurea (2 mM) for 45 min and allowed to recover. (C and D) Analysis of various histone modifications upon DNA damage by using 1 mM H2O2 or 6-Gy IR. Antibodies directed against the indicated specific histone modifications were used in Western assays. (E) DNA damage does not influence EGF-induced H3 phosphorylation. Cells were serum-starved for 24 h and then stimulated with EGF for 1 h as described in refs. 23-27, in conjunction with 1 mM H2O2 treatment or not.
The reduction of P-H3/Ser-10 levels in response to DNA damaging agents is not coupled to other histone H3 modifications (Ac-K14, Met-K9 and Met-K27; Fig. 1B) and occurs shortly before the ATM (ataxia telangiectasia mutated)/ATR (ATM- and Rad3-related)-induced phosphorylation of γ-H2AX. Importantly, phosphorylation of other histones is not altered (Ser-1 of H2A and Ser-1 of H4; Fig. 1B). These results demonstrate that the reduction in H3 Ser-10 phosphorylation in response to DNA damage is a specific event.
In addition to being a hallmark of mitotic chromatin condensation, H3 phosphorylation at Ser-10 has been associated with transcriptional activation at interphase (31). This event is stimulated by growth factors (e.g., EGF) and is mediated by a number of kinases, including Rsk-2, MSK1 (mitogen- and stress-activated protein kinase 1), and IκB kinase-α (31-33), depending on the cell type and signaling event. Here, we stimulated NIH 3T3 cells with EGF in a paradigm of early response, a treatment that results in the induction of early gene expression and transient H3 phosphorylation (34, 35). Importantly, the levels of EGF-induced H3 phosphorylation are not affected in response to DNA damage. Therefore, of the modifications assayed, mitotic H3 phosphorylation appears specifically sensitive to DNA damage (Fig. 1C). This result also implies that the decrease in H3 phosphorylation induced by DNA damage is likely to be independent from the activation of intracellular phosphatases.
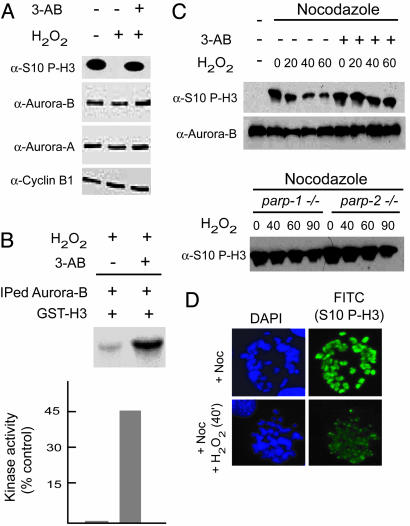
Oxidative DNA Damage Inhibits Aurora-B but only in the Presence of Active PARP-1. Interestingly, poly(ADP-ribosyl)ation is a cell cycle-regulated modification (36), and PARP-1 associates with centromeric proteins CENP-A, CENP-B, and Bub3 and poly(ADP-ribosyl)ates them in response to DNA damage (37). This notion and the finding of the decrease in mitotic H3 phosphorylation in response to DNA damage (Fig. 1) prompted us to explore the existence of a functional interplay between PARP-1 and Aurora-B. This possibility was supported by the observation that pretreatement of cells with 3-AB, a pharmacological inhibitor of PARP-1 enzymatic activity, efficiently blocks the effect elicited by H2O2 on H3 phosphorylation (Fig. 2A). Treatment with 3-AB directly affects Aurora-B catalytic activity, as demonstrated by kinase activity assays performed by using immunoprecipitated cellular Aurora-B from control and treated cells (Fig. 2B). Thus, activation of PARP-1 upon DNA damage appears to directly influence the activity of the endogenous Aurora-B kinase. Cells arrested in mitosis with nocodazole showed a reduction in H3 serine-10 phosphorylation upon H2O2 treatment, whereas it remained unchanged in cells similarly arrested in mitosis that were pretreated with 3-AB (Fig. 2C). Nocodazole-treated cells were exposed to H2O2, and P-H3/Ser-10 signals were analyzed by immunoflourescence with specific antibodies. A decrease in the levels of Ser-10 phosphorylation was seen in mitotic chromosomes in response to DNA damage by H2O2 (Fig. 2D), indicative of a specific event occurring during mitosis. To strengthen these results, we used MEFs derived from mice carrying a targeted mutation in the parp-1 and parp-2 genes (38) (Fig. 2C). In these mutant MEFs, which were stalled in mitosis by nocodazole, the effect of H2O2 is significantly abrogated compared with wild-type MEFs, providing genetic evidence of the regulatory role played by PARP-1 on H3 phosphorylation.
Fig. 2.
Functional relationship between Aurora-B and PARP-1 activation. (A) NIH 3T3 mouse fibroblasts were treated with H2O2 (1 mM) for the periods indicated in the presence or absence of the PARP-1 inhibitor 3-AB (cells were pretreated with 3 mM 3-AB for 3 h). (B) Kinase assay using immunoprecipitated Aurora-B from H2O2 (1 mM/60 min)-treated cells using GST-H3 as a substrate and [32P]-γATP. Pretreatment of cells with 3-AB (3 mM/3 h) increases in vitro Aurora-B kinase activity. Values were determined by densitometric analysis of the radioactivity (phospho-GST-H3) present in the substrate and are expressed as the percentage compared with a control sample (immunoprecipitated by using normal IgG). The lanes corresponding to + or - (3-AB) were taken from the same gel and sliced together as indicated. (C) Cells (Upper, NIH 3T3; Lower, MEFs from parp-1- and parp-2-null mice) were arrested in mitosis with nocodazole for 16 h and treated with 1 mM H2O2 as indicated in the presence of nocodazole. 3-AB (3 mM for 3 h) was added when necessary before H2O2 treatment. Western analysis was carried out by using antibodies directed against P-H3/Ser-10. The levels of Aurora-B were analyzed as a control. (D) NIH 3T3 cells were treated with nocodazole for 6-8 h and then with H2O2 for 40 min. Cells were harvested by mitotic shake-off, cytospun onto slides, and immunostained by using antibodies directed against P-H3/Ser-10. Cells untreated with H2O2 served as a control. DAPI was used as a counterstain.
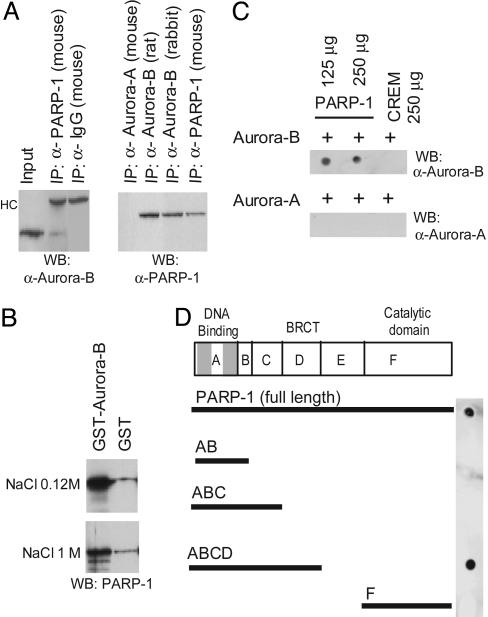
Aurora-B Interacts with PARP-1 Through the BRCA-1 C-Terminal (BRCT) Domain. The dependence of Aurora-B activity on PARP1 prompted us to investigate whether PARP-1 may physically interact with Aurora-B. This possibility was suggested by the fact that both proteins are found to be associated with nuclear chromatin (9, 37). Coimmunoprecipitations of endogenous proteins from GC-spg1 cells (Fig. 3A) and testis (not shown) with specific antibodies revealed that Aurora-B associates with PARP-1 (Fig. 3A). The interaction was further substantiated by reverse coimmuoprecipitations. Importantly, the interaction is highly specific; the similar kinase Aurora-A does not associate with PARP-1 (Fig. 3A). These findings were confirmed by GST pull-down assays (Fig. 3B) and far-Western analyses (Fig. 3C). Various deletions of PARP-1 were used to identify the domain required for interaction with Aurora-B (Fig. 3D). Interestingly, the BRCT domain (amino acids 373-524) is likely essential for association. The BRCT domain is an evolutionarily conserved module that is present in proteins that participate in DNA damage cell cycle checkpoint and/or DNA repair pathways (39, 40). Interestingly, BRCT domains have been identified as phosphoprotein-binding domains involved in cell cycle control (41, 42). Thus, the interaction of the mitotically regulated Aurora-B kinase with the BRCT in PARP-1 is appealing. Interestingly, the same BRCT domain in PARP-1 undergoes automodification (26) and directs the association with p53 (43) and the base excision repair protein XRCC1 (44). Further experiments will be needed to establish whether phosphorylation of the BRCT domain, perhaps by Aurora-B itself, could be involved in the interaction.
Fig. 3.
Aurora-B interacts specifically with PARP-1. (A) Extracts from GC-spg1 cells were immunoprecipitated with the indicated antibodies, and the immunocomplexes were probed with either anti-Aurora-B or anti-PARP-1 antibodies. Aurora-A does not interact with PARP-1. HC, heavy chain. (B) GST or GST-Aurora-B proteins coupled to glutathione beads were incubated with purified PARP-1 protein and washed with a low- or high-salt buffer. Western blotting with anti-PARP-1 antibodies was used to analyze the pulled-down products. (C) Far-Western analysis. Two aliquots of 125 μg and 250 μg of purified PARP-1 protein were spotted onto a nitrocellulose filter, hybridized with Aurora-A or Aurora-B, washed, and probed with anti-Aurora-A and/or anti-Aurora-B antibodies. (D) Equimolar aliquots of purified full-length PARP-1 and of truncated proteins corresponding to different functional domains were spotted onto a nitrocellulose filter, hybridized with Aurora-B, and revealed with anti-Aurora-B antibodies. The interaction between the kinase and PARP-1 involves the BRCT domain (amino acids 373-524), whereas no binding occurs between Aurora-B and the PARP-1 catalytic domain.
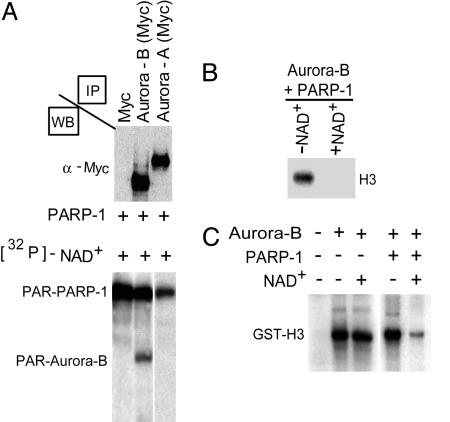
Poly(ADP-ribosyl)ation of Aurora-B by PARP-1 Inhibits Kinase Activity. Our results suggested that the activation of PARP-1 by DNA damage could negatively influence Aurora-B function. Therefore, we immunoprecipitated Aurora-B and Aurora-A and performed a poly(ADP-ribosyl)ation assay by using PARP-1 in the presence of [32P]-labeled NAD+. Strikingly, PARP-1 efficiently modifies Aurora-B but not Aurora-A, underscoring the specificity of this modification (Fig. 4A). To check the possible consequence of the poly(ADP-ribosyl)ation on Aurora-B kinase activity, we performed a kinase assay using histones as natural Aurora-B substrates for phosphorylation. We found a striking reduction in the phosphorylation of H3 incubated with a poly(ADP-ribosyl)ated Aurora-B in the presence of NAD+ (Fig. 4B). This effect is caused by a NAD+-dependent inactivation of Aurora-B kinase activity, as demonstrated by a kinase assay using GST-H3 in the presence of PARP-1 (Fig. 4C). Thus, PARP-1 associates with Aurora-B, induces poly(ADP-ribosyl)ation, and profoundly affects its capacity of phosphorylating histones. It also seems that Aurora-B could be regulated in a similar manner by PARP-2 (Fig. 7, which is published as supporting information on the PNAS web site). We have also found that PARP-1 is phosphorylated by Aurora-B, although this event does not seem to alter its enzymatic activity (not shown). Our observations strongly support a scenario where inhibition of Aurora-B kinase by poly(ADP-ribosyl)ation could be a critical step in cells that are undergoing DNA damage.
Fig. 4.
Poly(ADP-ribosyl)ation of Aurora-B blocks kinase activity. (A) Myc-tagged Aurora-A or Aurora-B kinases immunoprecipitated from COS-1 transfected cells were used as substrates in a poly(ADP-ribosyl)ation reaction catalyzed by PARP-1 using [32P]-NAD. Aurora-B is readily poly(ADP-ribosyl)ated by PARP-1, whereas Aurora-A is not. The reaction products were separated on SDS-polyacrylamide gel and visualized by autoradiography. Western blot analyses (WB) with an anti-Myc antibody revealed comparable amounts of expressed recombinant Aurora-A and Aurora-B. PAR, poly(ADP-ribosyl)ated. (B) Aurora-B immunoprecipitated from COS-1 cells was subjected to poly(ADP-ribosyl)ation in the presence or absence of NAD+, and its kinase activity was evaluated by using [32P]-γATP and a mix of histones as a substrate. Levels of phosphorylated H3 were detected by autoradiography. Only in the presence of NAD+ is the histone kinase activity of the poly(ADP-ribosyl)ated Aurora-B drastically inhibited, as shown by the absence of H3 phosphorylation. (C) GST-Aurora-B was similarly used for kinase assays using GST-H3 as a substrate in the presence or absence of NAD+ and/or PARP-1. Phosphorylation of GST-H3 was revealed by autoradiography. No difference was observed when the activity of Aurora-B was measured in the presence or absence of NAD+ alone. Kinase activity decreased after poly(ADP-ribosyl)ation by PARP-1.
Discussion
The essential role of Aurora-B during mitosis underscores the importance of the signaling pathways that control its function. The interplay between the molecular effectors of the DNA damage response and the mitotic machinery is likely to constitute a critical step in the control of cellular proliferation and metabolism. The present study indicates that Aurora-B is the target of the regulation exerted by PARP-1, a PARP that becomes activated in response to DNA damage (Fig. 5). Inhibition of Aurora-B kinase activity occurs in a NAD+-dependent manner and is specific because the highly related Aurora-A kinase does not associate with PARP-1 and is not poly(ADP-ribosyl)ated.
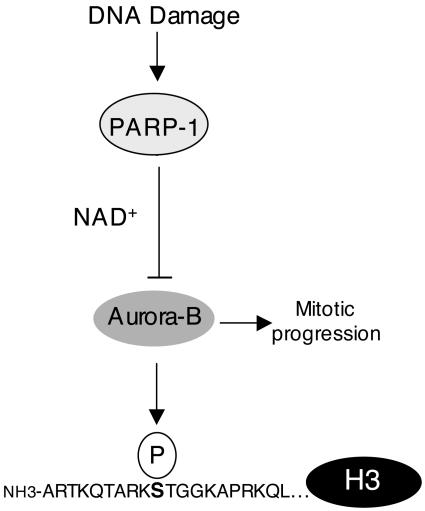
Fig. 5.
Regulation of the cell cycle-regulated Aurora-B kinase by poly(ADP-ribosyl)ation. Aurora-B, a chromosomal passenger protein implicated in mitotic progression, phosphorylates serine 10 of histone H3 during mitosis (6). Activation of PARP-1 by DNA damage results in a rapid, NAD+-dependent block of H3 phosphorylation. Aurora-B physically associates with PARP-1 and becomes highly poly(ADP-ribosyl)ated in response to DNA damage. This modification results in a striking inhibition of Aurora-B kinase activity. Specific inhibition of Aurora-B by PARP-1 may contribute to the physiological response to DNA damage.
Interestingly, PARP-1 interacts with Aurora-B through the BRCT domain (Fig. 3), a domain previously shown to constitute a phosphorylation-dependent protein-protein association interface (41, 42). Although it is still unclear whether the BRCT domain of PARP-1 needs to be phosphorylated to catalyze the interaction with Aurora-B, it is noteworthy that we have found that Aurora-B is able to phosphorylate PARP-1. Yet, this event does not seem to alter PARP-1 activity (data available on request). It is also worth stressing that PARP-1 mediates the DNA damage response through the interaction with other proteins by means of the BRCT domain (45). It has been shown that PARP-1 is present at centromeres during mitosis and that it associates with the centromeric proteins CENP-A, CENP-B, and Bub. Further, PARP-1 is also able to poly(ADP-ribosyl)ate these proteins after DNA damage induced by IR (37). Taken together, these observations indicate that PARP-1 is likely to play a crucial role in mediating the cellular stress response to the mitotic machinery.
What is the effect of DNA damage on mitotic chromatin? We have shown that the striking decrease in H3 Ser-10 phosphorylation is a highly specific event (Fig. 1), because other histone modifications remain unchanged. Importantly, histone H2B phosphorylation at Ser-14 (46) and phosphorylation of yeast H2A at Ser-129 have been implicated in genotoxic stress response (47). In the latter case, it has also been proposed that phosphorylation allows the recruiting of chromatin-modification complexes. Of course, mitotic chromatin represents a special case because of its high compaction. Poly(ADP-ribosyl)ation of histones elicited by PARP-1, and the potential interplay with other modifications, are central issues that deserve further study.
In conclusion, the central role played by Aurora-B in chromosomal dynamics (7-9) and the function of poly(ADP-ribosyl)ation in chromatin decondensation (48, 49) and DNA repair (27-29) provide an attractive framework for our results. Aurora-B was shown to interact with at least two other chromosomal passenger proteins, INCENP (inner-centromere protein) and Survivin/BIR-1 (7). It would be of interest to investigate whether poly(ADP-ribosyl)ation of Aurora-B may constitute a regulatory step to determine the interaction with either Survivin/BIR-1 or INCENP. Finally, it is important to emphasize that kinases of the Aurora family are highly expressed in various human cancers and have oncogenic potential (18-21), underscoring the importance that our results may have for human health.
Supplementary Material
Acknowledgments
We thank C. D. Allis and J. Toppari for help and discussions and Stéphanie Roux and Estelle Heitz for expert technical help. U.K.-S. is supported by a fellowship of Institut National de la Santé et de la Recherche Médicale (INSERM). This work was supported by Centre National de la Recherche Scientifique, INSERM, Centre Hospitalier Universitaire Régional, Fondation de la Recherche Médicale, Université Louis Pasteur, and Association pour la Recherche sur le Cancer. The P.S.-C. laboratory is an “Equipe Labelisée” of the Ligue Contre le Cancer.
Author contributions: L.M., U.K.-S., R.L., J.M.-d.M., G.d.M., and P.S.-C. designed research; L.M., U.K.-S., R.L., and J.M.-d.M. performed research; J.M.-d.M. and G.d.M. contributed new reagents/analytic tools; L.M., U.K.-S., R.L., J.M.-d.M., G.d.M., and P.S.-C. analyzed data; and L.M., U.K.-S., and P.S.-C. wrote the paper.
Abbreviations: PARP, poly(ADP-ribose) polymerase; MEF, mouse embryonic fibroblast; BRCT, BRCA-1 C-terminal; 3-AB, 3-aminobenzamide; IR, ionizing radiation.
References
- 1.O'Connell, M. J., Krien, M. J. & Hunter, T. (2003) Trends Cell Biol. 13, 221-228. [DOI] [PubMed] [Google Scholar]
- 2.Hendzel, M. J., Wei, Y., Mancini, M. A., Van Hooser, A., Ranalli, T., Brinkley, B. R., Bazett-Jones, D. P. & Allis, C. D. (1997) Chromosoma 106, 348-360. [DOI] [PubMed] [Google Scholar]
- 3.Cheung, P., Allis, C. D. & Sassone-Corsi, P. (2000) Cell 103, 263-271. [DOI] [PubMed] [Google Scholar]
- 4.Murnion, M. E., Adams, R. R., Callister, D. M., Allis, C. D., Earnshaw, W. C. & Swedlow, J. R. (2001) J. Biol. Chem. 276, 26656-26665. [DOI] [PubMed] [Google Scholar]
- 5.Hsu, J. Y., Sun, Z. W., Li, X., Reuben, M., Tatchell, K., Bishop, D. K., Grushcow, J. M., Brame, C. J., Caldwell, J. A., Hunt, D. F., et al. (2000) Cell 102, 279-291. [DOI] [PubMed] [Google Scholar]
- 6.Crosio, C., Fimia, G. M., Loury, R., Kimura, M., Okano, Y., Zhou, H., Sen, S., Allis, C. D. & Sassone-Corsi, P. (2002) Mol. Cell. Biol. 22, 874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmena, M. & Earnshaw, W. C. (2003) Nat. Rev. Mol. Cell. Biol. 4, 842-854. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff, J. R. & Plowman, G. D. (1999) Trends Cell Biol. 9, 454-459. [DOI] [PubMed] [Google Scholar]
- 9.Shannon, K. B. & Salmon, E. D. (2002) Curr. Biol. 12, R458-R460. [DOI] [PubMed] [Google Scholar]
- 10.Murata-Hori, M., Fumoto, K., Fukuta, Y., Iwasaki, T., Kikuchi, A., Tatsuka, M. & Hosoya, H. (2000) J. Biochem. (Tokyo) 128, 903-907. [DOI] [PubMed] [Google Scholar]
- 11.Goto, H., Yasui, Y., Kawajiri, A., Nigg, E. A., Terada, Y., Tatsuka, M., Nagata, K. & Inagaki, M. (2003) J. Biol. Chem. 278, 8526-8530. [DOI] [PubMed] [Google Scholar]
- 12.Kawajiri, A., Yasui, Y., Goto, H., Tatsuka, M., Takahashi, M., Nagata, K. & Inagaki, M. (2003) Mol. Biol. Cell 14, 1489-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaitna, S., Mendoza, M., Jantsch-Plunger, V. & Glotzer, M. (2000) Curr. Biol. 10, 1172-1181. [DOI] [PubMed] [Google Scholar]
- 14.Severson, A. F., Hamill, D. R., Carter, J. C., Schumacher, J. & Bowerman, B. (2000) Curr. Biol. 10, 1162-1171. [DOI] [PubMed] [Google Scholar]
- 15.Adams, R. R., Carmena, M. & Earnshaw, W. C. (2001) Trends Cell Biol. 11, 49-54. [DOI] [PubMed] [Google Scholar]
- 16.Hauf, S., Cole, R. W., LaTerra, S., Zimmer, C., Schnapp, G., Walter, R., Heckel, A., van Meel, J., Rieder, C. L. & Peters, J. M. (2003) J. Cell Biol. 161, 281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ditchfield, C., Johnson, V. L., Tighe, A., Ellston, R., Haworth, C., Johnson, T., Mortlock, A., Keen, N. & Taylor, S. S. (2003) J. Cell Biol. 161, 267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams, R. R., Eckley, D. M., Vagnarelli, P., Wheatley, S. P., Gerloff, D. L., Mackay, A. M., Svingen, P. A., Kaufmann, S. H. & Earnshaw, W. C. (2001) Chromosoma 110, 65-74. [DOI] [PubMed] [Google Scholar]
- 19.Ota, T., Suto, S., Katayama, H., Han, Z. B., Suzuki, F., Maeda, M., Tanino, M., Terada, Y. & Tatsuka, M. (2002) Cancer Res. 62, 5168-5177. [PubMed] [Google Scholar]
- 20.Zhou, H., Kuang, J., Zhong, L., Kuo, W. L., Gray, J. W., Sahin, A., Brinkley, B. R. & Sen, S. (1998) Nat. Genet. 20, 189-193. [DOI] [PubMed] [Google Scholar]
- 21.Bischoff, J. R., Anderson, L., Zhu, Y., Mossie, K., Ng, L., Souza, B., Schryver, B., Flanagan, P., Clairvoyant, F., Ginther, C., et al. (1998) EMBO J. 17, 3052-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieber, O. M., Heinimann, K. & Tomlinson, I. P. (2003) Nat. Rev. Cancer 3, 701-708. [DOI] [PubMed] [Google Scholar]
- 23.Hartwell, L. H. & Weinert, T. A. (1989) Science 246, 629-634. [DOI] [PubMed] [Google Scholar]
- 24.Shiloh, Y. (2003) Nat. Rev. Cancer 3, 155-168. [DOI] [PubMed] [Google Scholar]
- 25.Ame, J. C., Spenlehauer, C. & de Murcia, G. (2004) BioEssays 26, 882-893. [DOI] [PubMed] [Google Scholar]
- 26.Schreiber, V., Ame, J. C., Dolle, P., Schultz, I., Rinaldi, B., Fraulob, V., Menissier-de Murcia, J. & de Murcia, G. (2002) J. Biol. Chem. 277, 23028-23036. [DOI] [PubMed] [Google Scholar]
- 27.Burkle, A. (2001) BioEssays 23, 795-806. [DOI] [PubMed] [Google Scholar]
- 28.Shall, S. & de Murcia, G. (2000) Mutat. Res. 460, 1-15. [DOI] [PubMed] [Google Scholar]
- 29.Pieper, A. A., Verma, A., Zhang, J. & Snyder, S. H. (1999) Trends Pharmacol. Sci. 20, 171-181. [DOI] [PubMed] [Google Scholar]
- 30.Masutani, M., Nakagama, H. & Sugimura, T. (2003) Genes Chromosomes Cancer 38, 339-348. [DOI] [PubMed] [Google Scholar]
- 31.Nowak, S. J. & Corces, V. G. (2004) Trends Genet. 20, 214-220. [DOI] [PubMed] [Google Scholar]
- 32.Thomson, S., Clayton, A. L., Hazzalin, C. A., Rose, S., Barratt, M. J. & Mahadevan, L. C. (1999) EMBO J. 18, 4779-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto, Y., Verma, U. N., Prajapati, S., Kwak, Y. T. & Gaynor, R. B. (2003) Nature 423, 655-659. [DOI] [PubMed] [Google Scholar]
- 34.Mahadevan, L. C., Willis, A. C. & Barratt, M. J. (1991) Cell 65, 775-783. [DOI] [PubMed] [Google Scholar]
- 35.Sassone-Corsi, P., Mizzen, C. A., Cheung, P., Crosio, C., Monaco, L., Jacquot, S., Hanauer, A. & Allis, C. D. (1999) Science 285, 886-891. [DOI] [PubMed] [Google Scholar]
- 36.Tanuma, S. & Kanai, Y. (1982) J. Biol. Chem. 257, 6565-6570. [PubMed] [Google Scholar]
- 37.Saxena, A., Saffery, R., Wong, L. H., Kalitsis, P. & Choo, K. H. (2002) J. Biol. Chem. 277, 26921-26926. [DOI] [PubMed] [Google Scholar]
- 38.Menissier de Murcia, J., Ricoul, M., Tartier, L., Niedergang, C., Huber, A., Dantzer, F., Schreiber, V., Ame, J. C., Dierich, A., LeMeur, M., et al. (2003) EMBO J. 22, 2255-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bork, P., Hofmann, K., Bucher, P., Neuwald, A. F., Altschul, S. F. & Koonin, E. V. (1997) FASEB J. 11, 68-76. [PubMed] [Google Scholar]
- 40.Callebaut, I. & Mornon, J. P. (1997) FEBS Lett. 400, 25-30. [DOI] [PubMed] [Google Scholar]
- 41.Yu, X., Chini, C. C., He, M., Mer, G. & Chen, J. (2003) Science 302, 639-642. [DOI] [PubMed] [Google Scholar]
- 42.Manke, I. A., Lowery, D. M., Nguyen, A. & Yaffe, M. B. (2003) Science 302, 636-639. [DOI] [PubMed] [Google Scholar]
- 43.Wesierska-Gadek, J., Wojciechowski, J. & Schmid, G. (2003) J. Cell. Biochem. 89, 1260-1284. [DOI] [PubMed] [Google Scholar]
- 44.Masson, M., Niedergang, C., Schreiber, V., Muller, S., Menissier-de Murcia, J. & de Murcia, G. (1998) Mol. Cell. Biol. 18, 3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, S. (2001) Trends Biochem. Sci. 26, 174-179. [DOI] [PubMed] [Google Scholar]
- 46.Fernandez-Capetillo, O., Allis, C. D. & Nussenzweig, A. (2004) J. Exp. Med. 199, 1671-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Redon, C., Pilch, D. R., Rogakou, E. P., Orr, A. H., Lowndes, N. F. & Bonner, W. M. (2003) EMBO Rep. 4, 678-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poirier, G. G., de Murcia, G., Jongstra-Bilen, J., Niedergang, C. & Mandel, P. (1982) Proc. Natl. Acad. Sci. USA 79, 3423-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tulin, A. & Spradling, A. (2003) Science 299, 560-562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.