Abstract
Long-distance radical cation transport was studied in DNA condensates. Linearized pUC19 plasmid was ligated to an oligomer containing a covalently linked anthraquinone group and six regularly spaced GG steps, which serve as traps for the migrating radical cation. Treatment of the linear, ligated plasmid with spermidine results in formation of condensates that were detected by light scattering and observed by transmission electron microscopy. Irradiation of the anthraquinone group in the condensate causes long-distance charge migration, which is detected by reaction at the remote guanines. The efficiency of charge migration in the condensate is significantly less than it is for the corresponding oligomer in solution. This result is attributed to a lower mobility for the migrating radical cation in the condensate, which is caused by inhibited formation of charge-transfer-effective states.
Keywords: charge transfer, oxidative damage
It is well known that DNA is damaged by reactive chemical species. Extensive studies have shown that the one-electron oxidation of an oligonucleotide in solution creates a nucleobase radical cation (electron “hole”) that may migrate hundreds of angstroms through duplex DNA before it is irreversibly trapped by reaction with H2O or O2 (1-3). Trapping occurs most commonly at a guanine, because it is the base with the lowest ionization potential (4), and this reaction leads to mutagenic products such as 8-oxoguanine (5-8). DNA within cells is tightly packed and does not closely resemble oligonucleotides in solution. An examination of radical-cation hopping through DNA in loosely organized nucleosome core particles showed that histone binding does not affect the pattern and extent of oxidation (9). However, it is not known how the long-distance radical-cation migration that is observed in oligonucleotides is affected by conversion of the DNA to a well ordered structure. We began the systematic investigation of this question by examining the reactions of radical cations that were specifically introduced into DNA condensates.
Multivalent cations like spermine or spermidine and proteins such as polylysine initiate DNA condensation (10-14). DNA condensates are nanometer-scale particles formed by the collapse of extended DNA chains into compact, ordered structures containing a small number of molecules (15, 16). These structures are recognized as models that are useful for studying gene packing in viruses, bacteria, and eukaryotic cells (17).
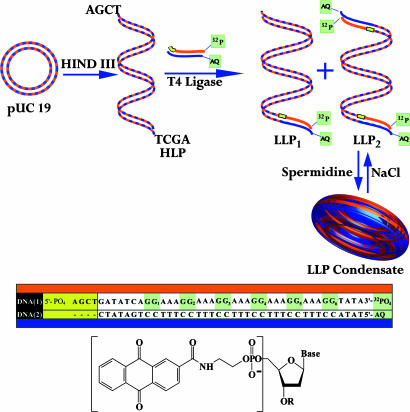
We prepared condensates formed from the linearized pUC19 plasmid (which contains 2,686 bp) that had been ligated to an oligonucleotide containing a covalently linked anthraquinone (AQ) group (for photoinitiated introduction of a radical cation) and a series of regularly spaced GG steps (as traps for the radical cation) (Fig. 1) (18). Dynamic light-scattering results and examination by transmission electron microscopy (TEM) shows that the ligated DNA sample is converted into condensates by the addition of spermidine. The condensates were irradiated with UV light (to introduce radical cations) and assayed by high-resolution PAGE. These studies show that long-distance radicalcation migration is less efficient in condensates than it is for DNA oligonucleotides in solution.
Fig. 1.
Schematic representation of the DNA oligomers, AQ structure, and the experimental design. The pUC19 plasmid is cut with HindIII to form the linear DNA structure (HLP) with two cohesive ends. HLP is treated with T4 ligase in the presence of the oligomer DNA-1/DNA-2, which has one blunt end and a cohesive end (yellow) that is complementary to HLP. DNA-1/DNA-2 is a duplex containing a covalently linked AQ group (structure shown) and a 32P radiolabel. Its ligation to HLP generates LLP1 and LLP2, which differ by being ligated at one and two ends of HLP, respectively. Treatment of LLP with spermidine leads to condensate formation, and treatment of the condensate with concentrated NaCl solution reverses the process.
Materials and Methods
The pUC19 plasmid, T4 DNA ligase, and HindIII restriction endonuclease were purchased from New England Biolabs. Terminal deoxynucleotidyltransferase (TdT) enzyme and [α-32P]ATP were purchased from Amersham Biosciences. Gel-extraction kits were purchased from Qiagen, and spermidine hydrochloride was obtained from Aldrich. DNA oligomers and the AQ-containing oligomer were synthesized by following standard procedures (1) on an Expedite 8909 DNA synthesizer. The oligonucleotides were purified by reverse-phase HPLC on a Hitachi (Tokyo) preparative HPLC system by using a Dynamax C18 column. Purified oligomers were desalted with Sep-Pak columns and characterized by mass spectroscopy. UV melting and cooling curves were recorded on a Cary 1E spectrophotometer (Varian) equipped with a multicell block, temperature controller, and sample transport accessory.
Restriction Endonuclease Cutting of Plasmid. pUC19 plasmid was treated with HindIII restriction endonuclease in a microcentrifuge tube containing 1× HindIII buffer and incubated overnight at 37°C. The restriction cut plasmid was identified by comparison with the uncut supercoiled and relaxed forms of the plasmid by electrophoresis using an analytical, nondenaturing, low-melting agarose gel.
Preparation of Radiolabeled DNA. The DNA-1 oligomer (Fig. 1) was radiolabeled with [α-32P]ATP and terminal deoxynucleotidyltransferase (TdT) enzyme and isolated by standard methods on a 20% denaturing polyacrylamide gel. The DNA was precipitated by addition of cold ethanol and glycogen, vortexed, placed on dry ice, and then centrifuged. The DNA sample was washed twice with 80% ethanol and air-dried.
Ligation of Plasmid with Radiolabeled Oligonucleotides. The radiolabeled DNA oligomers were passed through buffer-exchange spin columns to remove ammonium ions and other cations. Samples of the duplex oligomer for ligation were prepared by hybridizing radiolabeled DNA-1 with DNA-2 in sodium phosphate buffer solution at pH 7.0. The oligomer was ligated to the linearized pUC19 plasmid (4:1 molar ratio) with T4 DNA ligase, 1 mM ATP, and PEG 8000 in 1× ligase buffer at 16°C overnight in a Neslab RTE-7 incubator (Newington, NH). The reaction was analyzed by autoradiography on an analytical, nondenaturing low-melting agarose gel by following standard procedures (19). The DNA in the reaction mixture was isolated by precipitation with ethanol in the presence of glycogen, followed by centrifugation. The ligated plasmid DNA was isolated from a nondenaturing 0.8% preparative agarose gel in 1× TAE buffer (Tris-acetate-EDTA) containing ethidium bromide for visualization. The gel pieces were dissolved in high-ionic-strength buffer solution, incubated at 50°C for 20 min, and after repeated ethanol washes, extracted from spin columns (Qiagen) with 10 mM sodium phosphate buffer at pH 7.5.
Preparation of DNA Condensates. All solutions were filtered through Amicon UltraFree MC centrifugal filters (diameter, 0.22 μm; Millipore) before use in condensation reactions. A solution of linearized pUC19 plasmid (25 μg/ml) was added to an equal volume of spermidine solutions (50-400 μM at intervals of 50 μM). Condensations were performed in 10 mM sodium phosphate buffer (pH 7.8) by bringing the DNA to appropriate buffer concentration by using Microcon YM-30 centrifugal filter devices (Millipore). After 5 min of gentle agitation, the samples were analyzed by dynamic light scattering using a Dynapro MS/X dynamic light-scattering instrument (Proterion, Piscataway, NJ). The condensates were shown by light-scattering experiments to revert to the linear form when they were redissolved in a 1 M NaCl solution.
TEM. Condensates were prepared according to the standard procedure, deposited on a carbon-coated copper grid (Ted Pella, Inc., Redding, CA), and allowed to settle for 10 min. The grids were stained by the direct addition of 5 μl of 2% uranyl acetate, rinsed in 95% ethanol, and air-dried. Images of the DNA condensates at room temperature were collected on film by using a JEOL 100C transmission electron microscope at ×100,000 magnification.
UV Irradiation and Strand-Cleavage Analysis. Solutions (20 μl, 10 mM sodium phosphate buffer) of radiolabeled duplex oligomer, linear ligated plasmid, or freshly prepared condensates were irradiated in microcentrifuge tubes at ≈30°C in a Rayonet photoreactor (Southern New England Ultraviolet, Barnsford, CT) equipped with eight 350-nm lamps. After irradiation, the samples were precipitated with cold ethanol and glycogen, washed twice with 80% ethanol, dried, and treated with piperidine at 90°C for 30 min. After removal of the piperidine, the samples were suspended in denaturing loading dye and analyzed by PAGE, and the amount of strand cleavage was quantified by phosphorimagery.
Results
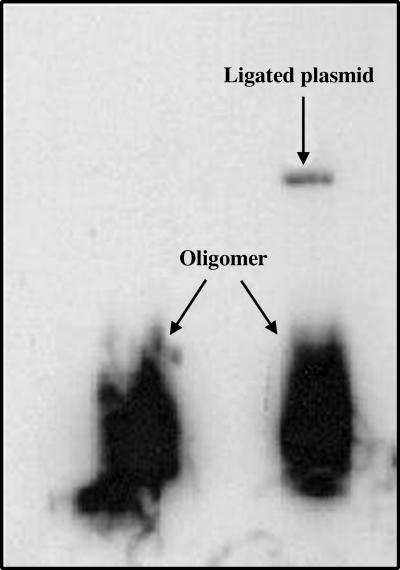
Preparation and Characterization of DNA Condensates. A single cut with HindIII restriction endonuclease linearizes the circular plasmid pUC19, leaving cohesive ends with 5′-AGCT-3′ overhangs. The radiolabeled AQ-containing DNA-1/DNA-2 duplex 42-mer (Tm = 49°C) has a phosphorylated 5′ overhang of four bases designed for cohesive ligation with the HindIII-cut linearized plasmid (HLP, Fig. 1). The reaction of HLP with the duplex oligomer in presence of T4 DNA ligase links the duplex oligomer once or twice (at opposite ends, Fig. 1) to the plasmid to form the ligated linearized plasmid (LLP; LLP1 and LLP2). Confirmation that ligation occurred successfully is based on the appearance of radioactivity from the oligomer in the linearized plasmid band on an agarose gel (Fig. 2). This synthesis provides a DNA sample of sufficient length to form condensates that contains an AQ group positioned for introduction of a radical cation, a series of guanines in the oligomer, and plasmid components that can act as traps for the radical cation, and a 32P label placed for analysis of reaction at these guanines by high-resolution PAGE.
Fig. 2.
Autoradiogram of a nondenaturing, low-melting agarose gel, showing LLP and the duplex oligomer DNA-1/DNA-2. The left lane is a control that contains only duplex oligomer, and the right lane is the product mixture from reaction of duplex oligomer and HLP with T4 DNA ligase. The dark lower band in both lanes corresponds to the duplex oligomer, and the upper band in the right lane is the product of ligation of the radiolabeled duplex oligomer to HLP. This upper band was excised from the gel, purified, and used in subsequent experiments.
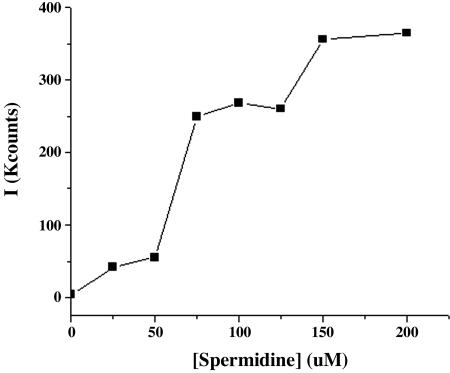
DNA condensates are formed from HLP and LLP by treatment, with spermidine as the condensing agent. In the absence of spermidine, experimental light-scattering autocorrelation curves for HLP and LLP in phosphate buffer solutions (12.5 μg/ml) are noisy and of low intensity, confirming the expected absence of significant numbers of light-scattering particles under these conditions (20). The addition of spermidine to these solutions results in the significant increase in light-scattering intensity, as shown in Fig. 3. The amount of scattering increases slowly when the spermidine concentration is raised in steps to 60 μM and then increases dramatically and reaches a plateau when the concentration of spermidine is 75-130 μM, which indicates the formation of stable, light-scattering particles over this concentration range. Further increases in spermidine concentration to >150 μM cause an additional increase in the light-scattering intensity, which signals the formation of aggregates. A summary of these experimental results is shown in Fig. 3. Analysis of these data indicates that the phosphate buffer solutions of HLP and LLP that contain spermidine in the concentration range of 50-130 μM form particles with an average radius of 100 ± 25 nm, which is consistent with the formation of a condensate composed of >90% of the DNA in solution. In support of this conclusion, we observe that these particles are disrupted when 1 M NaCl is added to the mixture.
Fig. 3.
Intensity of scattered light from a solution of LLP (12.5 μg/ml) in buffer solution at pH 7 as the concentration of spermidine is increased from 0 to 200 μM at intervals of 25 μM.
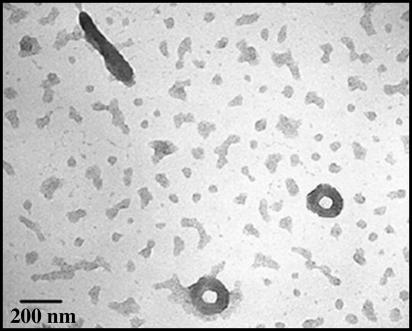
Confirmation of condensate formation under these conditions was obtained by TEM, in which toroids and rod-shaped objects are visible (Fig. 4) at ×100,000 magnification. The outer diameter of the toroid is ≈150 nm, which indicates that it is composed of 10-12 molecules of LLP. Particles that are formed in this way were used to study long-distance radical cation transfer in B-form DNA condensates.
Fig. 4.
TEM image of linear ligated plasmid condensed with 75 μM spermidine at room temperature taken at ×100,000 magnification, showing toroids and rod-shaped condensates. (Scale bar, 200 nm.)
Long-Distance Radical Cation Transfer in DNA Condensates. Intensive investigation has shown that irradiation of a DNA sample that contains a covalently linked AQ group at 350 nm, at which only the AQ absorbs, forms the excited singlet state of the AQ, which rapidly intersystem crosses to form its triplet state (21, 22). The triplet is a powerful one-electron oxidant that initiates electron transfer from an adjacent nucleobase to form the AQ radical anion and a base radical cation (23-25). The AQ radical anion is consumed by reaction with O2 to form superoxide, and the base radical cation can migrate through the DNA by hopping until it is trapped (usually at a guanine) by reaction with H2O and/or O2 (26, 27). This process forms lesions at guanines that yield strand breaks when the sample is treated with piperidine. These strand breaks are characterized by high-resolution PAGE in samples radiolabeled with 32P, and their relative yields are quantified by autoradiography. At low conversion (single-hit conditions, in which on average each DNA molecule reacts once or not at all), the distribution of strand breaks mirrors the distribution of the radical cation in the DNA oligomer. Numerous experiments of this sort that have been carried out on oligonucleotides in solution have revealed the mechanism of long-distance radical cation transfer and how the nucleobase sequence determines the efficiency of radical-cation migration. We applied these techniques to the study of long-distance radical cation transfer in the AQ-containing condensates formed from LLP.
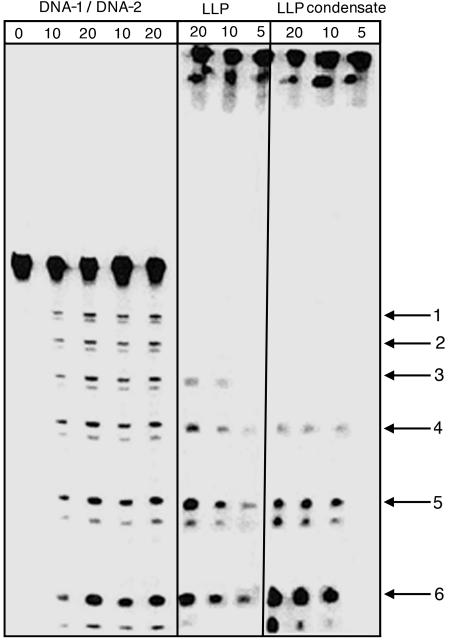
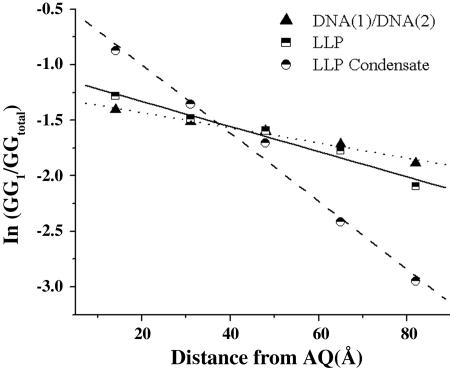
A sample containing condensates formed from AQ-linked LLP (5 μM, pH 7.0) by addition of spermidine was irradiated and subsequently treated with piperidine. Analysis by light scattering shows that the condensates are stable during and after the irradiation. The results of this experiment are shown in Fig. 5 as the autoradiogram of a high-resolution PAGE and in Fig. 6 as a semilog plot of strand cleavage efficiency with distance from the AQ. It is clear from these data that a radical cation injected into the DNA condensate leads to the formation of lesions at nearby guanines that are detected as strand cleavage. Moreover, the GG steps that are closer to the AQ react more frequently than those located farther away, which is characteristic of those cases where hopping of the radical cation from one GG step to the next (khop) is slower than its trapping at a GG step by reaction with H2O or O2 (ktrap) (22, 28-30). For the condensates, this distance dependence is rather steep, falling ≈10-fold over a distance of 70 Å.
Fig. 5.
Autoradiogram showing results of irradiation of DNA-1/DNA-2, LLP, and condensed plasmid. Numbered arrows on the right indicate the six GG steps of these DNA oligomers. The first five lanes correspond to the dark control, 10 and 20 min of irradiation of the oligomer duplex DNA-1/DNA-2, mixed with 75 μM spermidine and DNA-1/DNA-2, respectively. The next three lanes correspond to 20, 10, and 5 min of irradiation of the LLP, and the last three lanes to that of 20, 10, and 5 min of irradiation of the ligated condensed plasmid, respectively.
Fig. 6.
Semilog plot of the relative amount of strand cleavage detected at each of the five GG steps after piperidine treatment versus the distance from the AQ group based on the assumption of 3.4 Å per bp.
The effect of condensate formation on radical cation transfer in DNA is revealed by comparison with similar reactions carried out on the linear-linked plasmid and on the oligomeric duplex DNA-1/DNA-2 in the presence of spermidine. These results are also shown in Figs. 5 and 6, in which it is clear that both of these samples behave similarly but give results significantly different from that observed for the condensate. In particular, the semilog plots of reactivity with distance for LLP and DNA-1/DNA-2 fall off much more slowly than for the condensate. In a control experiment, we showed that piperidine-induced strand cleavage of DNA samples that had been condensed and then decondensed (by the addition of NaCl) is identical with DNA that had never been condensed. These findings indicate that long-distance radical cation transfer in the organized structure of the condensate is appreciably less efficient than it is for oligonucleotides in solution.
Discussion
The accepted mechanism for long-distance radical-cation migration in duplex DNA is phonon-assisted polaron hopping (31-33). In this model, the radical cation is stabilized by local distortions of the DNA structure and its environment, particularly tightly bound water molecules and associated counter ions. This self-trapped radical cation (the polaron) is usually located predominantly on guanines, but in appropriate sequences, it can also be delocalized onto adjacent adenines. Barriers to migration of the polaron from one site to another are overcome by thermal fluctuations (phonons) of the DNA and its environment that form charge-transfer effective states (34). For example, the duplex oligomer DNA-1/DNA-2 contains consecutive AGGA segments (identified as the polaronic site) separated by a single adenine (the barrier) (30). If the rate of hopping from one polaronic site to an adjacent site is much faster than the rate of its irreversible trapping in a chemical reaction, then the radical cation will be distributed approximately equally over all equivalent sites and there will be no apparent distance dependence for radical-cation migration. However, if the rate of irreversible trapping is much faster than hopping, then reaction will be detected only at those sites closest to the point of radical cation injection (the AQ, in this case).
The results from the investigation of DNA-1/DNA-2 reveal intermediate behavior. Strand cleavage is observed at all six of the reactive GG sites in this duplex oligomer when the irradiated sample is treated with piperidine, but the amount of cleavage at a particular site decreases as the distance from the AQ increases. Quantitatively, the slope of the semilog plot of cleavage efficiency against distance has a value of -0.007 Å-1 (Fig. 6). A kinetic analysis has been developed that permits characterization of the distance dependence of radical-cation migration with the dimensionless parameter kratio, which is equal to khop/ktrap (35, 36). The slope of the semilog plot for DNA-1/DNA-2 reveals that, in this case, kratio = 15, which indicates that hopping of the radical cation from site-to-site is 15 times faster than its irreversible consumption. The value of kratio for DNA-1/DNA-2 is unaffected by the addition of spermidine, and it does not change meaningfully when the oligomer is ligated to the linearized plasmid, but the value kratio decreases to 5 for the condensate, which shows that the efficiency of long-distance charge transfer decreases in this highly ordered structure.
Condensation of DNA may affect both the rate of hopping and the rate of trapping. One explanation for the observed decrease in charge transfer efficiency in the condensates is a reduction in khop. Ionization of the phosphate groups makes DNA a polyanion at normal physiological pH at which the negative charges are compensated by mobile counter ions (typically, sodium). Detailed quantum mechanical calculations have shown that motions of the sodium ions modulate the energy of a radical cation polaron in DNA (37). In some configurations, the sodium ions (and associated water molecules) form charge transfer effective orientations in which the energy of the polaron matches that of the barrier separating it from an adjacent site, which allows the polaron to hop (38, 39). In condensates, ≈90% of the mobile sodium ions are replaced by spermidine trications that will be far less mobile than a monovalent ion. In this circumstance, it will be more difficult to attain a charge transfer effective configuration, and hopping will be slower.
It is unlikely that condensate formation will increase the magnitude of ktrap, which would be required if condensation were the primary cause of the reduced charge efficiency. Spermidine wraps the DNA into a compact core having high peripheral positive charge that is resistant to hydrolytic cleavage by DNase I enzyme (40). This structure hinders access of oxygen and water molecules to the interior of the condensate, where reaction with the radical cation occurs. Consequently, the rate of trapping will be retarded, and if hindered access were the major consequence of condensate formation, then an increase in the efficiency of long-distance charge transfer is expected and not the decrease that is actually observed.
Conclusion
It is widely accepted that the one-electron oxidation of DNA is a major source of genetic damage leading to mutations. In particular, the conversion of a guanine to an 8-OxoG results in G-to-T transversion because the stable base pair that 8-OxoG forms with adenine causes failure of mismatch repair (41). Numerous experiments have shown that the nucleobase radical cations formed by electron loss from DNA oligomers in solution can migrate long distances before being trapped at a guanine. We have shown that this process also occurs in DNA condensates, which mimic genomic DNA in significant ways. However, the efficiency of radical cation transport in condensates is considerably less that that for an oligomer having an identical nucleobase sequence. The formation of DNA condensates with the multivalent cation spermidine inhibits the migration of radical cations and, consequently, DNA damage is more localized at the site of initial electron loss. This inhibition is attributed to the decreased probability for forming the charge-transfer-effective states that are required for radical-cation hoping when sodium counter ions of high mobility are replaced with the low-mobility polycationic agents used to form the condensate.
Acknowledgments
We thank Prof. Nicholas Hud and his students Igor Vilfran and Tumpa Sarkar (School of Chemistry and Biochemistry, Georgia Institute of Technology) for advice and assistance with the dynamic light-scattering and TEM experiments; Dr. Richard Redic for assistance with several experiments; and Ms. Janet Ziebell for assistance with the creation of the graphics. This work was supported by a grant from the National Science Foundation and by the Vassar Woolley Foundation.
Author contributions: G.B.S. designed research; and P.D. performed research.
Abbreviations: AQ, anthraquinone; TEM, transmission electron microscopy; HLP, HindIII-cut linearized plasmid; LLP, ligated linearized plasmid.
References
- 1.Gasper, S. M. & Schuster, G. B. (1997) J. Am. Chem. Soc. 119, 12762-12771. [Google Scholar]
- 2.Nunez, M., Hall, D. B. & Barton, J. K. (1999) Chem. Biol. 6, 85-97. [DOI] [PubMed] [Google Scholar]
- 3.Schuster, G. B. & Landman, U. (2004) Topics. Curr. Chem. 236, 139-162. [Google Scholar]
- 4.Steenken, S. & Jovanovic, S. V. (1997) J. Am. Chem. Soc. 119, 617-618. [Google Scholar]
- 5.Demple, B. & Harrison, L. (1994) Annu. Rev. Biochem. 63, 915-948. [DOI] [PubMed] [Google Scholar]
- 6.Loft, S. & Poulsen, H. E. (1996) J. Mol. Med. 74, 297-312. [DOI] [PubMed] [Google Scholar]
- 7.Giese, B. (2002) Annu. Rev. Biochem. 71, 51-70. [DOI] [PubMed] [Google Scholar]
- 8.Sugiyama, H. & Saito, I. (1996) J. Am. Chem. Soc. 118, 7063-7068. [Google Scholar]
- 9.Nunez, M. E., Noyes, K. T. & Barton, J. K. (2002) Chem. Biol. 9, 403-415. [DOI] [PubMed] [Google Scholar]
- 10.Bloomfield, V. A. (1996) Curr. Opinion Struct. Biol. 6, 334-341. [DOI] [PubMed] [Google Scholar]
- 11.Wilson, R. W. & Bloomfield, V. A. (1979) Biochemistry 18, 2192-2196. [DOI] [PubMed] [Google Scholar]
- 12.Eickbush, T. H. & Moudrianakis, E. N. (1976) Cell 13, 295-306. [DOI] [PubMed] [Google Scholar]
- 13.Chattoraj, D. K., Gosule, L. C. & Schellman, J. A. (1978) J. Mol. Biol. 121, 327-337. [DOI] [PubMed] [Google Scholar]
- 14.Koltover, I., Wager, K. & Safinya, C. R. (2000) Proc. Natl. Acad. Sci. USA 97, 14046-14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosule, L. C. & Schellman, J. A. (1976) Nature 259, 333-335. [DOI] [PubMed] [Google Scholar]
- 16.Bloomfield, V. A. (1997) Biopolymers 44, 269-282. [DOI] [PubMed] [Google Scholar]
- 17.Conwell, C. C., Vilfan, I. D. & Hud, N. V. (2003) Proc. Natl. Acad. Sci. USA 100, 9296-9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanisch, P. C., Vieira, J. & Messing, J. (1985) Gene 33, 103-119. [DOI] [PubMed] [Google Scholar]
- 19.Sugino, A., Goodman, H. M., Heyneker, H. L., Shine, J., Boyer, H. W. & Cozzarelli, N. R. (1977) J. Biol. Chem. 252, 3987-3994. [PubMed] [Google Scholar]
- 20.Allison, A. S., Herr, C. J. & Schurr, M. J. (1981) Biopolymers 20, 469-488. [DOI] [PubMed] [Google Scholar]
- 21.Navas, A. D. (1990) J. Photochem. Photobiol. A 53, 141-167. [Google Scholar]
- 22.Giese, B. & Biland, A. (2002) Chem. Commun. 667-672. [DOI] [PubMed]
- 23.Ly, D., Kan, Y., Armitage, B. & Schuster, G. B. (1996) J. Am. Chem. Soc. 118, 8747-8748. [Google Scholar]
- 24.Armitage, B., Yu, C. & Devadoss, C. (1994) J. Am. Chem. Soc. 116, 9847-9859. [Google Scholar]
- 25.Breslin, D. T. & Schuster, G. B. (1996) J. Am. Chem. Soc. 118, 2311-2319. [Google Scholar]
- 26.Kasai, H., Yamaizumi, Z., Berger, M. & Cadet, J. (1992) J. Am. Chem. Soc. 114, 9692-9694. [Google Scholar]
- 27.Schuster, G. B. (2000) Acc. Chem. Res. 33, 253-260. [DOI] [PubMed] [Google Scholar]
- 28.Giese, B., Amaudrut, J., Kohler, A., Sporman, M. & Wessely, S. (2001) Nature 412, 318-320. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, F. D., Liu, J., Zuo, X., Hayes, R. T. & Wasielewski, M. R. (2003) J. Am. Chem. Soc. 125, 4850-4861. [DOI] [PubMed] [Google Scholar]
- 30.Liu, C.-S., Hernandez, R. & Schuster, G. B. (2004) J. Am. Chem. Soc. 126, 2877-2884. [DOI] [PubMed] [Google Scholar]
- 31.Henderson, P. T., Jones, D., Hampikian, G., Kan, Y. & Schuster, G. B. (1999) Proc. Natl. Acad. Sci. USA 96, 8353-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giese, B. (2000) Acc. Chem. Res. 33, 631-636. [DOI] [PubMed] [Google Scholar]
- 33.O'Neill, M. A. & Barton, J. K. (2004) J. Am. Chem. Soc. 126, 11471-11483. [DOI] [PubMed] [Google Scholar]
- 34.Barnett, R. N., Cleveland, C. L., Joy, A., Landman, U. & Schuster, G. B. (2001) Science 294, 567-571. [DOI] [PubMed] [Google Scholar]
- 35.Kino, K., Saito, I. & Sugiyama, H. (1998) J. Am. Chem. Soc. 120, 7373-7374. [Google Scholar]
- 36.Giese, B. & Spichty, M. (2000) Angew. Chem. Int. Ed. Engl. 39, 3490-3491. [DOI] [PubMed] [Google Scholar]
- 37.Barnett, N. R., Cleveland, L. C., Landman, U., Boone, E., Kanvah, S. & Schuster, G. B. (2003) J. Phys. Chem. A 107, 3525-3537. [Google Scholar]
- 38.Liu, C.-S. & Schuster, G. B. (2003) J. Am. Chem. Soc. 125, 6098-6102. [DOI] [PubMed] [Google Scholar]
- 39.Leone, A. M., Tibodeau, J. D., Bull, S. H., W., F. S., Thorp, H. H. & Murray, R. W. (2003) J. Am. Chem. Soc. 125, 6784-6790. [DOI] [PubMed] [Google Scholar]
- 40.Adami, R. C., Collard, W. T., Gupta, S. A., Kwok, K. Y., Bonadio, J. & Rice, K. G. (2000) J. Pharm. Sci. 87, 678-683. [DOI] [PubMed] [Google Scholar]
- 41.Hsu, G. W., Ober, M., Carell, T. & Beese, L. (2004) Nature 431, 217-221. [DOI] [PubMed] [Google Scholar]