Abstract
During the evolution of metazoans and the rise of systemic hormonal regulation, the insulin-controlled class 1 phosphatidylinositol 3OH-kinase (PI3K) pathway was merged with the primordial amino acid-driven mammalian target of rapamycin (mTOR) pathway to control the growth and development of the organism. Insulin regulates mTOR function through a recently described canonical signaling pathway, which is initiated by the activation of class 1 PI3K. However, how the amino acid input is integrated with that of the insulin signaling pathway is unclear. Here we used a number of molecular, biochemical, and pharmacological approaches to address this issue. Unexpectedly, we found that a major pathway by which amino acids control mTOR signaling is distinct from that of insulin and that, instead of signaling through components of the insulin/class 1 PI3K pathway, amino acids mediate mTOR activation by signaling through class 3 PI3K, hVps34.
Keywords: insulin, nutrients, S6 kinase 1, endosomes, PI3P
Unicellular eukaryotes use the mammalian target of rapamycin (mTOR)/raptor/G-protein-β-subunit-like protein (GβL) pathway in a cell-autonomous manner (1); however, with the rise of metazoans and humoral systems, the insulin-controlled PI3K signaling pathway was merged with the mTOR signaling pathway to maintain cellular homeostasis. The clinical importance of mTOR has been underscored by the use of rapamycin and its derivatives in a number of pathological settings, including organ transplantation, restenosis, rheumatoid arthritis, and more recently the treatment of solid tumors (2, 3). Moreover, recent studies from this laboratory have led to a model whereby constitutive activation of the mTOR pathway acts as a key effector in the development of obesity and insulin resistance (4, 5). Earlier, mTOR was described to exist in an evolutionarily conserved, rapamycin sensitive, nutrient effector complex with two additional proteins, raptor and GβL (2). However, recent studies have shown that mTOR can also exist in a distinct rapamycin-resistant signaling complex with GβL and a protein termed AVO3 or rictor that acts to control the actin cytoskeleton (6, 7) and the activation of PKB (8). Over the last few years, studies have shown that insulin modulates mTOR/raptor/GβL signaling complex (mTOR/raptor/GβL) activity through the phosphorylation of IRS1 and the stimulation of PI3K, which in turn leads to the activation of PKB, followed by the phosphorylation and inactivation of the tumor suppressor complex made up of tuberous sclerosis complex proteins 1 and 2 (TSC1/2) (9, 10). The inhibitory effects of TSC1/2 on mTOR/raptor/GβL signaling are elicited through the GTPase stimulating activity of TSC2, which drives the small GTPase Rheb into the inactive GDP-bound state (11). It is thought that GTP-bound Rheb either acts directly on mTOR/raptor/GβL (12) or influences the ability of this complex to signal to downstream substrates such as S6 kinase 1 (S6K1) and the initiation factor 4E binding protein (4E-BP1) (10).
Although the insulin-stimulated pathway regulating mTOR/raptor/GβL signaling is largely elucidated, the mechanisms by which nutrients, such as amino acids, control this process are less defined. Initial studies showed that S6K1 activity, as a reporter for mTOR/raptor/GβL signaling, was largely protected from amino acid withdrawal in Drosophila cells where dTSC1 or dTSC2 levels were reduced or in TSC1- or TSC2-deficient rodent cells (13). This finding has led to the assumption that amino acids, especially branched chain amino acids, act through TSC1/2 to regulate mTOR activity (3, 14). However, the perception of the amino acid entry point has been put into question by a number of observations. First, tissue-specific overexpression of Rheb was initially found to drive Drosophila larval cell growth in a cell-autonomous manner in the absence of amino acids, favoring a model whereby the TSC1/2-Rheb signaling axis acts to drive amino acid uptake and mTOR signaling (15). In contrast, Zhang et al. (16) demonstrated that amino acid withdrawal had no effect on the GTP-bound levels of ectopically overexpressed Rheb, leading them to conclude that the amino acid input to mTOR/raptor/GβL resides on a parallel pathway to that of TSC1/2 and Rheb. Interestingly, during the completion of these studies, Smith et al. (17) showed that in TSC2-deficient cells, similar to those used by Gao et al. (13), amino acids regulate mTOR/raptor/GβL signaling. Moreover, unlike Zhang et al. (16), they found that amino acid withdrawal affected GTP-bound levels of ectopically overexpressed Rheb (17).
Here, we investigated which of these models is valid with respect to the control of mTOR/raptor/GβL signaling by amino acids. Next, we explored the potential role of Rheb in mediating the amino acid response independent of either TSC1 or TSC2. These studies led to the discovery that a major amino acid input to mTOR/raptor/GβL signaling is through a pathway parallel to that of the insulin-mediated TSC1/2-Rheb signaling axis mediated by class 3 PI3K, hVps34.
Materials and Methods
Reagents. The reagents used were insulin (Sigma-Aldrich); wortmannin and rapamycin (Calbiochem); anti-Rheb (C-19) antibody (Santa Cruz Biotechnology); anti-phospho-T389-S6K, -S473-PKB, -S939-TSC2, -S240/244-S6, and -T70-4E-BP1, PI3K, and PKB antibodies (Cell Signaling Technology, Beverly, MA); anti-tubulin (NeoMarkers, Fremont, CA); anti-TSC2 and TSC1 antibodies from M. Nellist (Rotterdam); anti-hVs34 antibody (Zymed); M1 anti-S6K1 antibody (18); horseradish peroxidase-coupled secondary antibodies (DAKO or Amersham Pharmacia Biosciences); 9E10 antibody (10); and enhanced chemoluminescence (ECL) reagents (Amersham Pharmacia Biosciences).
Cell Cultures and Transfections. All cell lines except for CACL-I-III were maintained as described in ref. 19. CACL-I-III cells were maintained in RPMI medium 1640 supplemented with 10% FCS. Plasmid transfections were carried out with FuGENE 6 (Roche). For amino acid measurements, 3 μg of plasmid per 10-cm dish per experiment were used to transfect with Lipofectamine 2000. Transfection of short interfering RNAs (siRNAs) was carried out as described in ref. 19. For amino acid deprivation, cells were first deprived of serum overnight and then for 2 additional hours without amino acids. Cells were then stimulated with either serum or amino acids, respectively.
Constructs, siRNAs, and Western Blot Analyses. The gst-Vps34 expression plasmid was from M. Wymann (Basel). All other constructs were as described in refs. 10 and 20. siRNAs were designed for targeting the coding sequence of human Rheb TCAGTGTAGTTTGTTGTTTAA, TSC1 CCGGACAGTGTTGGACAGCTA, TSC2 AAGGATTACCCTTCCAACGAA, human PI3Kα CAAGATATGCTAACACTTCAA, VPS34 (920) ATGGCTGAA-ACTACCAGTAAA, VPS34 (923) ACGGTGATGAATCATCTCCAA, or VPS15 CAAGCAATGCTGGGACTTTAA.
Amino Acid Quantification. Amino acid concentrations were determined as described in ref. 20. At least two independent measurements were carried out for each sample, and standard deviations are shown.
Radioactive GTP-Loading Assay for Rheb. Endogenous Rheb was pulled down with the C19 antibody (Santa Cruz Biotechnology), and the ratio of GTP/GDP-bound Rheb was measured as described in ref. 19.
Phosphatidylinositol 3-Phosphate (PI3P) Quantification. Cells were seeded on sterile coverslips and grown to 80% confluency. After treating cells as indicated in the text, we washed the cells twice in PBS and fixed them with 4% paraformaldehyde for 15 min. Permeabilization by -20°C methanol for 10 min was followed by quenching with 0.1% sodium borohydride and four washes with PBS. The preparations were blocked for 1 h in 1% PBS/10% BSA at room temperature. Monoclonal anti-PI3P (Echelon Biosciences, Salt Lake City) was used to label the fixed cells overnight (5 μg/ml in PBS/1% BSA) at 4°C. Cells were washed four times in PBS and treated with secondary antibody (20 μg/ml FITC-conjugated anti-mouse (Vector Laboratories) for 1 h at room temperature. Cells were washed again four times in PBS, and the coverslips were mounted on slides. Fluorescence imaging was done on a Zeiss Axioplan II confocal microscope and quantified on the accompanying software (axio vision 4.2) based on densitometry and area of punctuate signals.
hVps34 PI3K Assays. Cells were seeded in 10-cm dishes and grown to 80% confluency. Cells were treated as described in the text, then washed in cold PBS and lysed in 1% Nonidet P-40 (150 mM NaCl/50 mM Tris/10% glycerol/10 mM NaF/1 mM sodium pyrophosphate/protease and phosphatase inhibitors). Lysates were immunoprecipitated overnight at 4°C with 1 μg of C terminus anti-VPS34 antibody and then immobilized for 2 h on protein-A-conjugated Sepharose. Immunocomplexes were washed four times in 1% Nonidet P-40/1× PBS; four times in 100 mM Tris/500 mM LiCl; and three times in 10 mM Tris/100 mM NaCl/1 mM EDTA (TNE). Each immunocomplex was resuspended in 60 μl of TNE and incubated at 30°C for 30 min with 3 μl of 1 mM blocking peptide. After elution of immunocomplexes, 10 μl of 100 mM MnCl2 and 2 μg of sonicated phosphatidylinositol in 10 μl of 10 mM Tris/1 mM EGTA were added to each sample. Reactions were started with the addition of 5 μl of ATP mix (1 μl of 10 mM unlabeled ATP/1 μl of [γ-32P]ATP/3 μl of H2O) and allowed to proceed for 10 min. Reactions were terminated with the addition of 20 μlof8MHCl and then extracted with 160 μl of 1:1 chloroform/methanol. Dried silica, aluminum TLC plates were spotted with the organic phase of each extraction and run in a TLC chamber with 100 ml of mobile phase (60 ml of chloroform/47 ml of methanol/11.3 ml of H2O/2 ml of ammonium hydroxide). Plates were dried and exposed to autoradiography to visualize PI3P production.
Results
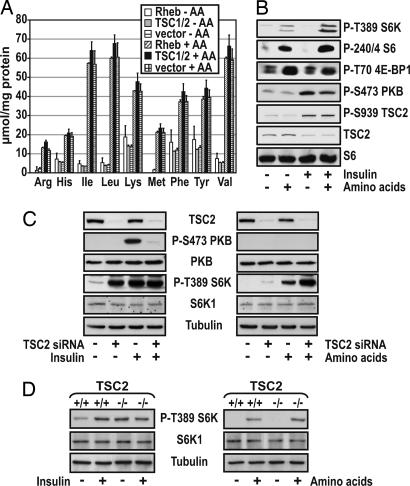
To determine whether TSC1/2 or Rheb regulates amino acid uptake, each was ectopically expressed in 293 cells, and the effect on steady-state intracellular amino acid pools was determined. Overexpression of TSC1/2 or Rheb had no effect on the steady-state levels of branched chain amino acids or arginine (Fig. 1A). Similarly, removal of amino acids lowered total amino acid content, but neither TSC1/2 nor Rheb overexpression increased or protected against the decline (Fig. 1 A). These findings do not support a model where the TSC1/2-Rheb axis is acting to increase intracellular steady-state amino acid levels to drive mTOR/raptor/GβL signaling, prompting us to examine the role of amino acids in regulating mTOR signaling through modulating TSC1/2 function. To study this question, the effect of amino acids and insulin on TSC2 S939 phosphorylation and TSC2 steady-state levels was first compared with that of S6K1 T389 phosphorylation in response to amino acid or insulin stimulation. Under these conditions, the readdition of amino acids alone had no effect on TSC2 S939 phosphorylation or on steady-state levels of TSC2, whereas they stimulated S6K1 T389 phosphorylation (Fig. 1B). Consistent with this finding, amino acids had no effect on PKB activation, whereas they increased S6 S240/244 phosphorylation (Fig. 1B). In contrast, insulin in the absence of amino acids led to a sharp increase in PKB S473 and TSC2 S939 phosphorylation, and a decrease in TSC2 steadystate levels, with no measurable impact on S6K1 T389 or S6 240/244 phosphorylation (Fig. 1B). Furthermore, the effect of insulin on PKB S473 was suppressed by amino acids (Fig. 1B), consistent with the additional rise in S6K1 T389 and S6 S240/244 phosphorylation (Fig. 1B) and the reported amino acid-induced negative feedback loop from mTOR/S6K1 to IRS1 (21). These results suggested that TSC1/2 may not be involved in regulating the amino acid input to mTOR/raptor/GβL. To test this possibility, we lowered TSC2 levels with siRNAs. Similar to what we have shown for TSC2-deficient cells (10), siRNA depletion of TSC2 led to constitutive S6K1 T389 phosphorylation in the absence of serum but did not effect PKB S473 phosphorylation (Fig. 1C). In contrast, insulin addition to untreated cells led to both increased S6K1 T389 and PKB S473 phosphorylation (Fig. 1C). Moreover, insulin had little effect on S6K1 T389 phosphorylation in TSC2 siRNA-treated cells (Fig. 1C), whereas PKB S473 phosphorylation was strongly suppressed (Fig. 1C), reflecting the negative feedback to IRS1 (4, 22, 23). Removal of amino acids from serum-deprived cells for an additional 2 h abolished S6K1 T389 phosphorylation in untreated cells (Fig. 1C). More striking, S6K1 T389 phosphorylation was not protected from amino acid withdrawal in TSC2 siRNA-treated cells (Fig. 1C), in agreement with the recent findings of Smith et al. (17). Under these conditions, there is no detectable effect on basal PKB S473 phosphorylation (Fig. 1C). Analogous effects were observed when we analyzed siRNA knockdown of TSC1 protein levels (data not shown). Moreover, we obtained similar results in either TSC2-/- MEFs (Fig. 1D) and in CACL-I-III cells (13) derived from a TSC1-/- mouse renal cell carcinoma (Fig. 6 A and B, which is published as supporting information of the PNAS web site). These results show that the effects of amino acids on S6K1 activation are not mediated directly through TSC1/2.
Fig. 1.
Amino acids and TSC1/TSC2 signaling. (A) HEK293 cells were transfected with either TSC1/TSC2 or Rheb 48 h before measuring the amino acid concentrations (as described in Materials and Methods). (B) HEK293 cells were deprived of serum overnight followed by a 2-h amino acid deprivation. Cells were stimulated with 200 nM insulin in the absence or presence of amino acids for 30 min. (C) HeLa cells were transfected with 16 nM TSC2 siRNA. Two days posttransfaction, cells were deprived of serum overnight followed by stimulation with 200 nM insulin for 30 min or, after serum withdrawal, cells were deprived of amino acids for 2 h and then stimulated with 2× amino acids for 30 min. (D) TSC2+/+ and TSC2-/- MEFs were treated as in C.In B-D, cell lysates were analyzed by Western blot analysis with the indicated antibodies.
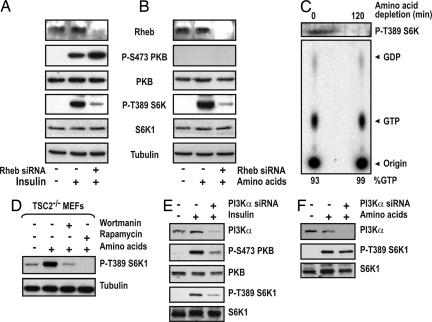
The target of TSC1/2 tumor suppressor complex is the small GTPase Rheb, and we previously showed that overexpression of Rheb protects S6K1 T389 from dephosphorylation by amino acid withdrawal (19). This result argues that the amino acid input to mTOR/raptor/GβL requires Rheb. To test this possibility, we knocked down Rheb with siRNAs before treatment with either insulin or amino acids (Fig. 2A). As previously shown (19), Rheb siRNA blocks insulin induction of S6K1 T389 phosphorylation and in parallel potentiates PKB S473 phosphorylation (Fig. 2 A), consistent with the results observed with TSC1 and TSC2 siRNAs (data not shown and Fig. 1). As with insulin, Rheb siRNA also blocks the amino acid-induced increase in S6K1 T389 phosphorylation, with no effect on basal PKB S473 phosphorylation (Fig. 2B). Amino acid deprivation leads to S6K1 inactivation in a TSC1/2-independent manner (Fig. 1), and Rheb is required for the amino acid input to S6K1 (Fig. 2B), suggesting that amino acids may regulate Rheb GTP levels through a distinct GAP or GTPase exchange factor (GEF), whose function is bypassed through overexpression of Rheb (19). To test this model, we measured endogenous Rheb GTP-loading in either TSC2-/- MEFs or in TSC1-/- CACL-I-III cells. Surprisingly, the results show that S6K1 T389 phosphorylation is rapidly lost in both cell types, with no effect on high Rheb-GTP levels (Figs. 2C and 6C, respectively). Thus, elevated levels of endogenous Rheb-GTP are necessary, but not sufficient, for amino acid-induced mTOR/raptor/GβL signaling.
Fig. 2.
Rheb GTP and a wortmannin-sensitive pathway. (A and B) HeLa cells were transfected with 8 nM Rheb siRNA, and, 2 days posttransfection, cells were deprived of serum overnight followed by stimulation with 200 nM insulin for 30 min or, after serum withdrawal, cells were deprived of amino acids for 2 h and then stimulated with 2× amino acids for 30 min. (C Upper) Serum-deprived TSC2-/- MEFs were depleted of amino acids for 2 h and subjected to Western blot analysis. (C Lower) Parallel dishes were [32P]orthophosphate-labeled, and the ratio of endogenous GTP to GDP-bound Rheb was determined as described in ref. 19. (D) TSC2-/- MEFs were serum-starved overnight and deprived of amino acids for 2 h before being stimulated with 2× amino acids for 30 min in the presence of 100 nM wortmannin or 20 nM rapamycin for 15 min before amino acid stimulation. (E and F) HeLa cells were transfected with 16 nM PI3Kα siRNA, and 2 days posttransfection, cells were deprived of serum overnight followed by stimulation with 200 nM insulin for 30 min or, after serum withdrawal, were deprived of amino acids for 2 h and then stimulated with 2× amino acids for 30 min. (A-E) Cell lysates were analyzed by Western blot analysis with the indicated antibodies.
The above findings imply that the major route by which amino acids signal to the mTOR/raptor/GβL complex may reside on a parallel pathway to that of insulin-mediated class 1 PI3K. However, wortmannin, a potent class 1 PI3K inhibitor, has been reported to block amino acid-induced mTOR/raptor/GβL signaling, as judged by decreased S6K1 activity (24, 25). To study this issue, we measured the effect of wortmannin and rapamycin on amino acid-induced S6K1 T389 phosphorylation in TSC2-/- MEFs. Consistent with the findings of others (24, 25), wortmannin abolished amino acid-induced S6K1 T389 phosphorylation to almost the same extent as rapamycin (Fig. 2D). One possibility is that amino acids activate a unique pool of class 1 PI3K that does not signal to PKB. Therefore, we examined insulin and amino acid-induced S6K1 T389 phosphorylation in cells treated with siRNAs against class 1 PI3Kα. The results show that such a siRNA effectively lowers class 1 PI3Kα levels and inhibits insulin-induced PKB S473 and S6K1 T389 phosphorylation (Fig. 2E). However, such treatment has no effect on amino acid-induced S6K1 T389 phosphorylation (Fig. 2F), arguing that the wortmannin-sensitive amino acid input to mTOR/raptor/GβLis not via class 1 PI3Kα.
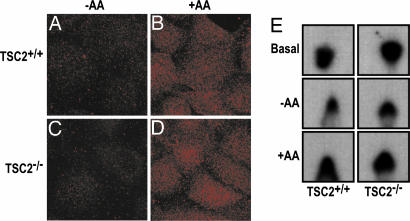
Although the above studies do not exclude other class 1 PI3Ks, they suggested that the wortmannin-sensitive amino acid input to mTOR/raptor/GβL was through an element not yet implicated in mTOR/raptor/GβL signaling. Among the other such targets, it is known that the yeast orthologue of human class 3 PI3K (hVps34), which is represented by a single gene, is as sensitive to wortmannin as the class 1 PI3Ks (26). Moreover, recent studies in yeast have shown that GβL, or LST8, localizes to endosomes, where Vps34 regulates endocytic membrane trafficking (27). The product of hVps34, PI3P, is involved in recruiting proteins to endosomal membranes containing FYVE or PX domains (28), with these PI3P-rich microdomains serving as platforms to build signaling complexes (29-31). If hVps34 is responsible for mediating the amino acid response, then a similar effect should be observed for PI3P production and hVps34 activity in both TSC2+/+ or TSC2-/- MEFs as observed for regulation of S6K1. Using confocal microscopy and a PI3P-specific antibody, we found in both cell types that in the absence of amino acids, little PI3P is visualized, whereas amino acid addition led to a sharp increase in PI3P staining as bright punctate spots (Fig. 3 A-D), similar to those reported for endosomal markers. Consistent with these data, wortmannin largely abolished all PI3P staining (data not shown). More importantly, the changes in PI3P staining are paralleled by changes in hVps34 activity, such that in both cell types immunoprecipitated hVps34 activity is low in the absence of amino acids and increases on their readdition (Fig. 3E). These findings are consistent with hVps34 being the wortmannin-sensitive target for amino acid input to mTOR/raptor/GβL.
Fig. 3.
Amino acid-induced PI3P production and hVps34 activation. (A-D) TSC2+/+ and TSC2-/- MEFs were deprived of serum overnight and then deprived of amino acids for 2 h before being stimulated with 2× amino acids for 30 min. Staining with PI3P antibody was carried out as described in Materials and Methods. (E) In vitro PI3P production in TSC2+/+ and TSC2-/- MEFs was assayed under conditions of serum deprivation, amino acid deprivation, and amino acid stimulation.
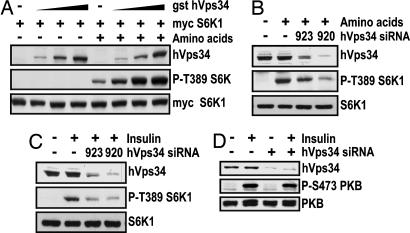
To determine whether hVps34 is critical in regulating the amino acid input to mTOR/raptor/GβL, we tested the ability of a GST-epitope-tagged hVps34 (GST-hVps34) cDNA to induce myc-epitope-tagged S6K1 (myc-S6K1) T389 phosphorylation. In the absence of amino acids, increased expression of GST-hVps34 had no effect on myc-S6K1T389 phosphorylation, whereas in the presence of amino acids, GST-hVps34 led to a marked dose-dependent increase in this response (Fig. 4A), placing hVps34 downstream of amino acids. Consistent with this hypothesis, knockdown of hVps34 protein to different levels by two distinct hVps34 siRNAs showed a coordinate reduction in amino acid-induced S6K1 T389 phosphorylation (Fig. 4B). Moreover, knockdown of hVps34 protein levels in the presence of amino acids blocks insulin-induced S6K1 T389 phosphorylation (Fig. 4C), in agreement with amino acids being required for insulin-induced activation of mTOR/raptor/GβL (Fig. 1B). In contrast, knockdown of hVps34 with the most potent hVps34 siRNA has no effect on insulin-induced PKB S473 phosphorylation (Fig. 4D), consistent with the results obtained in Fig. 1 in the absence of amino acids. These data argue that the amino acid input to mTOR/raptor/GβL is mediated by hVps34.
Fig. 4.
hVps34 is required for amino acid-induced S6K1 activation. (A) HEK293 cells were transfected with myc-S6K1 and increasing amounts of gst-hVps34 expression vectors. Two days posttransfection, cells were serum deprived overnight and further deprived of amino acids for 2 h and then stimulated with 2× amino acids for 30 min. (B and C) HeLa cells were transfected with 16 nM of either hVps34 siRNA 920 or 923, and 2 days posttransfection, cells were deprived of serum overnight followed by stimulation with 200 nM insulin for 30 min or, after serum withdrawal, cells were deprived of amino acids for 2 h and then stimulated with 2× amino acids for 30 min. (D) As in C, except cells were only transfected with 16 nM hVps34 920 siRNA. (A-D) Cell lysates were analyzed by Western blot analysis with the indicated antibodies.
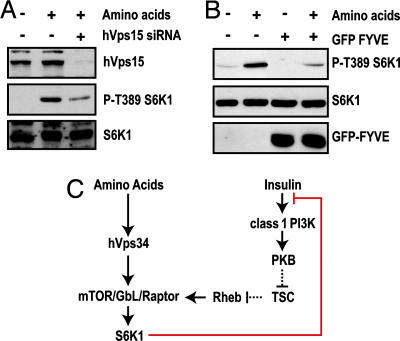
In yeast, Vps34 is associated with Vps15, a protein kinase, which recruits Vps34 to endosomal membranes and whose kinase activity is required for Vps34 activity (32). The mammalian homologue of Vps15, termed p150 or hVps15, is also associated with hVps34 and is known to stimulate its lipid kinase activity in vitro (33). Consistent with this observation, siRNA knockdown of hVps15 protein levels also inhibits amino acid-induced S6K1 T389 phosphorylation (Fig. 5A). Taken together, these findings argue that the increase in hVps34 activity and the production of PI3P is responsible for mediating this response. To test this possibility, a vector containing two Fab1/YOTB/ZK632.12/Vac1/EEA1 (FYVE) domain repeats was cotransfected together with the myc-S6K1 reporter. The products of such vectors are known to function as dominant interfering proteins, by competing for intracellular PI3P-docking sites. The results show that the FYVE domain construct inhibits both basal and amino acid-induced S6K1 T389 phosphorylation (Fig. 5B). The simplest explanation for these observations is that wortmannin blocks amino acid-induced mTOR/raptor/GβL signaling by inhibiting hVps34 production of PI3P. Taken together, these findings argue that, whereas the wortmannin-sensitive insulin input to mTOR/raptor/GβL is mediated by class 1 PI3Ks, the amino acid input is controlled by hVps34 (see model in Fig. 5C).
Fig. 5.
PI3P is required for mTOR signaling, and hVps34 associates with mTOR. (A) HeLa cells were transfected with 16 nM hVps15 siRNA and analyzed as in Fig. 4C.(B) HEK293 cells were transfected with myc-S6K1 and a GFP-FYVE expression vector and analyzed as in Fig. 4A.(C) Model depicting two signaling pathways converging on the mTOR/raptor/GβL signaling complex.
Discussion
In mammals, 10 of the 20 amino acids are essential, i.e., they must be acquired from the external milieu, whereas the remaining 10 are produced by the organism. The link between amino acids and mTOR signaling initially arose from studies in autophagy, where it was shown that this process was blocked by amino acids, but in parallel, amino acids induced S6K1 activation (34). Subsequently, it was demonstrated that the amino acid-dependent S6K1 activation is mTOR-dependent (24), with some studies pointing to the importance of branched-chain amino acids, especially leucine. Nevertheless, it is clear that nonessential amino acids can also affect mTOR signaling (24, 35), consistent with both essential and nonessential amino acid alcohols, which act as competitive inhibitors of tRNA aminoacylation, inhibiting mTOR signaling (25). These findings led to the suggestion that deacylated tRNA may negatively regulate the pathway, similar to GCN2 kinase activation and initiation factor eIF2α phosphorylation in Saccharomyces cerevisiae by deacylated histidyl-tRNA (36). However, amino acid withdrawal leads to S6K1 and 4E-BP1 dephosphorylation with no immediate effect on aminoacylated tRNA levels (20). Instead, amino acid deprivation resulted in a rapid decrease in the levels of essential amino acids, particularly the branched-chain amino acids (20), indicating that intracellular pools of free amino acids are critical. Branchedchain amino acid levels fall the most rapidly, so that their depletion may be the first to affect mTOR/raptor/GβL signaling by a common upstream element, such as hVps34.
In initial studies in which either TSC1 or TSC2 was absent or reduced, S6K1 activity was largely protected from amino acid withdrawal (13). These findings led to two models, one where TSC1/2 functions as an obligatory component between amino acids and mTOR/raptor/GβL in a linear pathway, or where TSC1/2 act on a parallel pathway to mediate the amino acid input to mTOR (13). Subsequent studies showed that ectopically expressed Rheb-GTP loading is unchanged after amino acid withdrawal, favoring the second model (16). However, ectopic expression of Rheb protects dS6K1 from inactivation by amino acid withdrawal (15), similar to its effects in the absence of growth factors (19). Thus, overexpression of Rheb combined with its low intrinsic GTPase activity may simply override the effects of amino acid withdrawal, putting the latter interpretation into question. In contrast, based on the observation that ectopic expression of Rheb drove cell autonomous growth in the absence of nutrients, Saucedo et al. (15) favored a model in which TSC1/2-Rheb acted to stimulate amino acid uptake, driving mTOR signaling. Our data suggested a parallel pathway mediates the amino acid input to mTOR/raptor/GβL, which is distinct from that controlled by insulin through TSC1/2 and Rheb (Figs. 1 and 2). It should be noted that during the completion of these studies others also found that amino acids regulate mTOR/raptor/GβL signaling independent of TSC1/2 (17). Although we could not exclude a role for amino acids in regulating Rheb through a distinct mechanism, the search for the amino acid input to mTOR/raptor/GβL led to a parallel wortmannin-sensitive pathway to that of TSC1/2 and Rheb, mediated by hVps34 (Figs. 4 and 5). Consistent with this finding, withdrawal of amino acids in TSC2-/- MEFs has no effect on endogenous Rheb GTP levels but leads to a drop in S6K1 T389 phosphorylation, PI3P levels, and hVps34 kinase activity (Figs. 2, 3, 4, 5). Consistent with these findings, others have demonstrated that removal of amino acids causes a decrease in hVps34 activity, which parallels S6K1 T389 dephosphorylation (37). In mammalian cells, PI3P is primarily found in early endosomes and the internal vesicles of multivesicular endosomes (30). Recent studies have shown that PI3P-rich microdomains serve as platforms to build signaling complexes (29, 30). Because PI3P contains little recognition information, these larger signaling complexes are generated through distinct protein-protein interaction motifs that are found in FYVE and PX domain-containing proteins (28, 38). Interestingly, in yeast, TOR1, which is only found in the rapamycin-sensitive TOR complex 1 (39), localizes to endosomes through GβL (27). Moreover, in preliminary studies, we have found that hVps34 coimmunoprecipitates with mTOR, a finding that needs more rigorous analysis. It is now important to determine whether the role of hVps34 is to generate a mTOR/raptor/GβL signaling platform and whether hVps34 is a constituent of such a platform.
An important function for hVps34 is to control macroautophagy, a process in which portions of cytoplasm are sequestered within double-membrane vesicles known as autophagosomes for delivery to the lysosomes for degradation and recycling of cellular components (40). The autophagy link to hVps34 is through studies demonstrating that decreasing hVps34 levels with antisense oligonucleotides inhibits the rate of macroautophagy and that one of the autophagic genes, Atg6 in yeast (41, 42) and beclin1 in mammals (43), interacts directly with hVps34 (44). However, under conditions of amino acid withdrawal, macroautophagy is thought to be under the negative control of mTOR (40). Given the results shown here, the two findings would appear at odds. One possibility is that hVps34 operating through distinct complexes is differentially regulating autophagy and signaling through the mTOR/raptor/GβL. Such a model would fit with identification of at least two distinct Vps34 protein complexes (45) and the finding that whereas all of the cellular beclin is associated with hVps34, 50% of hVps34 is beclinfree (44). Indeed, by indirect immunofluorescence microscopy, the beclin/hVps34 complex is localized to the TGN, but hVps34 is also separately localized to endosomes (44), such that distinct hVps34 complexes may be involved in regulating these responses. The importance of elucidating the underlying molecular mechanisms by which hVps34 function is used is underscored by the role of the mTOR signaling pathway in a number of specific pathogenic responses, including cancer (3) and the development of insulin resistance (4).
Supplementary Material
Acknowledgments
We thank B. Fowler for amino acid measurements; P. Dennis for his critical reading of the manuscript; and P. Dennis, H. Stenmark, and S. Emr for critical discussions. We also thank R. S. Yeung for providing Eker REFs, D. Kwiatkowski for TSC MEFs, T. Kobayashi for CACL-I-III cells, M. Nellist for TSC1 and TSC2 antibodies, and M. Wymann for the hVPS34 construct. M.J. is supported by the Swiss Cancer League; P.G. is supported by the National Cancer Institute's Mouse Models for Human Cancer Consortium; S.G.D. is supported by an Air Force Office of Scientific Research Fellowship through the Oak Ridge Institute for Science and Education; M.R. is supported by the Netherlands Genomics Initiative; and T.N. and G.T. are supported in part by the Collaborative Cancer Research Project of the Swiss Cancer League.
Author contributions: M.P.B., J.M.B., J.L.B., F.J.T.Z., and G.T advised research; J.L.B., F.J.T.Z., and G.T. guided research; T.N., M.J., M.R., S.G.D., S.Y.K., and P.G. performed research; F.N. contributed new reagents/analytical tools; and G.T. wrote the paper.
Abbreviations: PI3K, phosphatidylinositol 3OH-kinase; PI3P, phosphatidylinositol 3-phosphate; mTOR, mammalian target of rapamycin; GβL, G-protein-β-subunit-like protein; TSC, tuberous sclerosis complex; S6K1, S6 kinase 1; siRNA, short interfering RNA; 4E-BP1, initiation factor 4E binding protein; FYVE, Fab1/YOTB/ZK632.12/Vac1/EEA1.
References
- 1.Matsuo, T., Kubo, Y., Watanabe, Y. & Yamamoto, M. (2003) EMBO J. 22, 3073-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hay, N. & Sonenberg, N. (2004) Genes Dev. 18, 1926-1945. [DOI] [PubMed] [Google Scholar]
- 3.Bjornsti, M. A. & Houghton, P. J. (2004) Nat. Rev. Cancer 4, 335-348. [DOI] [PubMed] [Google Scholar]
- 4.Um, S. H., Frigerio, F., Watanabe, M., Picard, F., Joaquin, M., Sticker, M., Fumagalli, S., Allegrini, P. R., Kozma, S. C., Auwerx, J. & Thomas, G. (2004) Nature 431, 200-205. [DOI] [PubMed] [Google Scholar]
- 5.Patti, M. E. & Kahn, B. B. (2004) Nat. Med. 10, 1049-1050. [DOI] [PubMed] [Google Scholar]
- 6.Sarbassov, D. D., Ali, S. M., Kim, D. H., Guertin, D. A., Latek, R. R., Erdjument-Bromage, H., Tempst, P. & Sabatini, D. M. (2004) Curr. Biol. 14, 1296-1302. [DOI] [PubMed] [Google Scholar]
- 7.Jacinto, E., Loewith, R., Schmidt, A., Lin, S., Ruegg, M. A., Hall, A. & Hall, M. N. (2004) Nat. Cell Biol. 6, 1122-1128. [DOI] [PubMed] [Google Scholar]
- 8.Sarbassov, D. D., Guertin, D. A., Ali, S. M. & Sabatini, D. M. (2005) Science 307, 1098-1101. [DOI] [PubMed] [Google Scholar]
- 9.Marygold, S. J. & Leevers, S. J. (2002) Curr. Biol. 12, R785-R787. [DOI] [PubMed] [Google Scholar]
- 10.Jaeschke, A., Hartkamp, J., Saitoh, M., Roworth, W., Nobukuni, T., Hodges, A., Sampson, J., Thomas, G. & Lamb, R. (2002) J. Cell Biol. 159, 217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Y., Corradetti, M. N., Inoki, K. & Guan, K. L. (2004) Trends Biochem. Sci. 29, 32-38. [DOI] [PubMed] [Google Scholar]
- 12.Inoki, K., Li, Y., Zhu, T., Wu, J. & Guan, K. L. (2002) Nat. Cell Biol. 4, 648-657. [DOI] [PubMed] [Google Scholar]
- 13.Gao, X., Zhang, Y., Arrazola, P., Hino, O., Kobayashi, T., Yeung, R. S., Ru, B. & Pan, D. (2002) Nat. Cell Biol. 4, 699-704. [DOI] [PubMed] [Google Scholar]
- 14.Kimball, S. R. & Jefferson, L. S. (2004) Curr. Opin. Clin. Nutr. Metab. Care 7, 39-44. [DOI] [PubMed] [Google Scholar]
- 15.Saucedo, L. J., Gao, X., Chiarelli, D. A., Li, L., Pan, D. & Edgar, B. A. (2003) Nat. Cell Biol. 5, 566-571. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, Y., Gao, X., Saucedo, L. J., Ru, B., Edgar, B. A. & Pan, D. (2003) Nat. Cell Biol. 5, 578-581. [DOI] [PubMed] [Google Scholar]
- 17.Smith, E. M., Finn, S. G., Tee, A. R., Browne, G. J. & Proud, C. G. (2005) J. Biol. Chem. 280, 18717-18727. [DOI] [PubMed] [Google Scholar]
- 18.Lane, H. A., Fernandez, A., Lamb, N. J. C. & Thomas, G. (1993) Nature 363, 170-172. [DOI] [PubMed] [Google Scholar]
- 19.Garami, A., Zwartkruis, F. J., Nobukuni, T., Joaquin, M., Roccio, M., Stocker, H., Kozma, S. C., Hafen, E., Bos, J. L. & Thomas, G. (2003) Mol. Cell 11, 1457-1466. [DOI] [PubMed] [Google Scholar]
- 20.Dennis, P. B., Jaeschke, A., Saitoh, M., Fowler, B., Kozma, S. C. & Thomas, G. (2001) Science 294, 1102-1105. [DOI] [PubMed] [Google Scholar]
- 21.Tremblay, F. & Marette, A. (2001) J. Biol. Chem. 276, 38052-38060. [DOI] [PubMed] [Google Scholar]
- 22.Harrington, L. S., Findlay, G. M., Gray, A., Tolkacheva, T., Wigfield, S., Rebholz, H., Barnett, J., Leslie, N. R., Cheng, S., Shepherd, P. R., et al. (2004) J. Cell Biol. 166, 213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah, O. J., Wang, Z. & Hunter, T. (2004) Curr. Biol. 14, 1650-1656. [DOI] [PubMed] [Google Scholar]
- 24.Hara, K., Yonezawa, K., Weng, Q. P., Kozlowski, M. T., Belham, C. & Avruch, J. (1998) J. Biol. Chem. 273, 14484-14494. [DOI] [PubMed] [Google Scholar]
- 25.Iiboshi, Y., Papst, P. J., Kawasome, H., Hosoi, H., Abraham, R. T., Houghton, P. J. & Terada, N. (1999) J. Biol. Chem. 274, 1092-1099. [DOI] [PubMed] [Google Scholar]
- 26.Fruman, D. A., Meyers, R. E. & Cantley, L. C. (1998) Annu. Rev. Biochem. 67, 481-507. [DOI] [PubMed] [Google Scholar]
- 27.Chen, E. J. & Kaiser, C. A. (2003) J. Cell Biol. 161, 333-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmon, M. A. (2003) Traffic 4, 201-213. [DOI] [PubMed] [Google Scholar]
- 29.Miaczynska, M., Pelkmans, L. & Zerial, M. (2004) Curr. Opin. Cell Biol. 16, 400-406. [DOI] [PubMed] [Google Scholar]
- 30.Burda, P., Padilla, S. M., Sarkar, S. & Emr, S. D. (2002) J. Cell Sci. 115, 3889-3900. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Gaitan, M. & Stenmark, H. (2003) Cell 115, 513-521. [DOI] [PubMed] [Google Scholar]
- 32.Stack, J. H., Herman, P. K., Schu, P. V. & Emr, S. D. (1993) EMBO J. 12, 2195-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volinia, S., Dhand, R., Vanhaesebroeck, B., MacDougall, L. K., Stein, R., Zvelebil, M. J., Domin, J., Panaretou, C. & Waterfield, M. D. (1995) EMBO J. 14, 3339-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blommaart, E. F., Luiken, J. J., Blommaart, P. J., van Woerkom, G. M. & Meijer, A. J. (1995) J. Biol. Chem. 270, 2320-2326. [DOI] [PubMed] [Google Scholar]
- 35.Xu, G., Kwon, G., Marshall, C. A., Lin, T. A., Lawrence, J. C., Jr., & McDaniel, M. L. (1998) J. Biol. Chem. 273, 28178-28184. [DOI] [PubMed] [Google Scholar]
- 36.Hinnebusch, A. G. & Natarajan, K. (2002) Eukaryot. Cell 1, 22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byfield, M. P., Murray, J. T. & Backer, J. M. (2005) J. Biol. Chem., in press. [DOI] [PubMed]
- 38.Gruenberg, J. & Stenmark, H. (2004) Nat. Rev. Mol. Cell Biol. 5, 317-323. [DOI] [PubMed] [Google Scholar]
- 39.Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J. L., Bonenfant, D., Oppliger, W., Jenoe, P. & Hall, M. N. (2002) Mol. Cell 10, 457-468. [DOI] [PubMed] [Google Scholar]
- 40.Klionsky, D. J. & Emr, S. D. (2000) Science 290, 1717-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petiot, A., Ogier-Denis, E., Blommaart, E. F., Meijer, A. J. & Codogno, P. (2000) J. Biol. Chem. 275, 992-998. [DOI] [PubMed] [Google Scholar]
- 42.Tsukada, M. & Ohsumi, Y. (1993) FEBS Lett. 333, 169-174. [DOI] [PubMed] [Google Scholar]
- 43.Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H. & Levine, B. (1999) Nature 402, 672-676. [DOI] [PubMed] [Google Scholar]
- 44.Kihara, A., Kabeya, Y., Ohsumi, Y. & Yoshimori, T. (2001) EMBO Rep. 2, 330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kihara, A., Noda, T., Ishihara, N. & Ohsumi, Y. (2001) J. Cell Biol. 152, 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.