Abstract
HslVU is a bacterial homolog of the proteasome, where HslV is the protease that is activated by HslU, an ATPase and chaperone. Structures of singly and doubly capped HslVU particles have been reported, and different binding modes have been observed. Even among HslVU structures with I-domains distal to HslV, no consensus mode of activation has emerged. A feature in the Haemophilus influenzae HslVU structure, insertion of the C termini of HslU into pockets in HslV, was not seen in all other structures of the enzyme. Here we report site-directed mutagenesis, peptide activation, and fluorescence experiments that strongly support the functional relevance of the C terminus insertion mechanism: we find that mutations in HslV that disrupt the interaction with the C termini of HslU invariably lead to inactive enzyme. Conversely, synthetic peptides derived from the C terminus of HslU bind to HslV with 10−5 M affinity and can functionally replace full HslU particles for both peptide and casein degradation but fail to support degradation of a folded substrate. Thus, the data can be taken as evidence for separate substrate unfoldase and protease stimulation activities in HslU. Enhanced HslV proteolysis could be due to the opening of a gated channel or allosteric activation of the active sites. To distinguish between these possibilities, we have mutated a series of residues that line the entrance channel into the HslV particle. Our mutational and fluorescence experiments demonstrate that allosteric activation of the catalytic sites is required in HslV, but they do not exclude the possibility of channel opening taking place as well. The present data support the conclusion that the H. influenzae structure with I-domains distal to HslV captures the active species and point to significant differences in the activation mechanism of HslV, ClpP, and the proteasome.
HslVU (ClpQY) is the eubacterial counterpart of eukaryotic proteasomes (1–6). HslU, a member of the Clp/Hsp100 family of proteins (7), is an ATPase (6) and is also known to possess a chaperone activity (2, 8). It interacts with HslV, a protease, for activation in a manner that is not yet well understood. Other members of the Hsp 100 family include ClpA and ClpX, which activate ClpP, their common protease (9). It is generally believed that there is a symmetry mismatch in the ClpAP/ClpXP systems, with ClpA and ClpX forming a hexamer and ClpP forming a heptamer (10–12). No such symmetry mismatch occurs in HslVU: both HslU and HslV are hexamers (4, 6, 13).
The preferred orientation of the HslU particle vis-à-vis HslV has been a matter of debate recently (6, 14–17), not least because of its implications in the enzyme mechanism of the Clp/Hsp100 family of proteins. X-ray analysis (4, 6) had revealed the structures of both components, HslV and HslU, in detail and showed the complex in an arrangement with the I-domains proximal to HslV. Earlier electron microscopy studies (13, 18), interpreted (16) in the light of our crystal structures (4, 6), suggested that the I-domains of Escherichia coli HslU were located distal to HslV. This conclusion was supported by later structures of the singly (HslU/HslV = 1:1) and doubly (HslU/HslV = 2:1) capped E. coli HslVU complex (17) and the doubly capped Haemophilus influenzae HslVU structure (19).
Interestingly, no single mode of association between HslU and HslV has emerged from this series of x-ray structures. Among the x-ray structures with I-domains distal to HslV, symmetric and asymmetric complexes have been observed. Complexes differ in the azimuth of HslU relative to HslV (17, 20) and also in the arrangement of the C-terminal residues of HslU. Whereas the C terminus is found to be buried inside HslU in the E. coli structures, it inserts into a cleft between adjacent HslV subunits in the H. influenzae structure. It is currently not clear whether the various x-ray structures represent different modes of association or whether they can be regarded as snapshots at different stages of the functional cycle. The existence of a cation-binding site near the proteolytic site of H. influenzae HslV was reported recently (21). It was suggested that the cation (Na+, K+) might influence the catalytic activity of the protease, but more experiments are required to clarify its role.
Experimentally, binding between HslV and HslU is found to be very labile, especially for the E. coli enzyme. In contrast, studies from this laboratory (14) have shown that the functional interaction between HslV and HslU is quite robust. Mutations in HslU involving the deletion of the entire I-domain as well as the introduction of pentaglycine segments on the HslU surface that would interact with HslV according to the electron microscopy studies could not abolish amidolytic and caseinolytic activities. This observation was taken as an indication that no precise complex was required for these activities, at least in the E. coli enzyme. Results are very different for folded substrates: they require interactions of both surfaces, possibly for different reasons (14). As our study of HslU mutants could not convincingly identify any one complex as the only functional complex for all substrates, we are now complementing this work with the characterization of a series of HslV mutants.
In a first set of experiments, we targeted the α-helical segment that lines the channel into the HslV particle and reversed or neutralized charges in this segment. These mutants were designed to probe the role of a largely conserved set of positive charges at the entrance of HslV and to find out whether the recently characterized “gated channel” mechanism of proteasomes (22) was operating in the HslVU system as well.
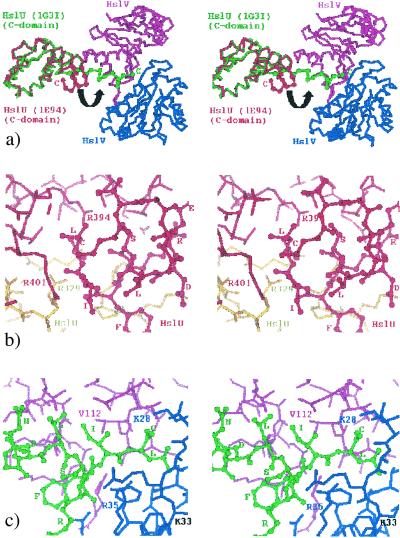
In a second set of experiments, we tested the functional significance of a highly unanticipated feature in the x-ray structure of the H. influenzae complex solved by Sousa et al. (19). These authors reported that the C-terminal helix that is buried in free HslU particles distends and inserts into a cleft between adjacent HslV subunits in the HslVU complex (Fig. 1). This insertion was coupled with significant conformational changes in the active site region of HslV. Intriguingly, no such distension was found in two recent E. coli HslVU structures (17), although the orientation of HslU (I domains distal to HslV) is similar in the H. influenzae complex. On the basis of the structure of the H. influenzae enzyme, we designed mutants of E. coli HslV to probe the functional relevance of the C terminus insertion mechanism. The role of the C-terminal residues of HslU in activating HslV was probed through “peptide activation” experiments involving peptides derived from the C terminus of HslU and their variants. Additionally, we measured dissociation constants of complexes of free and covalently inhibited HslV with one of the peptides using fluorescence spectroscopy. We also report direct evidence for allosteric interaction between the binding of an activator peptide and calpain inhibitor I, a substrate analog aldehyde inhibitor of HslV.
Figure 1.
(a) Stereo view (Cα-trace) of the superposition of the C domain of an HslU subunit (red) from the original E. coli HslVU complex (6) onto that of the H. influenzae HslU subunit (green) from its complex (19). Two HslV subunits (pink and blue) from the H. influenzae complex are also shown to illustrate the binding of the C-terminal segment of H. influenzae HslU to the pocket between the HslV subunits (indicated also by a black curved arrow). Color of the labels follows that of the respective subunits. Protein Data Bank codes are also indicated. (b) Stereo view of the close-up of the C-terminal residues of an E. coli HslU subunit (red) from the complex (6). The carboxylate of the terminal leucine residue forms salt bridges with R394 of the same subunit and with R329 of an adjacent (yellow) HslU subunit that is not illustrated in a for clarity. (c) Stereo view of the close-up of the C-terminal residues of an HslU subunit from the H. influenzae HslVU complex (19). Color coding is the same as in a. Residues of HslV that have been mutated in the present study in this region are indicated. K33, an essential residue of the catalytic apparatus of HslV is indicated in black.
Experimental Procedures
Detailed experimental procedures are published as supporting information on the PNAS web site (www.pnas.org). All mutants were checked by DNA sequencing and/or electron spray ionization mass spectroscopic analysis. Unless otherwise indicated, all mutants migrated similar to wild type on sizing columns.
Carbobenzoxy-Gly-Gly-Leu-7-amido-4-methylcoumarin (Z-Gly-Gly-Leu-AMC; Bachem) and FITC-casein were typically digested with 1 μg of HslV/2.5 μg of HslU/6 μg of HslV/15 μg of HslU, respectively. Assays with 14C-methyl casein (24) and MBP-SulA were essentially carried out as reported earlier (6, 14, 25, 26). For the peptide and caseinolytic assays, ATP-γS was used instead of ATP to avoid ADP contamination of samples.
Fluorescence spectroscopy was performed with Dansyl-EDLSRFIL at emission and excitation wavelengths of 335 and 533 nm, respectively. We found enhancement of fluorescence (positive ΔF533) in the presence of protein compared with the experiment in its absence. For quantitative measurements, dansylated peptide activator (Dansyl-EDLSRFIL) was added in aliquots of 1.5/2.5 μl from a 1,250 μM stock solution to a 1.5-ml solution of HslV (monomeric concentration of 26 μM). Kd and ΔFmax were fitted to maximize agreement with the law of mass action that can be expressed (27) as ΔF533 = (ΔFmax/[L]tn)[A − sqrt(A2 − [L]tn[R]t)], where A = ([R]t + [L]tn + Kd)/2. Here, [L]t is the total concentration of peptide, [R]t the total concentration of protein, ΔFmax is ΔF533 of the complex at infinite ligand concentration, n is the number of binding sites per monomer, and Kd is the dissociation constant of the HslV−Dansyl-EDLSRFIL complex.
Results
Substrate Access to the Proteolytic Chamber.
HslV and proteasomes exhibit only residual proteolytic activity in the absence of their respective activators. In proteasomes, access to the proteolytic chamber has been reported to be the rate-limiting step for the hydrolysis of small chromogenic peptides (22, 28). A 20S core particle mutant with an opened axial channel shows much enhanced activity, comparable to the activity of wild-type 26S proteasomes. The term “gated channel” has been coined to describe this situation.
Here, we address the question whether access to the central proteolytic chamber is rate-limiting in HslV. Specifically, three mutations, the R89A/K90A, Δ86–91, and Δ83–92, were designed, which are expected to widen the axial pore in HslV. In all cases, the mutations did not interfere with subunit folding and assembly, because mutant proteins migrated like wild-type on a gel filtration column. In contrast to the findings for proteasomes, all HslV mutants were largely inactive against the chromogenic peptide substrates if assayed in the absence of the natural activator HslU (see Table 1). Failure to increase the peptide hydrolysis activity either can be attributed to subtle structural perturbations or can be taken as evidence that substrate access to the central proteolytic chamber is not rate-limiting in HslV. To distinguish between these two possibilities, the behavior of the HslV mutants was subsequently assayed in the presence of HslU. Somewhat disappointingly, the R89A/K90A and Δ83–92 mutants remained inactive in the presence of HslU, and thus did not allow any clear-cut conclusions. In contrast, the Δ86–91 mutant turned out to be interesting: this mutant was indistinguishable from wild type, inactive in the absence of HslU, and fully active against peptide substrates, casein, and MBP-SulA in its presence. Results for this mutant suggest that HslU does more than simply open a gate into the HslV particle.
Table 1.
Summary of HslV mutants
| Peptide | Casein | MBP-SulA | |
|---|---|---|---|
| Wild type | ++++ | ++++ | ++++ |
| Mutants involving residues at the entrance | |||
| R86D | − | − | − |
| R89D | ++ | ++ | ++ |
| R89A/K90A | − | − | − |
| R89D/K90E | +++++ | − | − |
| Δ86–91 | ++++ | ++++ | ++++ |
| Δ83–92 | − | − | − |
| Mutants designed to probe activation mechanism | |||
| K28A | − | − | − |
| R35A | − | − | − |
| V112D | − | − | − |
The values are relative to the wild-type HslVU activity as a 100% (+++++, 100–120%; ++++, 80–100%; +++, 60–80%; ++, 40–60%; +, 20–40%; −, <20%; NA, not applicable). Note: Only residual activities were observed for wild-type HslV and its mutants in the absence of its activator, HslU.
HslV contains a prominent accumulation of positive charges around the central axial channel, a feature that is conserved across bacterial species and appears to have its counterpart in an accumulation of negative charge around the axis of symmetry in HslU. We wanted to probe the role of these positive charges and engineered a set of charge reversal mutations, namely R86D, R89D, and R89D/K90E. We found widely different results for the different mutations: the R86D mutant exhibited no peptidase and protease activities, and the R89D mutant was partially active in all assays. The most surprising behavior was observed for the double mutant R89D/K90E: it was at least as active as wild type when the chromogenic peptide was used for the assay. In contrast, no activity was observed with casein or MBP-SulA as the substrate. The crystal structure of this mutant (as also R89D) could not explain this observation, because the mutated residues and the neighboring region were disordered in them (data not shown).
The Activation Mechanism: Mutations in HslV.
In free HslU particles, the C-terminal helices are buried in the interior of the particle. It was a major surprise from the structure of the H. influenzae complex that these helices distend and insert into grooves between subunits in the HslV particle (19). This feature was of course absent from the first E. coli HslV-HslU structure (6) with I-domains proximal to HslV, and intriguingly, it was also not present in later structures of the E. coli enzyme with I-domains distal to HslV (17). We therefore set out to independently test the functional relevance of the C terminus insertion mechanism for the HslV particle through mutations in the E. coli enzyme.
In the H. influenzae HslVU structure, the C-terminal segment of HslU forms a salt bridge with K28 of HslV, whereas R35 forms hydrogen bonds to the peptide backbone. Furthermore, it was stabilized by hydrophobic interactions, which involve V112 of a neighboring HslV subunit (see Fig. 1c). To disrupt these interactions, K28 and R35 were each mutated to an alanine, whereas V112 was mutated to an aspartate. The mutants were prepared and expressed as soluble protein. We found that all three mutants had no amidolytic, caseinolytic, or MBP-SulA activities (Table 1). These results are consistent with the strict conservation of K28 and R35 in all known HslV sequences and the nearly strict conservation of V112, which is conservatively replaced with Ile in the Aquifex aeolicus sequence.
The Activation Mechanism: Synthetic Peptides.
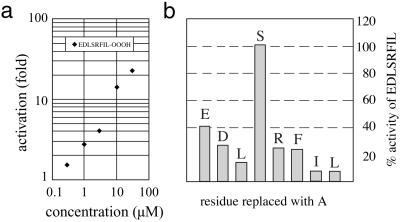
In another approach to probe the functional significance of the activation mechanism, we attempted to truncate the C terminus of HslU with various carboxy peptidases. We observed a loss of activity, but because we also observed a loss of solubility of the enzyme, these experiments were inconclusive. We also note that mutations to the C-terminal segment of HslU resulted invariably in insoluble protein (14), probably because the mutants disrupt its intimate integration and multiple van der Waals and electrostatic interactions with the core of the C domain (6) (Fig. 1b). We next synthesized a peptide, NH2-EDLSRFIL-COOH, with an amino acid sequence derived from the C-terminal eight residues of E. coli HslU. This peptide turned out to be a potent activator of HslV activity as assayed with Z-Gly-Gly-Leu-AMC as substrate. With the small peptide chromogenic substrate, more than 20-fold activation could be achieved (Fig. 2a). Although the peptide is of course less potent than the natural activator if used in identical stoichiometry, we were surprised to find that a 10- to 100-fold excess of peptide over the number of HslU subunits in the physiological complex was sufficient to achieve equivalent activation with Z-GGL-AMC as substrate. The activator peptide showed some activity with 14C-labeled casein as substrate as well. In 100 μM concentration, the peptide lead to a 4-fold increase of radioactivity in the supernatant in the 14C-degradation assay. The peptide could not stimulate the degradation of the MBP-SulA substrate, presumably because of the unfolding step necessary in this case, a step that we associate with the intact HslU (6, 14).
Figure 2.
(a) Activation (fold) as a function of the concentration of the peptide activator EDLSRFIL. The decapeptide (ADEDLSRFIL) exhibited similar activation as the octapeptide, namely about 20 and 50% activation as wild-type HslU at 25 and 100 μM concentrations, respectively, whereas the dodecapeptide (LVADEDLSRFIL) and dansylated octapeptide (Dansyl-EDLSRFIL) exhibited 50 and 100% activation at these concentrations, respectively, compared with wild-type HslU. Note that the curve represents activation in our assay and is not a determination of Kd. A determination of Kd is presented in Fig. 3. (b) Result of an alanine scan of the activation octapeptide (EDLSRFIL). Activity of the “mutant” peptides is stated as percent activity of the wild-type peptide. Bars representing the activity of “mutant” peptides are labeled with the residue that has been replaced with alanine.
To exclude an unspecific effect of this octapeptide on HslV, we synthesized eight peptides where each residue of the “parent” octapeptide was replaced by an alanine in turn. With the exception of serine, all side chains of all residues were found to contribute to the activity (Fig. 2b). Circular dichroism experiments indicated no significant secondary structure in the octapeptide in the absence of HslV. Cocrystallization of the peptide with HslV yielded diffracting crystals, but unfortunately no density for the peptide could be observed (data not shown).
Encouraged by the results of the experiments with the octapeptide, we synthesized deca (ADEDLSRFIL) and dodeca (LVADEDLSRFIL) peptides corresponding to residues in the C terminus of HslU. Although the decapeptide was only as potent as the octapeptide, the dodecapeptide, at higher concentrations, was almost as potent as HslU in activating HslV toward Z-Gly-Gly-Leu-AMC (see Fig. 2a legend). The activation of HslV against FITC–casein also followed similar trends, with dansylated octapeptide and the dodecapeptide activators being as potent as wild-type HslU and the octapeptide being about half as potent.
The Activation Mechanism: Fluorescence Spectroscopy Experiments.
The dose–response curves for peptide activation presented in Fig. 2a represent the complexities of the three-component system of HslV, Z-GGL-AMC, and activator peptide. Their interpretation could be further complicated by interactions between subunits (cooperativity) or limiting substrate access to the active sites. To avoid these complications, we decided to measure binding of activator peptide to HslV by fluorescence spectroscopy, independent of its effect on substrate hydrolysis. Therefore, we synthesized the dansylated derivative of the octapeptide (Dansyl-EDLSRFIL). The dansyl label did not interfere with the biological activity of the peptide. Control experiments showed that this peptide, like the dodecapeptide activator, was as potent as wild-type HslU in activating HslV toward peptide (see legend to Fig. 2) and FITC–casein substrates. Like its nontagged counterparts, Dansyl–EDLSRFIL did not activate HslV for MBP-SulA degradation.
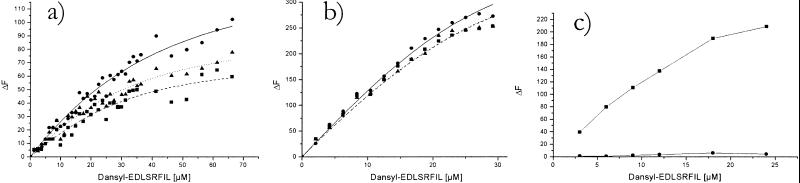
Binding of activator peptide to HslV enhances the fluorescence of the danysl moiety. For a fixed total amount of peptide, the difference in fluorescence from samples with and without HslV is therefore directly proportional to the amount of HslV–activator peptide complex (see Fig. 3) and thus depends on the binding equilibrium. In preliminary analyses, we generalized the interpolation equation given in Experimental Procedures to allow for either systematically fewer than 12 binding sites on the HslV particle (HslV is a dimer of hexamers) or cooperativity or anticooperativity in binding. In no case did we find significant deviation from the simple structurally plausible assumption that activator peptides bind independently of each other. We therefore assumed one binding site per subunit of HslV and analyzed the fluorescence data as explained under Experimental Procedures. (For further details, see Supporting Information on the PNAS web site.) Three independent experiments gave a weighted Kd of 1.7 ± 0.6 × 10−5 M for the complex of free HslV with dansylated peptide (Fig. 3a). This figure is in the right range to be compatible with the dose–response curve for hydrolysis of chromogenic substrates (see Fig. 2a).
Figure 3.
Determination of the dissociation constant of (a) the free HslV−Dansyl-EDLSRFIL complex and (b) the inhibited HslV−Dansyl-EDLSRFIL complex. The complex formation is accompanied by a fluorescence change that is plotted on the y axis. The dissociation constants were obtained as explained in Experimental Procedures. Data of three independent experiments are plotted as filled circles, squares, and triangles, whereas the regression analysis is plotted as continuous, dotted, and dashed lines, respectively. (c) Fluorescence change between free and inhibited HslV samples (filled squares) measured in the presence of various concentrations (3–24 μM) of Dansyl-EDLSRFIL. The substrate analog inhibitor, acetyl-Leu-Leu-Norleucinal, was added directly to the cuvette containing HslV and peptide activator for measurements involving inhibited protein. This procedure ensured that pipetting errors were eliminated. The average of 15 fluorescence scans was taken for each measurement. Filled circles represent the control experiment performed in the absence of protein.
We next attempted to use the fluorescence technique to obtain independent information about the mechanism of HslV activation. If our peptides act through allosteric activation of HslV active sites, we expect a corresponding allosteric “back-action” on activator binding, if substrates or inhibitors are bound to the active site. Conversely, if the activator peptides act as mere gate openers, the dissociation constant for activator binding should not depend on inhibitor binding to the active sites. Our measurements with calpain inhibitor I support allosteric activation of HslV, but the larger error limits on Kd do not allow for detailed conclusions: we obtain the best fit for our data in the presence of calpain inhibitor I with an activator peptide dissociation constant of 0.5 × 10−5 M (Fig. 3b) and can state an upper limit of 1.2 × 10−5 M with confidence. Unfortunately, we cannot give a lower limit for the binding constant. In the tight binding regime (Kd ≪ enzyme concentration), practically all available inhibitor binds to the enzyme independent of Kd. Measurements at lower enzyme concentration were hampered by too-low absolute fluorescence differences.
The errors in the dissociation constants of the activator peptide–HslV complex in the presence and absence of inhibitor include a large contribution from the uncertainty in ΔFmax, the enhancement in fluorescence corresponding to fully saturated enzyme. We therefore compared the fluorescence enhancement for a fixed amount of activator peptide and enzyme in the presence and absence of inhibitor. As would be expected from the dissociation constant measurements, we found that it is directly proportional to the extra amount of activator peptide that binds to HslV on addition of inhibitor (Fig. 3c). To exclude an unspecific effect of the inhibitor on activator peptide fluorescence, we repeated the same experiment in the absence of enzyme as a control (Fig. 3c). Throughout the range of tested concentrations, we find enhanced activator peptide binding on adding the inhibitor.
Discussion
Substrate Access to the Proteolytic Chamber.
The low activity of 20S proteasomes in their “latent” state has been attributed to a lack of access to the interior of the particle (22, 28). In the present work, we have addressed the question whether this explanation could also account for the low activity of HslV in the absence of HslU and nucleotide. Experimentally, we followed the approach that was used to demonstrate the existence of a rate-limiting gated channel into the 20S core particle. For this particle, it had been shown (22) that the deletion of a nonapeptide fragment from the N terminus of the α3-subunit leads to an opening of the central channel and to a concomitant enhancement of the hydrolysis activity from residual levels to the maximal levels that are normally observed only in the presence of the natural activators.
By analogy, we generated HslV mutants that were expected to have a wider entrance channel than wild-type particles. An enhancement of amidolytic activity would demonstrate that substrate access was rate-limiting in the HslVU system as well. Conversely, failure to activate HslV in this way could be taken as evidence that HslU did not merely act through opening a gate into HslV. For this conclusion to be warranted, unintended structural perturbations of the active site had to be ruled out. This demonstration was convincingly possible for the Δ86–91 construct. Addition of HslU restored all activities to wild-type levels for this mutant. Although it is currently not clear why this was not possible for the seemingly more conservative mutation R89A/K90A, it came as less of a surprise that the very drastic Δ83–92 mutation resulted in inactive HslV.
Our results show that lack of access to the proteolytic chamber cannot be the exclusive explanation for the low residual activity of HslV in the absence of HslU. However, they are perfectly consistent with the possibility that the pore in HslV is widened as a result of binding to HslU. In fact, a comparison of the structures of free and bound HslV from H. influenzae suggests that HslU facilitates access to HslV. The most prominent change is a complete reorientation of R86. In free HslV, this residue is well ordered and pointing almost straight toward the central axis, thus blocking part of the entrance channel into HslV. In the H. influenzae complex structure, this residue has a very high temperature factor, is no longer obstructing the pore, and has been modeled as pointing toward a loop in HslU. Superficially, this loop appears to be the functional equivalent of the “activation” loop in PA26, which has been shown to pull the occluding N termini of 20S proteasomes away from the central channel (28). However, we point out that the loop is poorly conserved, disordered in the H. influenzae structure, and not required for degradation of chromogenic peptide substrates and casein, as shown by our peptide activation experiments. Less drastic changes on binding of HslU are observed for the other basic residues that line the entrance into HslV. The side chain of R89 is anchored to the carbonyl oxygen of R83 through hydrogen bonding in free HslV. The crystal structure of bound HslV shows this residue anchored to the carbonyl oxygens of any of residues 82, 83, and 84, not always in ideal hydrogen bonding geometry. K90 does not seem to have a defined interaction partner in either free or bound HslV.
The above observations from the H. influenzae crystal structure provide a framework to understand some of our results for the charge reversal mutations. Given the central role of R86 in the crystal structure, it is not surprising that the charge reversal mutation R86D abolishes all activity, including any activity in the presence of HslU. Interestingly, the activity in the presence of HslU is recovered in the HslV mutant Δ86–91, where four residues in addition to R86 are replaced with a triglycine linker. This successful “rescue” could indicate that the defect in the R86D mutation may be because of a block in substrate access that may be maintained in the presence of HslU. According to this interpretation, deletion of further residues around the pore in HslV restores substrate access to the particle, and the continued requirement for HslU in the HslV Δ86–91 mutant points to one or more additional essential activities of HslU. In light of results from our peptide activation experiments, it is very likely that one such additional activity of HslU is allosteric activation of the active sites in HslV.
The Activation Mechanism.
An essential role of the HslU C terminus distension and insertion mechanism was suggested by the H. influenzae crystal structure. Our present experiments provide strong support for this mechanism. They go beyond the previous results, in that they demonstrate that HslU C terminus insertion is actually sufficient for the degradation of small chromogenic substrates and even casein. As we are able to stimulate protease activity against chromogenic substrates to levels normally seen only in the presence of the natural activator HslU, we conclude that unfolding and translocation play no role for these substrates.
The results of our peptide activation experiments agree well with the H. influenzae crystal structure. Although this crystal structure shows some flexibility in the binding mode of the H. influenzae HslU C-terminal segment to HslV, all eight residues, with the exception of Ser-440, are engaged in interactions with HslV (typically L444 to V20, I443 to L72/V112, F442 to V76, R441 to E61 or R36 O, L439 to L72, D438 to K73, and E437 to K73 in H. influenzae) (see also Fig. 1c). Consistent with these crystallographic results, our alanine scans of the activator peptide single out Ser-440 as the only residue that is not required for activation.
In the crystal structure of H. influenzae HslVU, the terminal carboxylate of the activation peptide forms a salt bridge with K28, which is close to K33, an essential component of the catalytic apparatus of HslV (4) (Fig. 1c). It was therefore proposed (19) that active site residues (which include Thr-1) may be suboptimally arranged in free HslV. According to this suggestion, these residues would be allosterically rearranged during activation (19), a proposal that had previously been advanced on the basis of findings with the HslU-dependent vinyl sulfone inhibitors of HslV (29). Our peptide activation experiments, together with the lack of HslV activation in the channel opening mutants, agree with such an allosteric mechanism.
An independent more direct demonstration is provided by our fluorescence experiments. Enhanced activator peptide binding in the presence of inhibitor is thermodynamically equivalent with enhanced inhibitor binding in the presence of activator peptide. As the inhibitor mimics either the substrate or the transition state of the proteolysis reaction, the binding constant or turnover rate for substrate should increase in the presence of activator peptide. We expect, therefore, similar allosteric effects in the activator peptide–HslV and HslU-HslV systems.
It was previously reported that vinyl sulfone-based inhibitors could covalently bind to HslV only in the presence of HslU and ATP (29). We therefore wanted to examine whether our activator peptides could support the binding of such inhibitors to HslV. In preliminary experiments using electron spray ionization mass spectroscopy as a tool to probe the binding of such an inhibitor (Ac-PRLN-VinylSulphone), we unexpectedly found that our activator peptides do not support its binding (unpublished data), although we were able to reproduce the earlier finding that vinyl sulfone-based inhibitors do bind to HslV in the presence of HslU and ATP.
Functional Orientation of HslU in Its Interaction with HslV.
The present mutagenesis, peptide activation, and fluorescence studies strongly support the C terminus distention and insertion mechanism and thus suggest that the H. influenzae HslVU crystal structure represents the active complex. The considerable affinity of our peptides to HslV contrasts sharply with the known labile nature of the interaction between HslV and its natural activator HslU. This discrepancy could be due to varying affinity of HsIU for HsIV in different steps of the mechanistic cycle. It must, however, be pointed out that the affinity of HslU for HslV is governed by factors in addition to those for the peptide–HslV complex. There are favorable factors such as multivalency (hexa), interactions at segments other than the C-terminal segment, and unfavorable ones like the disruption of the intramolecular interactions of the C-terminal segment required for functional complex formation. The known lability of the E. coli HslVU complex suggests balanced positive and negative interactions of the components possibly linked to the nucleotide bound/unbound states. We believe that such a model, and not a crystallographic reanalysis of the original HslVU crystals (30) that we have shown to be in error (20), accounts for the existence of alternative E. coli HslVU structures in crystals where lattice forces may be of equal magnitude as biological interactions.
Comparison of HslVU with Related Systems.
The essential role of the C-terminal segment of the activators appears to be a recurrent theme in mechanistic studies of HslV and proteasome activation (31, 32). X-ray studies of the 11S regulator (PA26) complexed with 20S proteasome have demonstrated that it activates the proteasome by inserting its C-terminal tails into proteasomal pockets (28). As in the HslVU system, proteasomal hydrolysis activity against chromogenic substrates can be stimulated with peptides derived from the C terminus of PA28 (33). Despite these similarities, there are important differences: in proteasomes, where the PA26/PA28 activators bind to the antechambers of 20S proteasomes and remain separated in space from the central proteolytic chamber, they seem to act as “gate openers” only. Allosteric activation appears unlikely, and there is no experimental evidence for it. In contrast, our studies confirm that allosteric activation of the active sites is essential in the HslVU system. Another difference relates to the chemistry of optimal synthetic activator peptides. Whereas peptide alcohols were found to be effective in the proteasome system (33, 34), they were less potent than the carboxylic acids in the HslVU system. This observation is explained with the salt bridge between the terminal carboxylate of HslU and the ɛ-amino group of K28 (see Fig. 1c) in the H. influenzae crystal structure.
Major modification of the C terminus insertion mechanism would be required for it to operate in the ClpAP and ClpXP systems. First, it is generally assumed that ClpAP and ClpXP are symmetry mismatched systems, compatible with an insertion mechanism only if the interactions are transient and need not happen simultaneously. Second, sequence comparisons show that the C termini of both ClpA and ClpX are very poorly conserved (data not shown). Their functional equivalent may be a loop in ClpX that has recently been shown to be a strong predictor of ClpP binding (35). On the basis of these findings, it has been argued that the coiled-coil domains in ClpX are distal to the protease in the functional ClpXP complex.
A similar situation arises in the complex of archaebacterial 20S proteasome with its activator PAN. Whereas the protease component has firmly established 7-fold symmetry, PAN is a AAA-protein and is thus expected to be hexameric. Sequence comparisons reveal a lower degree of sequence conservation in the C termini than in the rest of the protein. The situation in eukaryotic proteasomes, which contain six rather than seven AAA-ATPases in the 19S cap, is unclear. The C termini of the individual ATPases are moderately conserved between different species. Consensus sequences for individual ATPases of the 19S cap from various species can be defined, but the degree of sequence conservation in the C termini is not higher than in the rest of the protein if the poorly conserved N termini are excluded from the comparison. There is no consensus for the C termini of the different ATPases in the 19S cap of any particular species. This lack of consensus could be explained with the need to bind to different local environments in pseudo-7-fold 20S proteasomes or could be taken as an indication that the C termini of the proteasomal AAA-ATPases play no particular role in complex formation. Clearly, further studies are required to distinguish between these two possibilities.
Supplementary Material
Acknowledgments
We thank M. Boicu for DNA sequencing, S. Körner for mass spectroscopic analysis, F. Siedler for peptide synthesis, M. Bogyo for the gift of inhibitor, R. Behrendt for HPLC analysis, P. Zwickl (Max-Planck-Institut für Biochemie) for 14C-labeled casein, A Higashitani (National Institute of Genetics, Japan) for MBP-SulA fusion protein clone, and M. Zylicz and A. Wawrzynow for critical and helpful comments to an early version of the manuscript. R.R. is a recipient of a fellowship from the Alexander von Humboldt Foundation.
References
- 1.Chuang S E, Burland V, Plunkett G D, Daniels D L, Blattner F R. Gene. 1993;134:1–6. doi: 10.1016/0378-1119(93)90167-2. [DOI] [PubMed] [Google Scholar]
- 2.Rohrwild M, Coux O, Huang H C, Moerschell R P, Yoo S J, Seol J H, Chung C H, Goldberg A L. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoo S J, Seol J H, Shin D H, Rohrwild M, Kang M S, Tanaka K, Goldberg A L, Chung C H. J Biol Chem. 1996;271:14035–14040. doi: 10.1074/jbc.271.24.14035. [DOI] [PubMed] [Google Scholar]
- 4.Bochtler M, Ditzel L, Groll M, Huber R. Proc Natl Acad Sci USA. 1997;94:6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. Annu Rev Biophys Biomol Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- 6.Bochtler M, Hartmann C, Song H K, Bourenkov G, Bartunik H D, Huber R. Nature (London) 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 7.Schirmer E C, Glover J R, Singer M A, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 8.Seong I S, Oh J Y, Lee J W, Tanaka K, Chung C H. FEBS Lett. 2000;477:224–229. doi: 10.1016/s0014-5793(00)01808-1. [DOI] [PubMed] [Google Scholar]
- 9.Missiakas D, Schwager F, Betton J M, Georgopoulos C, Raina S. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 10.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 11.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 13.Kessel M, Wu W, Gottesman S, Kocsis E, Steven A C, Maurizi M R. FEBS Lett. 1996;398:274–278. doi: 10.1016/s0014-5793(96)01261-6. [DOI] [PubMed] [Google Scholar]
- 14.Song H K, Hartmann C, Ravishankar R, Bochtler M, Behrendt R, Moroder L, Huber R. Proc Natl Acad Sci USA. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bochtler M, Hartmann C, Song H K, Ravishankar R, Huber R. Nature (London) 2000;408:668. [Google Scholar]
- 16.Ishikawa T, Maurizi M R, Belnap D, Steven A C. Nature (London) 2000;408:667–668. doi: 10.1038/35047165. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Song J J, Franklin M C, Kamtekar S, Im Y J, Rho S H, Seong I S, Lee C S, Chung C H, Eom S H. Structure (London) 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 18.Rohrwild M, Pfeifer G, Santarius U, Muller S A, Huang H C, Engel A, Baumeister W, Goldberg A L. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 19.Sousa C M, Trame C B, Tsuruta H, Wilbanks S M, Reddy V S, McKay D B. Cell. 2000;103:663–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 20.Bochtler M, Song H K, Hartmann C, Ravishankar R, Huber R. J Struct Biol. 2001;135:281–293. doi: 10.1006/jsbi.2001.4413. [DOI] [PubMed] [Google Scholar]
- 21.Sousa M C, McKay D B. Acta Crystallogr D. 2001;57:1950–1954. doi: 10.1107/s090744490101575x. [DOI] [PubMed] [Google Scholar]
- 22.Groll M, Bajorek M, Kohler A, Moroder L, Rubin D M, Huber R, Glickman M H, Finley D. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 23.Twining S S. Anal Biochem. 1984;143:30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- 24.Driscoll J, Goldberg A L. J Biol Chem. 1990;265:4789–4792. [PubMed] [Google Scholar]
- 25.Higashitani A, Ishii Y, Kato Y, Horiuchi K. Mol Gen Genet. 1997;254:351–357. doi: 10.1007/s004380050426. [DOI] [PubMed] [Google Scholar]
- 26.Seong I S, Oh J Y, Yoo S J, Seol J H, Chung C H. FEBS Lett. 1999;456:211–214. doi: 10.1016/s0014-5793(99)00935-7. [DOI] [PubMed] [Google Scholar]
- 27.Sondermann P, Jacob U, Kutscher C, Frey J. Biochemistry. 1999;38:8469–8477. doi: 10.1021/bi982889q. [DOI] [PubMed] [Google Scholar]
- 28.Whitby F G, Masters E I, Kramer L, Knowlton J R, Yao Y, Wang C C, Hill C P. Nature (London) 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 29.Bogyo M, McMaster J S, Gaczynska M, Tortorella D, Goldberg A L, Ploegh H. Proc Natl Acad Sci USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J. J Struct Biol. 2001;134:15–24. doi: 10.1006/jsbi.2001.4347. [DOI] [PubMed] [Google Scholar]
- 31.Dubiel W, Pratt G, Ferrell K, Rechsteiner M. J Biol Chem. 1992;267:22369–22377. [PubMed] [Google Scholar]
- 32.Ma C P, Willy P J, Slaughter C A, DeMartino G N. J Biol Chem. 1993;268:22514–22519. [PubMed] [Google Scholar]
- 33.Wilk S, Chen W E. Mol Biol Rep. 1997;24:119–124. doi: 10.1023/a:1006851428691. [DOI] [PubMed] [Google Scholar]
- 34.Ma C P, Slaughter C A, DeMartino G N. J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 35.Kim Y I, Levchenko I, Fraczkowska K, Woodruff R V, Sauer R T, Baker T A. Nat Struct Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.