Abstract
The [Het-s] infectious element of the filamentous fungus Podospora anserina is a prion. We have recently reported that recombinant HET-s protein aggregates in vitro into amyloid fibers. In vivo, the protein aggregates specifically in the [Het-s] prion strains. Here, we show that biolistic introduction of aggregated recombinant HET-s protein into fungal cells induces emergence of the [Het-s] prion with a high frequency. Thus, we demonstrate that prion infectivity can be created de novo, in vitro from recombinant protein in this system. Although the amyloid filaments formed from HET-s could transmit [Het-s] efficiently, neither the soluble form of the protein nor amorphous aggregates would do so. In addition, we have found that (i) [Het-s] infectivity correlates with the ability to convert HET-s to amyloids in vitro, (ii) [Het-s] infectivity is resistant to proteinase K digestion, and (iii) HET-s aggregates formed in vivo in [Het-s] strains have the ability to convert the recombinant protein to aggregates. Together, our data designate the HET-s amyloids as the molecular basis of [Het-s] prion propagation.
It is now widely accepted that an altered form of the prion protein (PrPSc) is the infectious element in spongiform encephalopathies (1). Yet the “protein-only” hypothesis still awaits formal demonstration. Thus far, generation of infectious material from purified natural PrPC or from recombinant sources has been unsuccessful (2–4). Prions (infectious proteins) have also been identified in fungi (yeast and filamentous fungi) (5, 6). The yeast [URE3] and [PSI] prions represent valuable models to explore the mechanism of prion emergence and propagation (for recent reviews see refs. 7–9). Numerous reports suggest that the yeast prions propagate as infectious amyloids (10–16). However, the molecular nature of the template for prion replication in vivo is still open for debate (17).
The [Het-s] infectious element of the filamentous fungus Podospora anserina represents the prion form of the HET-s protein (6). [Het-s] designates the prion state whereas the prion-free state is termed [Het-s*]. [Het-s] strains differ from [Het-s*] (prion-free) strains by their reactivity toward strains expressing the HET-S protein. HET-S is a polymorphic variant that differs from HET-s by 13 amino acid residues (18). Filamentous fungi spontaneously undergo vegetative cell fusions (19). Cell fusions between [Het-s*] (prion-free) and [Het-S] strains are viable, but a cell death reaction occurs when [Het-s] (prion) and [Het-S] cells fuse. This cell death phenomenon is termed heterokaryon incompatibility (19, 20). Macroscopically, this cell death reaction can readily be detected because it leads to the formation of an abnormal contact line (“barrage”) when [Het-s] and [Het-S] strains are confronted on solid medium. In vivo, a HET-s-GFP fusion protein (GFP, green fluorescent protein) is soluble in [Het-s*] strains but forms aggregates after transition to the [Het-s] prion state (21). In vitro, recombinant HET-s protein undergoes a transition from a soluble to an aggregated state. These aggregates are typical amyloid fibers and catalyze precipitation of the soluble protein (22). These results lead to the suggestion that, as proposed for the yeast prions, HET-s amyloid aggregation in vitro mirrors [Het-s] propagation in vivo.
To determine whether [Het-s]-prion infectivity can be created from recombinant HET-s protein and explore the connection between amyloid formation and prion propagation, we have developed a method to introduce the recombinant HET-s protein into fungal cells. In contrast to yeast, filamentous fungi such as Podospora do not grow as individual cells but as a syncytial structure (the mycelium) composed of a network of filaments divided in articles with incomplete crosswalls. In other words, in filamentous fungi there is a cytoplasmic continuity throughout the mycelium. Consequently, when the [Het-s] prion emerges in one fungal “cell,” it propagates easily through the entire mycelium. [Het-s] is known to propagate within the mycelium very rapidly (up to 70 mm per day) (23). Therefore, this biological system is well suited to detect even very rare conversion events to the prion state. Thus, we chose to introduce the HET-s prion protein into whole mycelium rather than into isolated cells as described for the yeast system (15). Microprojectile bombardment (biolistic) is an efficient way to introduce DNA into various cell types including those displaying a cell wall (24, 25). We successfully used this method to introduce recombinant HET-s into Podospora. We demonstrate that in this system, prion infectivity can be created in vitro from recombinant protein and provide direct support for the hypothesis that amyloid aggregates represent the molecular basis of [Het-s]-prion infectivity.
Materials and Methods
Preparation of HET-s Fibrils and Other Protein Aggregates.
Recombinant HET-s protein expressed as a C-terminal histidine-tagged construct in Escherichia coli was purified under denaturing conditions and renatured in buffer A (150 mM NaCl/100 mM Tris⋅HCl, pH 8/1 mM DTT) as described (22). For spontaneous aggregation, HET-s at 1 mg⋅ml−1 in buffer A was incubated at 4°C for 72 h. For HET-s conversion (induced aggregation), soluble protein at 1 mg⋅ml−1 in buffer A was incubated for 4 h at 4°C after inoculation in a 1:10 ratio with preformed HET-s fibers. Amorphous HET-s aggregates were obtained by adding 10% trichloroacetic acid (wt/vol) to soluble HET-s protein (1 mg⋅ml−1 in buffer A) or by heating at 100°C for 5 min. Aggregates were recovered by centrifugation (10,000 × g), and the pellets were washed in buffer A and sonicated. For light microscopy, fluorescein-labeled aggregated HET-s protein was obtained by incubating soluble HET-s protein (1 mg⋅ml−1) in sodium phosphate buffer (50 mM, pH 8) for 5 min with a 20-fold molar excess of fluorescein isothiocyanate (Sigma). The labeled protein was purified by size exclusion chromatography, and aggregation was induced for 4 h by adding sonicated preformed HET-s aggregates in a 1:10 ratio. Alzheimer β-amyloid peptide (Aβ40) was purchased from Sigma, and fibrils were prepared as described (26). Electron microscopy of protein aggregates was performed on 10,000 × g pellet fractions as described (22).
Biolistic Procedure.
[Het-s*] (prion-free) strains were grown for 2 days at 26°C on solid corn meal agar containing 0.8 M sorbitol. Four strains were grown per Petri dish. HET-s and control proteins were overlaid onto each mycelium in a final volume of 100 μl of buffer A. After evaporation of the buffer, plates were bombarded with 2 mg of tungsten particles (0.4 μm) in a Bio-Rad PDS-1000/He system by using 1,100 psi (1 psi = 6.89 kPa) rupture disks. Plates were incubated for 2 days at 26°C, and two mycelial fragments were sampled from each strain and inoculated on corn meal agar in confrontation with a [Het-S] tester strain. If at least one of the two subcultures produced a “barrage” reaction (abnormal contact line resulting from the incompatibility cell death reaction) with the tester, the strain was scored as positive for the [Het-s] prion phenotype. The [Het-s] prion state of these strains was confirmed by testing their ability to propagate the infectious element to [Het-s*] (prion-free) strains by contact.
Proteinase K Digestion of HET-s Fibrils.
HET-s fibers (100 μg) were digested for 15 min at room temperature with 10 μg of proteinase K. After digestion the resistant fragment was recovered by centrifugation for 15 min at 10,000 × g. The pellet was resuspended in buffer A and analyzed by electron microscopy. After SDS/PAGE, no remaining full-length HET-s protein was detected by Coomassie blue staining or by Western blot.
In Vitro Conversion of the Recombinant HET-s Protein.
For in vitro aggregation experiments, freshly renatured HET-s protein (1 mg⋅ml−1 in buffer A) was inoculated with different HET-s samples in buffer A with a 1:10 ratio. Protein aggregation was analyzed at different time points by centrifugation (50 μl aliquots for 15 min at 10,000 × g). Supernatant and pellet fractions were analyzed by SDS/PAGE followed by Coomassie blue staining, and protein concentration in the supernatant fraction was measured with the Bio-Rad protein assay reagent.
Preparation of HET-s Aggregates from Crude P. anserina Extracts.
A crude protein extract from a [Het-s] expressing HET-s under control of a strong constitutive promoter (21) or of the Δhet-s knockout strain was obtained by osmotic lysis of protoplasts in buffer A and submitted to ultracentrifugation for 2 h at 200,000 × g on 30% sucrose (wt/vol) as described (16). The pellet was resuspended in buffer A and sonicated. By Western blot, the amount of HET-s protein in this sucrose pellet fraction was estimated to be about 1% of the total protein.
Results
Biolistic Introduction of HET-s into Living Cells.
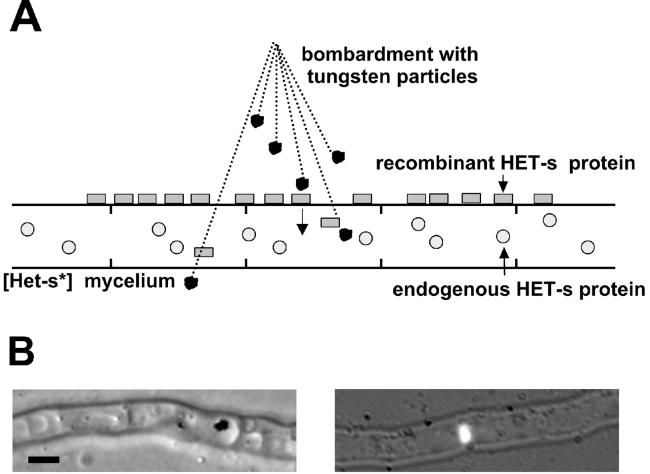
A schematic representation of the biolistic procedure is given Fig. 1A. We have modified the biolistic method by overlaying the protein on the mycelium rather than binding it to the microprojectiles. Microprojectile bombardment was performed on Podospora mycelium grown on solid medium containing 0.8 M sorbitol to try to limit cytoplasmic bleeding. After bombardment, intracellular tungsten particles can be detected in a limited number of cells by light microscopy and are often located in the vacuole (Fig. 1B). We have verified that DNA-mediated transformation of P. anserina with a vector conferring hygromycine resistance can be achieved with this method (not shown). We then used recombinant HET-s labeled with fluorescein to confirm that the protein can be introduced into the mycelium. Again, in a limited number of cells HET-s aggregates can be detected intracellularly by fluorescence microscopy (Fig. 1B). After the biolistic experiment, samples from each bombarded strain were tested for presence of the [Het-s] prion by determining their incompatibility phenotype by confrontation with a [Het-S] tester (Fig. 2)
Figure 1.
Biolistic introduction of recombinant HET-s protein into [Het-s*] mycelium. (A) Schematic representation of the biolistic assay. Mycelium from a [Het-s*] (prion-free) strain is overlaid with recombinant HET-s protein and bombarded with tungsten particles. (B) Light micrographs of mycelium after bombardment. (Left) Tungsten particle located within a fungal vacuole. (Right) Fluorescein-labeled HET-s aggregates detected within a fungal cell. (Bar = 2 μm.)
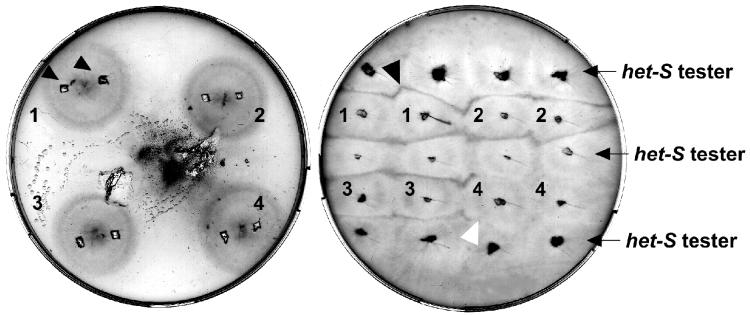
Figure 2.
Phenotypic detection of the [Het-s] prion after microprojectile bombardment. Petri dish after bombardment (Left). The tungsten particles appear clustered in one spot but are actually dispersed over the entire Petri dish. Two inoculates are sampled from each strain (Left, arrowheads) and subcultured in confrontation with a [Het-S] tester strain for incompatibility tests (Right). The black arrow indicates a cell death reaction (“barrage”), and the white arrow indicates a normal contact line. In this example, strains 1–3 have acquired the [Het-s] prion whereas strain 4 has remained in the [Het-s*] (prion-free) state.
Recombinant HET-s Induces the [Het-s] Prion.
First, in control experiments with no protein we have verified that the biolistic procedure per se does not significantly increase spontaneous emergence of [Het-s] (Table 1, from 1 to 3%). Then, when converted or spontaneously aggregated HET-s protein was introduced by biolistics, induction of [Het-s] state was very high: 87–99% of the bombarded strains had acquired the [Het-s] prion state (Table 1). Decreasing the amount of HET-s aggregated protein from 6 μg to 0.04 μg per mycelium did not strongly affect the level of prion induction (Table 1). [Het-s] induction was still unambiguously detected with as little as 10 ng of protein per mycelium. However, when freshly renatured HET-s protein was used in the biolistic assay, only 15 of 149 strains (9%) acquired the [Het-s] prion (Table 1). Because after renaturation HET-s is initially soluble and monomeric (22), these data suggested that [Het-s]-inducing activity is associated with the presence of HET-s aggregates.
Table 1.
[Het-s] induction after biolistic introduction of the recombinant HET-s into [Het-s*] (prion-free) strains
| Protein | Amount of protein | [Het-s] mycelia | % of prion induction |
|---|---|---|---|
| Controls | |||
| No shot | — | 1/126 | 1 |
| No protein | — | 12/416 | 3 |
| BSA | 4.5 | 1/32 | 3 |
| Aggregated HET-s (converted) | 4 | 215/216 | 99 |
| Aggregated HET-s (spontaneous) | 6 | 59/68 | 87 |
| 1 | 54/70 | 77 | |
| 0.3 | 46/64 | 72 | |
| 0.1 | 42/60 | 70 | |
| 0.04 | 24/32 | 75 | |
| 0.01 | 8/32 | 25 | |
| 0.004 | 4/68 | 6 | |
| Freshly renatured HET-s | 3.5 | 16/181 | 9 |
| Proteinase K-digested aggregates | 2† | 56/64 | 87 |
| Amorphous HET-s aggregates | |||
| TCA-precipited | 4 | 3/56 | 5 |
| Heat-denatured | 4 | 2/52 | 4 |
| Aβ40 peptide amyloids | 1.4 | 2/48 | 4 |
The first column gives the protein used in biolistic. The second column gives the amount of protein in micrograms per mycelium. The third column gives the number of strains that acquired the [Het-s] prion over the total number of strains. The last column gives the corresponding percentage of [Het-s] induction. TCA, trichoracetic acid.
Amount of HET-s prior to digestion.
Aggregated but Not Soluble HET-s Induces the [Het-s] Prion.
To assess further whether prion induction was caused by the soluble or aggregated forms of HET-s, a spontaneously aggregating HET-s sample—containing both soluble and aggregated forms—was submitted to 100,000 × g ultracentrifugation. Although the supernatant was still containing about half the amount of total HET-s protein, no infectivity remained in this soluble fraction (Table 2, 5% of [Het-s] induction). [Het-s]-inducing activity was found only in the pellet fraction, indicating that infectivity is associated to the insoluble material. After low-speed centrifugation (2,000 × g), both pellet and supernatant fractions shared a high [Het-s]-inducing activity (Table 2). This result suggests that [Het-s] infectivity is not associated to HET-s aggregates of a specific size. Apparently, various sizes of HET-s aggregates can be infectious.
Table 2.
[Het-s] induction after biolistic introduction of the recombinant HET-s into [Het-s*] (prion-free) strains after centrifugation of the HET-s protein sample
| Centrifugation speed (× g) | 100,000 | 10,000 | 2,000 | |||
|---|---|---|---|---|---|---|
| Fractions | S | P | S | P | S | P |
| Amount of HET-s protein | 2.7 | 3 | 1.6 | 2.3 | 1 | 2.6 |
| No. of [Het-s] strains | 7/132 | 54/60 | 38/224 | 105/144 | 21/32 | 59/69 |
| % of [Het-s] induction | 5 | 90 | 17 | 73 | 67 | 86 |
The first line gives the centrifugation speed (in g). The second line gives the amount of the protein used in micrograms per mycelium for the supernatant (S) and the pellet (P) fractions. The third line gives the number of strains that acquired the [Het-s] prion over the total number of strains and the corresponding percentage of [Het-s] induction.
Amorphous HET-s Aggregates Do Not Induce [Het-s].
Because only aggregates of HET-s are infectious, we propose that HET-s amyloids induce [Het-s] prion by templating aggregation of the endogenous HET-s protein. It was, however, conceivable that HET-s aggregates introduced by biolistics simply increase the rate of spontaneous emergence of [Het-s]. For example, recombinant HET-s aggregates might be dissociated by cellular chaperones (27) thus creating a pool of soluble HET-s. Because it is known that overexpression of HET-s favors emergence of [Het-s] (6), this possible local increase in HET-s concentration might promote de novo generation of [Het-s]. Alternatively, HET-s amyloid aggregates might “saturate” the cellular machinery that serves to prevent protein misfolding and aggregation (chaperones and proteasome) (28) and thus favor spontaneous emergence of [Het-s]—in this case—from endogenous HET-s. If, as implied by these two hypotheses, HET-s amyloids simply increase the rate of spontaneous emergence of [Het-s], then nonamyloid HET-s aggregates or unrelated amyloids should also induce [Het-s]. Amorphous HET-s aggregates were obtained by trichloroacetic acid precipitation or heat denaturation. By electron microscopy, these aggregates were clearly distinct from typical HET-s amyloids (Fig. 3A and data not shown). None of these amorphous HET-s aggregates was able to induce the [Het-s] prion state in the biolistic assay (Table 1, 4 and 5% of induction). We also tested the possibility that an unrelated amyloid aggregate could posses [Het-s]-inducing activity. Amyloid fibers of the Aβ40 were prepared and fibril formation was verified by electron microscopy (not shown). These Aβ40 peptide fibrils did not induce [Het-s] (Table 1). The above experiments strongly suggest that [Het-s] induction does not result from an increase in the rate of spontaneous [HET-s] emergence and that HET-s amyloids indeed template prion replication in vivo.
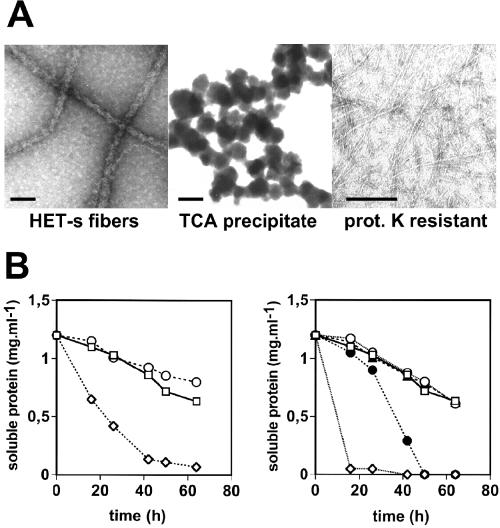
Figure 3.
Electron micrographs of HET-s aggregates used in the biolistic assay and their ability to convert recombinant HET-s. (A) HET-s aggregates were prepared and analyzed by electron microscopy after negative staining. (Bar = 50 nm.) (B) In vitro aggregation assays of recombinant soluble HET-s were inoculated in a 1:10 ratio with various HET-s samples. (Left) Control (○); 100,000 × g supernatant of spontaneously aggregating HET-s (□); 100,000 × g pellet of spontaneously aggregating HET-s (◊). (Right) Control (○); trichloroacetic acid (TCA)-precipitated HET-s (□); heat-denatured HET-s (▴); proteinase K-digested HET-s fibers (●); sonicated HET-s fibers (◊). Aggregation assays were performed as described.
[Het-s]-Inducing Activity Is Resistant to Proteinase K Digestion.
Resistance to proteolysis is a relevant feature of amyloid aggregates and limited proteolysis can be used to identify the highly hydrogen-bonded “core” of amyloid aggregates (29). We therefore determined whether [Het-s]-inducing activity of HET-s aggregates was resistant to proteinase K digestion. The resistant material generated after proteinase K digestion of full-length HET-s fibers remains insoluble and is composed of a fragment of about 7 kDa (22). We have analyzed the resistant material by electron microscopy (Fig. 3A). In contrast to HET-s fibers, the proteinase K-treated fibers display a reduced diameter (about 3–4 nm as opposed to 15–20 nm for the undigested fibers). Proteinase K-digested insoluble material was used in biolistic experiments and was found to display a high level of [Het-s]-inducing activity (Table 1, 87% of induction). Acquisition of proteinase K resistance is a common feature of different prion proteins. Because amyloids are partially resistant to proteolysis, this experiment further supports the notion that the molecular basis of [Het-s] infectivity is an amyloid structure.
[Het-s]-Inducing Activity and Ability to Trigger HET-s Aggregation in Vitro Are Correlated.
To determine whether [Het-s]-inducing activity in the biolistic assay and the ability to convert HET-s into amyloids in vitro are correlated, different HET-s samples used in the biolistic experiments were analyzed for their ability to induce HET-s aggregation in vitro. As expected, HET-s samples displaying [Het-s]-inducing activity (100,000 × g pellet and proteinase K-resistant material) were able to convert soluble HET-s, whereas HET-s samples lacking infectivity (100,000 × g supernatant and amorphous HET-s aggregates) did not (Fig. 3B). Reciprocally, if HET-s aggregates formed in vivo are amyloids, they should be able to seed aggregation of recombinant HET-s. For the yeast [PSI] prion, aggregated Sup35 protein from [PSI+] strains was found to convert the recombinant prion domain of Sup35 into amyloids in vitro (12, 16). Protein extracts of a [Het-s] strain overexpressing HET-s (21) and of a Δhet-s knockout strain were prepared and added to soluble recombinant HET-s. Only the extract containing HET-s aggregates was able to catalyze aggregation of recombinant HET-s protein (Fig. 4). Thus, similar to recombinant HET-s amyloids formed in vitro, HET-s aggregates formed in vivo convert HET-s to aggregates. Together these experiments clearly associate infectivity to converting activity.
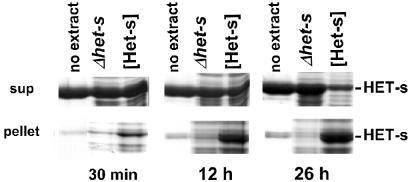
Figure 4.
Extracts from a [Het-s] strain containing HET-s aggregates convert soluble recombinant HET-s into aggregates. Recombinant soluble HET-s protein (1 mg⋅ml−1) was inoculated with an equal amount (wt/wt) of a sucrose pellet fraction from a [Het-s] strain containing HET-s aggregates ([Het-s] lane), or from the het-s-knockout strain (Δhet-s lane), or with buffer A alone (“no extract” lane) and kept at 4°C. The reaction mixture was centrifuged at 10,000 × g for 15 min after 30 min, 12 h, and 26 h, and supernatant and pellet fractions were analyzed by SDS/PAGE and Coomassie blue staining. The position of recombinant HET-s protein, which migrates at 32 kDa, is marked.
Discussion
The present study shows that HET-s aggregates formed in vitro induce the [Het-s] prion when introduced into [Het-s*] (prion-free) strains. This result indicates that prion infectivity can be obtained de novo in vitro from recombinant material. R. B. Wickner (5) defined the expected genetic properties of a prion, and [Het-s] was identified as a prion primarily based on genetic criteria (6). The genetic demonstration of the prion hypothesis for [Het-s] was, however, weaker than for [URE3] and [PSI], because [Het-s] fails to meet one of the relevant genetic criteria (30). Namely, the presence of the prion should produce the same phenotype as mutations altering the gene required for its propagation. The present results now represent a formal demonstration that the [Het-s] element is the prion form of the HET-s protein. Thus far, de novo generation of prion infectivity had been reported only for the [PSI] yeast prion (15). In that work, the prion domain of the Sup35 protein was introduced with a liposome-based transformation procedure. [PSI] induction was increased ≈100-fold; however, the [PSI+] convertants represented only about 1–2% of the transformed strains. The biolistic procedure used here leads to induction of the [Het-s] prion with a very high efficiency—up to 99% of the strains become [Het-s]. There is a fundamental physiological difference between the [Het-s] system and yeast prion systems, because filamentous fungi grow as a syncytial multicellular structure. Biolistic conversion in a single viable “cell” is sufficient to infect the whole mycelium. Because of this cytoplasmic continuity within the mycelium, the biolistic assay proved very efficient and allowed us to clearly demonstrate the infectious character of the recombinant HET-s protein. The present work offers further direct support of the “protein-only” hypothesis. Although the protein-only concept originated from studies on mammalian prions (31, 32), it was the discovery of prions in eukaryotic microorganisms (yeast and filamentous fungi) that allowed formal demonstration of that hypothesis.
We show that [Het-s] infectivity is strictly associated with insoluble material. A soluble HET-s fraction isolated by ultracentrifugation does not induce [Het-s] in our assay. In our previous studies, we could show that in vivo acquisition of the [Het-s] prion is correlated with aggregation of HET-s (21). This aggregation, however, could be shown only by using strains overexpressing HET-s. No HET-s aggregates were detected in wild-type [Het-s] strains (21). We have suggested that the amount of aggregated HET-s protein in wild-type strains was too low to be detected. The present work now confirms that in vivo and in vitro, [Het-s] prion infectivity is based on HET-s aggregate formation. The HET-s aggregates used in the biolistic assay have been identified as amyloids (22). The 10,000 × g pellet fraction analyzed by electron microscopy is essentially composed of fibrillar aggregates; we do not detect any amorphous nonamyloid aggregates. Importantly, we show that [Het-s] infectivity resists proteinase K digestion—a relevant feature of amyloids—and that this proteinase K-digested material remains fibrillar. Moreover, only amyloid HET-s aggregates induce [Het-s], and [Het-s]-inducing activity is correlated with the ability to convert HET-s in vitro. Together these results indicate that the HET-s amyloid aggregates are responsible for [Het-s] induction. Although, ultracentrifugation completely removes infectious HET-s material, [Het-s]-inducing activity is found in the 10,000 × g and 2,000 × g supernatant fractions. This observation suggests that aggregates of a large range of sizes are capable of inducing [Het-s]. We propose that HET-s amyloids induce [Het-s] by templating prion replication in vivo. Recombinant HET-s aggregates introduced in vivo presumably recruit soluble HET-s monomers. Subsequent, breakage of this prion seed would then allow reproduction of the infectious particle (33).
The mechanism of HET-s fibrillization awaits detailed characterization but it is know that amyloid formation occurs by means of the formation of ordered nuclei and oligomeric prefibrillar intermediates (34). Such oligomeric intermediates have in particular been described during the in vitro polymerization of the prion domain of yeast Sup35 (35). The role of mature fibrils and oligomeric intermediates in in vivo infectivity is an unresolved question. It has been suggested that in vivo, large amyloid aggregates could represent secondary “dead-end” products in the prion replication process with no active role in infectivity (17, 36). We find no significant difference in the level of [Het-s]-inducing activity for samples undergoing spontaneous aggregation and presumably containing aggregation intermediates and converted HET-s samples aggregated to completion. Moreover, a significant fraction of the infectious material can be sedimented at low-speed centrifugation. We therefore suggest that “mature” fibrils generated in vitro can be infectious. In the case of the Aβ40 peptide, direct addition of peptide monomers to the fiber end without formation of intermediates has been reported (37). We find that HET-s aggregates formed in vivo are able to convert recombinant soluble HET-s, suggesting as reported for the yeast prions that HET-s amyloid or at least amyloid-like aggregates exist in vivo (10, 16, 38). Whether such aggregates represent bona fide infectious material in vivo remains to be clarified. We hypothesized that the [Het-s]-associated cell death reaction could be caused by oligomeric aggregation intermediates as proposed in the case of human protein deposition diseases (21). In that hypothesis, HET-s amyloid formation may have a protective role. Our results suggest that amyloid fibrils generated in vitro are able to template prion replication in vivo and are not simple dead-end products, at least in terms of prion propagation. It is conceivable that in vivo, large HET-s aggregates are still capable of propagating [Het-s] but are not active in the incompatibility cell death reaction.
For the Ure2p and Sup35 yeast prion proteins, a prion domain can be delimited to an N-terminal N/Q-rich region (see ref. 39 for a review). This region is responsible both for prion propagation in vivo and amyloid formation in vitro. HET-s lacks such an N/Q-rich region. We show that the 7-kDa proteinase K-resistant HET-s fragment displays prion infectivity in the biolistic assay. This result suggests that the prion properties of HET-s are also the results of a subregion of the protein. We propose that the HET-s prion domain could correspond to the proteinase K-resistant “core” of the HET-s amyloid aggregate. Preliminary experiments indicate that the corresponding 7-kDa peptide is able to form amyloids in vitro and propagate [Het-s] in vivo (S.D.R. and S.J.S., unpublished data).
There is ever-growing evidence that amyloid formation and prion propagation (at least in the case of fungal prions) are intimately linked. However, the molecular nature of the bona fide infectious material that propagates in vivo remains unknown. Further studies directly connecting in vitro aggregation and in vivo propagation are needed to clarify this issue. Based on its sensitivity, this experimental procedure described here might represent a useful tool to assess further the role of potential aggregation intermediates and mature fibrils in [Het-s] prion infectivity. This system could also be used to assess infectivity of other amyloid proteins, namely fungal N/Q-rich proteins or polypeptides involved in protein deposition diseases. One could for instance determine whether biolistic introduction of an amyloid protein is able to seed the self-propagating aggregation of the corresponding protein expressed as a green fluorescent protein fusion in Podospora.
Acknowledgments
We thank J. Bégueret for his suggestions and support, M. Sicault-Sabourin for technical assistance, O. Cuvillier for critical reading of the manuscript, J. P. DiRago for the use of the biolistic equipment, C. Cullin for helpful discussion, and the referees for valuable comments. This research is supported by the Centre National de la Recherche Scientifique and by the Ministère de la Recherche (Groupement d'Intérêt Scientifique Prion). M.-L.M. is a recipient of a fellowship from the Fondation pour la Recherche Médicale. S.D.R. and S.D.C. are fellows of the Ministère de la Recherche et l'Enseignement Supérieur.
Note About the Terminology
The het-s locus exists as two alleles designated het-s and het-S. HET-s and HET-S designate the corresponding protein products. Strains expressing the HET-s protein display two alternate phenotypes : [Het-s] and [Het-s*]. Strains expressing the HET-S protein display the [Het-S] phenotype. After confrontation, [Het-s] and [Het-S] strains produce a barrage reaction (i.e., are incompatible) whereas [Het-s*] and [Het-S] strains are compatible (for a review see ref. 20).
References
- 1.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Caughey B. Nature (London) 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 3.Kaneko K, Peretz D, Pan K M, Blochberger T C, Wille H, Gabizon R, Griffith O H, Cohen F E, Baldwin M A, Prusiner S B. Proc Natl Acad Sci USA. 1995;92:11160–11164. doi: 10.1073/pnas.92.24.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill A F, Antoniou M, Collinge J. J Gen Virol. 1999;80:11–14. doi: 10.1099/0022-1317-80-1-11. [DOI] [PubMed] [Google Scholar]
- 5.Wickner R B. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 6.Coustou V, Deleu C, Saupe S, Begueret J. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caughey B. Nat Med. 2000;6:751–754. doi: 10.1038/77476. [DOI] [PubMed] [Google Scholar]
- 8.Serio T R, Lindquist S L. Trends Cell Biol. 2000;10:98–105. doi: 10.1016/s0962-8924(99)01711-0. [DOI] [PubMed] [Google Scholar]
- 9.Wickner R B, Taylor K L, Edskes H K, Maddelein M L, Moriyama H, Roberts B T. J Struct Biol. 2000;130:310–322. doi: 10.1006/jsbi.2000.4250. [DOI] [PubMed] [Google Scholar]
- 10.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 11.Glover J R, Kowal A S, Schirmer E C, Patino M M, Liu J J, Lindquist S. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 12.DePace A H, Santoso A, Hillner P, Weissman J S. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 13.Liu J J, Lindquist S. Nature (London) 1999;400:573–576. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- 14.Taylor K L, Cheng N, Williams R W, Steven A C, Wickner R B. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 15.Sparrer H E, Santoso A, Szoka F C, Jr, Weissman J S. Science. 2000;289:595–599. doi: 10.1126/science.289.5479.595. [DOI] [PubMed] [Google Scholar]
- 16.Uptain S M, Sawicki G J, Caughey B, Lindquist S. EMBO J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Bellot E, Guillemet E, Ness F, Baudin-Baillieu A, Ripaud L, Tuite M, Cullin C. EMBO Rep. 2002;3:76–81. doi: 10.1093/embo-reports/kvf011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deleu C, Clave C, Begueret J. Genetics. 1993;135:45–52. doi: 10.1093/genetics/135.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass N L, Jacobson D J, Shiu P K. Annu Rev Genet. 2000;34:165–186. doi: 10.1146/annurev.genet.34.1.165. [DOI] [PubMed] [Google Scholar]
- 20.Saupe S J. Microbiol Mol Biol Rev. 2000;64:489–502. doi: 10.1128/mmbr.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coustou-Linares V, Maddelein M L, Begueret J, Saupe S J. Mol Microbiol. 2001;42:1325–1335. doi: 10.1046/j.1365-2958.2001.02707.x. [DOI] [PubMed] [Google Scholar]
- 22.Dos Reis S, Coulary-Salin B, Forge V, Lascu I, Begueret J, Saupe S J. J Biol Chem. 2002;277:5703–5706. doi: 10.1074/jbc.M110183200. [DOI] [PubMed] [Google Scholar]
- 23.Beisson-Schecroun J. Ann Genet. 1962;4:3–50. [PubMed] [Google Scholar]
- 24.Johnston S A. Nature (London) 1990;346:776–777. doi: 10.1038/346776a0. [DOI] [PubMed] [Google Scholar]
- 25.Lorito M, Hayes C K, Di Pietro A, Harman G E. Curr Genet. 1993;24:349–356. doi: 10.1007/BF00336788. [DOI] [PubMed] [Google Scholar]
- 26.Holm Nielsen E, Nybo M, Svehag S E. In: Amyloid, Prions, and Other Protein Aggregates. Wetzel R, editor. Vol. 309. San Diego: Academic; 1999. pp. 491–495. [Google Scholar]
- 27.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 28.Wickner S, Maurizi M R, Gottesman S. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 29.Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 30.Wickner R B. Proc Natl Acad Sci USA. 1997;94:10012–10014. doi: 10.1073/pnas.94.19.10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffith J S. Nature (London) 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 32.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 33.Jackson G S, Clarke A R. Curr Opin Struct Biol. 2000;10:69–74. doi: 10.1016/s0959-440x(99)00051-2. [DOI] [PubMed] [Google Scholar]
- 34.Rochet J C, Lansbury P T., Jr Curr Opin Struct Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 35.Serio T R, Cashikar A G, Kowal A S, Sawicki G J, Moslehi J J, Serpell L, Arnsdorf M F, Lindquist S L. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 36.Tuite M F. Science. 2000;289:556–557. doi: 10.1126/science.289.5479.556. [DOI] [PubMed] [Google Scholar]
- 37.Tseng B P, Esler W P, Clish C B, Stimson E R, Ghilardi J R, Vinters H V, Mantyh P W, Lee J P, Maggio J E. Biochemistry. 1999;38:10424–10431. doi: 10.1021/bi990718v. [DOI] [PubMed] [Google Scholar]
- 38.Speransky V V, Taylor K L, Edskes H K, Wickner R B, Steven A C. J Cell Sci. 2001;153:1327–1336. doi: 10.1083/jcb.153.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickner R B, Taylor K L, Edskes H K, Maddelein M L. Curr Biol. 2000;10:335–337. doi: 10.1016/s0960-9822(00)00460-7. [DOI] [PubMed] [Google Scholar]