Abstract
BRAF35, a structural DNA-binding protein, initially was identified as a component of a large BRCA2-containing complex. Biochemical analysis revealed the presence of a smaller core–BRAF35 complex devoid of BRCA2. Here we report the isolation of a six-subunit core–BRAF35 complex with the capacity to deacetylate histones, termed the BRAF–histone deacetylase complex (BHC), from human cells. BHC contains polypeptides reminiscent of the chromatin-remodeling complexes SWI/SNF and NuRD (nucleosome remodeling and deacetylating). Similar to NuRD, BHC contains an Mi2-like subunit, BHC80, and a PHD zinc-finger subunit as well as histone deacetylases 1/2 and an MTA-like subunit, the transcriptional corepressor CoREST. We show that BHC mediates repression of neuron-specific genes through the cis-regulatory element known as the repressor element 1 or neural restrictive silencer (RE1/NRS). Chromatin-immunoprecipitation experiments demonstrate the recruitment of BHC by the neuronal repressor REST. Expression of BRAF35 containing a single point mutation in the HMG domain of the protein abrogated REST-mediated transcriptional repression. These results demonstrate a role for core–BRAF35-containing complex in the regulation of neuron-specific genes through modulation of the chromatin structure.
The genome of eukaryotes is packaged into chromatin, the fundamental unit of which is the nucleosome. The higher order chromatin structure is formed by arrangement of nucleosomes into an array. Such higher order chromatin structure presents a barrier to cellular processes such as transcription, DNA replication, and DNA repair. Therefore, controlling accessibility to the nucleosomal DNA provides an important regulatory point in these processes (1).
Recent genetic and biochemical studies have culminated in the discovery of a host of multisubunit complexes that, in an ATP-dependent manner, are able to alter the structure of the nucleosome. The first of such multiprotein complexes, the SWI/SNF complex, was discovered initially through genetic studies in yeast, and its catalytic subunit, SWI2/SNF2, was identified as the DNA-dependent ATPase (2–4). A complex similar to that of the SWI/SNF complex was identified recently in yeast (RSC), which unlike the SWI/SNF complex is essential for growth (5). Complexes homologous in polypeptide composition and biochemical activity to that of SWI/SNF have been identified in other organisms (6–10). More recently, a number of groups reported the isolation and characterization of a complex termed NuRD (nucleosome remodeling and deacetylating, also NURD and NRD), that not only contains a DNA-dependent ATPase subunit but also histone deacetylase (HDAC) 1/2 (11–13).
In addition to such chromatin-remodeling complexes a number of transcriptional regulatory complexes have been identified that contain histone acetylation or deacetylation activities. It was shown previously that the hyperacetylated chromatin correlates with active genes, whereas the repressed genes exhibit a pattern of hypoacetylation (14, 15). This contention was strengthened by the discovery of the association of a number of transcriptional corepressors with histone deacetylation activity. In particular, the transcriptional corepressor Sin3 was shown to be in a multiprotein complex containing HDAC activity (16–18). This complex was shown to act as a transcriptional corepressor for a number of DNA-binding repressors including Mad, the nuclear hormone receptors, and the RE1-binding silencer protein, REST (16, 19–22). It was further shown that the Sin3 protein interacts with the N-terminal repression domain of REST (20, 21). REST (also called NRSF) is a multi-zinc-finger protein that repress the expression of number of neuronal-specific genes in nonneuronal cells (23, 24).
In the course of purifying the BRCA2–BRAF35 complex (25), we noticed that a portion of BRAF35 is in a complex devoid of BRCA2. Here, we describe the isolation and a complete identification of the components of a core–BRAF35 complex termed BHC for BRAF–HDAC complex. The realization that the neuronal corepressor CoREST (26) was an integral component of this complex prompted us to investigate the role of BHC in mediating repression of neuronal-specific genes. We show that BHC interacts with the promoter of the synapsin gene and mediates its RE1-dependent repression.
Materials and Methods
Conventional Chromatographic Purification of BHC.
BHC was purified from 2 g of HeLa nuclear extract. Nuclear extract was loaded on a 250-ml column of phosphocellulose (P11, Whatman) and fractionated stepwise by the indicated KCl concentrations in buffer A (20 mM Tris⋅HCl, pH 7.9/0.2 mM EDTA/10 mM βME/10% glycerol/0.2 mM PMSF). The P11 0.5 M KCl fraction (250 mg) was loaded on a 45-ml DEAE-Sephacel column (Amersham Pharmacia) and eluted with 0.35 M KCl. The 0.35 M KCl elution (140 mg) was dialyzed to 700 mM NH4SO4 in buffer HB (20 mM Hepes, pH 7.6/4 mM DTT/0.5 mM EDTA/10% glycerol/0.5 mM PMSF/1 μg/ml aprotinin/1 μg/ml leupeptin/and 1 μg/ml pepstatin) and loaded on butyl-Superose (Amersham Pharmacia). The column was resolved by using a linear 10-column volume gradient of 700–0 mM NH4SO4 in buffer HB. BHC-containing fractions were dialyzed to 10 mM KxPO4 in buffer HA (5 mM Hepes, pH 7.6/1 mM DTT/0.5 mM PMSF/10 μM CaCl2/10% glycerol/40 mM KCl/1 μg/ml apro-tinin/1 μg/ml leupeptin/1 μg/ml pepstatin) and loaded on a BioScale CHT5-I column (Bio-Rad). The column was resolved by using a linear 15-column volume gradient of 10–600 mM KxPO4 in buffer HA. Fractions containing BHC were dialyzed to 100 mM KCl in buffer A containing 1 μg/ml aprotinin, leupeptin, and pepstatin and loaded on heparin-5PW (TosoHaas, Montgomeryville, PA). The column was resolved by using a linear 20-column volume gradient of 100–450 mM KCl in buffer A containing 1 μg/ml aprotinin, leupeptin, and pepstatin. BHC-containing fractions were fractionated on a MonoS 5/5 (Amersham Pharmacia) equilibrated in 0.1 M KCl in buffer A and resolved by using a linear 10-column volume of 100–500 mM KCl and 1 μg/ml aprotinin, leupeptin, and pepstatin.
Immunoaffinity Purification of BHC.
HeLa nuclear extract (1.2 g) was fractionated according to the protocol described above by using P11 and DEAE-Sephacel columns. The DEAE-Sephacel pool then was dialyzed to 150 mM KCl in buffer D (20 mM Hepes, pH 7.9/0.25 mM EDTA/20% glycerol/0.1% Tween 20). Anti-BRAF35 antibodies (300–500 μg, C-terminal) were cross-linked to protein A-Sepharose (1 ml, Repligen) by using standard techniques (27) for affinity purification of BHC. The fractionated nuclear extract (DEAE-Sephacel, 10 mg) was incubated with 1 ml of antibody-protein A beads for 4–5 h in buffer D at 4°C. The beads were washed first with 0.5 M KCl in buffer D followed by a wash with 0.5 M KCl buffer D and 0.2 M guanidine hydrochloride. The beads then were washed with 100 mM KCl in buffer D, and the proteins were eluted with 0.1 M glycine, pH 2.5, and neutralized with a 1:10 volume of 1.0 M Tris⋅HCl, pH 8.0.
Affinity Purification of Flag-BRAF35.
Flag-BRAF35 and a selectable marker for puromycin resistance were cotransfected into 293 human embryonic kidney cells by calcium-phosphate coprecipitation. Transfected cells were grown in the presence of 10 μg/ml puromycin, and individual colonies were isolated and analyzed for Flag-BRAF35 expression. A cell line expressing Flag-tagged BRAF35, FBRAF35-4, were used for the affinity purification of Flag-BHC as described previously for the Flag-Ini 1–11 cell line (10, 28).
Immunoblot Analysis.
Anti-BRAF35 and CoREST antibodies have been described (25, 26). Anti-HDAC2 antibodies were obtained from Zymed. Immunoblotting with alkaline phosphatase was performed as described (10). Polyclonal antibodies to BHC110 and BHC80 were generated against an N-terminal 20 amino acids of each protein.
Transient Transfection Analysis.
The minimal type II promoter or the minimal type II promoter with the RE1 site has been described (29). These promoters were cloned into pGL2-Basic (Promega) to create the type II and type II-RE1 luciferase reporter vectors. BHC80 and BRAF35 were cloned in pFlag-CMV2 (Sigma) by using standard PCR techniques. BRAF35 was subcloned into pFlag-CMV2 (Sigma) to create the expression vector Flag-BRAF35. Flag-BRAF35(K116I) was constructed by using the PCR-based overlap extension method of oligonucleotide-directed mutagenesis (30) by using Flag-BRAF35 as template.
Twenty-four hours before transfection, the cells were plated (80% confluence) in 6-well tissue-culture dishes. Transient transfection experiments were performed by the calcium-phosphate coprecipitation method according to Chen and Okayama (31). A total of 5 μg of DNA, including 1 μg of reporter plasmid, 0.5 μg of β-galactosidase expression plasmid (pSV-β-gal, Promega) as an internal control, with or without various amounts of expression plasmid of the effector gene, and pUC18 DNA, was cotransfected into 293 cells. After 48 h of incubation, cells were harvested and extracted in 100 μl of lysis buffer by freezing and thawing. Cell extraction, luciferase assays, and β-galactosidase assays were performed according to manufacturer instructions (Promega). Luciferase activity was normalized to β-galactosidase activity, and all experiments were performed six times in at least three separate experiments.
Chromatin Immunoprecipitation (ChIP).
ChIPs were performed by using a modified protocol from Upstate Biotechnology (Lake Placid, NY). Cells in a 10-cm dish (70% confluent) were treated for 10 min with 1% formaldehyde at room temperature. The cells were lysed in cell lysis buffer (5 mM Hepes, pH 8.0/85 mM KCl/0.5% Triton X-100), and the nuclei were resuspended in nuclei lysis buffer (50 mM Tris⋅HCl, pH 8.0/10 mM EDTA/1% SDS). The lysate was sonicated under conditions yielding fragments ranging from 200 to 1,000 bp. Samples were subsequently precleared at 4°C with recombinant protein G agarose beads (GIBCO) coated in salmon-sperm DNA and yeast tRNA for 1 h. Precleared lysate (100 μl) diluted in immunoprecipitation buffer (0.01% SDS/1.1% Triton X-100/1.2 mM EDTA/16.7 mM Tris⋅HCl, pH 8.0/167 mM NaCl) was used for a 12-h immunoprecipitation with 5 μg of specific antibody at 4°C. Complexes were collected for 4 h by using recombinant protein G agarose beads (GIBCO) coated in salmon-sperm DNA and yeast tRNA. After washing and elution, formaldehyde cross-linking was reversed with a 12-h incubation at 65°C. Samples were purified through PCR purification kit columns (Qiagen, Chatsworth, CA) and used as a template in PCRs to detect specific templates.
Results
Affinity Purification of BHC.
We previously described BRAF35 as a component of at least two complexes, one of which did not contain BRCA2 (25). To purify the core–BRAF35 complex, we used affinity purification with polyclonal anti-BRAF35 antibodies following the protocol outlined in Fig. 1a. To obtain a core–BRAF35 complex, the affinity matrix was subjected to a high-salt wash (0.5 M KCl) followed by washing with 0.2 M guanidine hydrochloride. This purification resulted in the isolation of a complex containing six polypeptides termed BHC (Fig. 1b).
Figure 1.
Affinity purification of BHC. (a) Purification scheme. HeLa nuclear extract (NE) was fractionated by using P11 chromatography. The 0.5 M KCl elution was concentrated on a DEAE-Sephacel column and purified further by using an anti-BRAF35 antibody column (α-BRAF35). The bound proteins were washed with buffer containing 0.5 M KCl and 0.2 M guanidine hydrochloride and eluted by using 0.2 M glycine, pH 2.5. (b) Polypeptide composition of the affinity-purified BHC. Affinity-purified BHC was analyzed by SDS/PAGE followed by silver staining or Colloidal blue. α-IgG represents the control preimmune IgG eluate. Molecular mass markers are shown to the left of the figure. (c) Primary amino acid sequence of BHC80. Peptides sequenced by ion-trap mass spectrometry are double-underlined. The acidic Q/P-rich region is underlined, and the leucine zipper and the PHD domains are represented by black and gray shadings, respectively. (d) Diagrammatic representation of BHC80 structural domains. (e) Reverse transcription–PCR representing the expression profile of BRAF35 and BHC80 in different human tissues. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Identification of the BHC Subunits Indicates that BHC80 Is a PHD Zinc-Finger Protein.
Subunits of BHC were cut from a colloidal stained gel, digested with trypsin, and subjected to sequencing by ion-trap mass spectrometry (10). Seventeen tryptic peptides identified BHC110 as the protein encoded by KIAA0601. Five peptide sequences identified BHC80 as a previously uncharacterized protein (Fig. 1c). Interestingly, BHC80 contains a PHD zinc-finger domain most similar to that of the Mi2 protein. In addition, BHC80 contains a C-terminal and an N-terminal leucine zipper (Fig. 1d), one of which has high homology with the leucine zipper contained in the transcriptional activator c-Myc (32). BHC60, BHC57, BHC55, and BHC35 were identified as the transcriptional corepressor CoREST (26), HDAC 1/2, and BRAF35, respectively. Analysis of BRAF35 expression levels revealed a nearly ubiquitous pattern of expression in different tissues, with liver and heart displaying lower levels of expression (Fig. 1e). In contrast to BRAF35, BHC80 expression is highly tissue-specific, with brain expressing the highest level of BHC80 analyzed (Fig. 1e).
Purification of BHC by Conventional Chromatography.
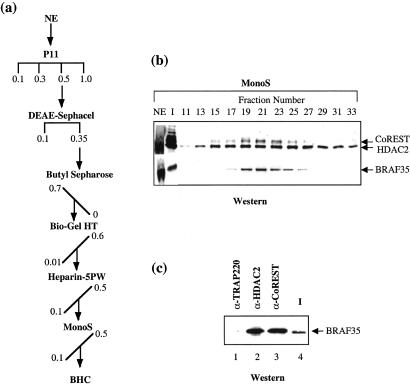
To establish that BHC is a stable multisubunit complex we followed CoREST, HDAC2, and BRAF35 immunoreactivity through six chromatographic steps (Fig. 2a). Analysis of the final fractionation step revealed the coelution of BRAF35, CoREST, and HDAC2 (Fig. 2b). However, HDAC2 eluted with a broader peak, indicating that the later fractions (fraction 25) may contain a second HDAC2-containing complex. Moreover, immunoprecipitation of BHC by anti-HDAC2 antibodies or anti-CoREST antibodies specifically precipitated BRAF35 (Fig. 2c).
Figure 2.
Conventional purification of BHC. (a) Purification scheme. HeLa nuclear extract (NE) was fractionated by chromatography as described in Materials and Methods. The horizontal and diagonal lines indicate stepwise and gradient elution, respectively. Concentrations are given as molar. (b) Western blot analysis of the Mono S fractions (15 μl) using antibodies shown to the right of the figure. (c) Western blot analysis of immunoeluates of antibodies shown on the top of the figure using anti-BRAF35 antibodies. Input (I) is the BHC pool from the 0.5 M P11 fractions (10 μl). P11 (100 μl of 0.5 M) was used for immunoprecipitation.
Purification of BHC from a Stable Flag-Tagged BRAF35 Cell Line.
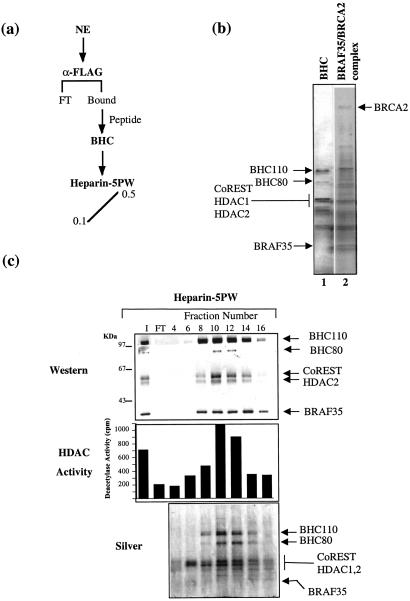
We also purified BHC from a 293-derived cell line expressing Flag-BRAF35, FBRAF35-4. Flag-BRAF35 was affinity-purified from FBRAF35-4 nuclear extract by using anti-Flag antibodies followed by elution of the bound material with Flag peptide (Fig. 3a). Interestingly, the early elutions from the Flag-affinity matrix contained the core–BRAF35 complex, whereas the later fractions contained the BRAF35–BRCA2 complex (Fig. 3b). To rigorously demonstrate the association of all six polypeptides, the BHC-containing fractions were chromatographed further on a heparin-5PW column. Analysis of the column fractions revealed the coelution of all six polypeptides with HDAC activity (Fig. 3c). However, a small fraction of BRAF35 elutes at a higher salt concentration (fraction 14), indicating that a portion of Flag-BRAF35 seems to be in a monomeric form.
Figure 3.
Affinity purification of human BHC from Flag-BRAF35 cells. (a) Purification scheme. Nuclear extract (NE) from FBRAF35-4 cells was fractionated by using an anti-Flag M2 affinity column. Bound proteins were analyzed further by chromatography on a heparin-5PW column. (b) Silver-stain analysis of the anti-Flag affinity eluates (15 μl) corresponding to early (lane 1) and late (lane 2) eluting fractions. (c) Western blot, HDAC activity, and silver-stain analysis of the heparin-5PW column fractions (15 μl). Western blotting was performed by using antibodies shown to the right of the figure.
BHC Mediates RE1-Dependent Transcriptional Repression.
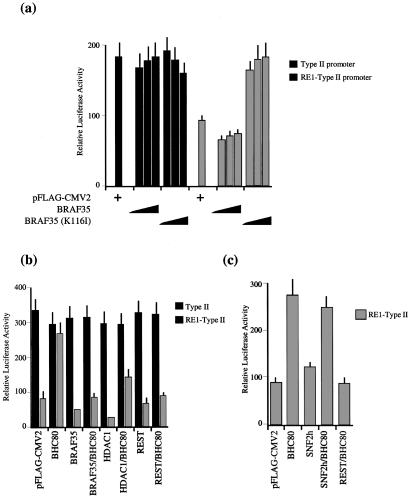
CoREST was shown to interact with the transcriptional repressor REST and function as a corepressor (26). REST binds to RE1 elements and represses the transcription of neuronal genes in nonneuronal cells (23, 24). Consistent with a previous report (21), the addition of HDAC inhibitor tricostatin A to 293 cells resulted in the derepression of endogenous RE1-containing promoter (data not shown). Therefore, we next asked whether BHC is required for RE1-mediated repression of transcription. To assess the role of BHC in RE1-mediated transcriptional repression, we analyzed the effect of BRAF35 and a mutant form of BRAF35 that contains a single point mutation in the HMG domain, BRAF35(K166I). Similar mutations in other sequence-nonspecific HMG domains are shown to abrogate the DNA-binding ability of the HMG domain (33, 34). Although titration of BRAF35 slightly enhanced the repression from the RE1-containing reporter in 293 cells, titration of BRAF35(K116I) completely abrogated the transcriptional repression from the RE1-containing reporter and restored transcription to levels obtained from the promoter lacking the RE1 sites (Fig. 4a). These results reveal that BRAF35(K116I) functions as a dominant negative and point to the role of the HMG domain of BRAF35 in mediating RE1-dependent transcriptional repression.
Figure 4.
BHC mediates RE1-dependent transcriptional repression. (a) Graph of relative luciferase activity after cotransfection of 293 cells. Transfection and luciferase assay were carried out as described in Materials and Methods. The cells were cotransfected with the reporter plasmid of either type II promoter lacking the RE1 element (black) or RE1-containing type II promoter (gray) and various concentrations of BRAF35 (0.3, 1, or 2 μg of DNA) and BRAF35(K116I) (0.3, 1, or 2 μg of DNA) or with pFlag-CMV2 control vector (2 μg of DNA). The standard errors are indicated by the thin vertical lines. Each point represent six measurements, and the experiment was repeated six independent times. (b) Graph of relative luciferase activity after cotransfection of 293 cells with the reporter construct of RE1-containing type II sodium-channel promoter or the type II promoter lacking RE1 sites in addition to pFlag-CMV2 (2 μg of DNA), BHC80 (1 μg of DNA), BRAF35 (1 μg of DNA), HDAC1 (1 μg of DNA), and REST (1 μg of DNA). The standard errors are indicated by the thin vertical lines. Each point represents six measurements. (c) Graph of relative luciferase activity after cotransfection of 293 cells with the RE1-containg type II sodium-channel promoter and the following constructions: pFlag-CMV2 (2 μg of DNA), BHC80 (1 μg of DNA), SNF2h (1 μg of DNA), and REST (1 μg of DNA). The standard errors are indicated by the thin vertical lines.
To analyze the contribution of BHC80 to RE1-mediated transcription, we first tested the effect of wild-type BHC80 on RE1-dependent transcriptional repression. In contrast to BRAF35, transfection of BHC80 completely abrogated the RE1-dependent transcriptional repression (Fig. 4b). Because this effect of BHC80 may be caused by squelching of competent repressive complexes, we reasoned that the addition of increasing concentrations of other components of the complex might rescue the BHC80 effect. Indeed, the derepression activity of BHC80 could be reversed by addition of BRAF35 or HDAC1, although HDAC1 only partially restored the repression (Fig. 4b). The partial effect of overexpressing HDAC1 on overcoming the BHC80-mediated antirepression may represent an HDAC1-independent mechanism of repression by BHC (for example, through the HMG domain of BRAF35). Moreover, although addition of exogenous REST in 293 cells, which already contained REST, only slightly increased the RE1-mediated repression, it completely overcame the antirepressory effect of BHC80 on the RE1-mediated repression (Fig. 4b). This effect of the BHC components and REST is specific, because the addition of other chromatin-modifying proteins does not effect BHC80-mediated antirepression (Fig. 4c). These results support a role for BHC in REST-mediated transcriptional repression.
BHC Is Recruited to RE1 Sites Occupied by Transcriptional Repressor REST.
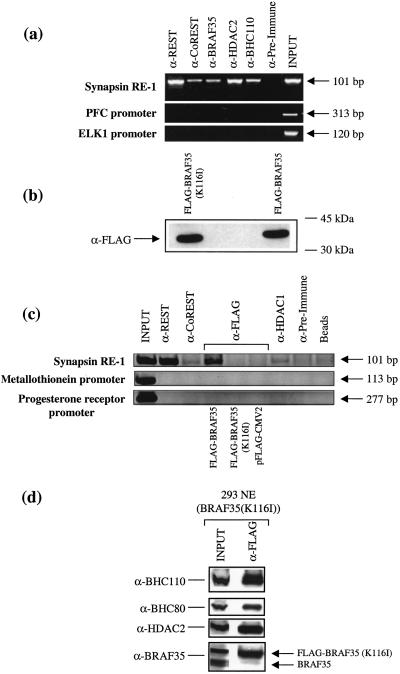
These results prompted us to ask whether BHC is recruited to the endogenous REST-binding sites in the synapsin promoter by using ChIP. This analysis revealed the specific association of CoREST, HDAC2, BHC110, and BRAF35 with DNA containing REST-binding sites in 293 cells (Fig. 5a). It is noteworthy that anti-REST antibodies brought down a greater amount of synapsin RE1 as compared with the components of BHC, which may reflect the fact that BHC components associate with other transcriptional repressor proteins bound to other sites. Therefore the presence of these other sites may dilute the amount of synapsin RE1 fragment that is brought down by anti-BHC antibodies.
Figure 5.
BHC is recruited to the endogenous synapsin gene. (a) ChIP in 293 cells using antibodies shown on the top of the figure. PCR products corresponding to synapsin, PFC, and ELK1 promoters are displayed as 101-, 313-, and 120-bp bands on an ethidium bromide-stained agarose gel, respectively. (b) Western blot analysis using anti-Flag antibodies after transfection of Flag-BRAF35 or Flag-BRAF35(K116I) in 293 cells. (c) ChIP was performed by using formaldehyde-treated HeLa or 293 cells, respectively. After transfection with either Flag-BRAF35 or Flag-BRAF35(K116I), antibodies to REST, CoREST, HDAC2, and the Flag epitope were used to immunoprecipitate complexes associated with the synapsin RE1 sites. The PCR products are displayed as 101-, 113-, and 277-bp bands on an ethidium bromide-stained agarose gel. (d) Western blot analysis using antibodies shown to the left of the figure of Flag-affinity eluate purified using 293 cells stably expressing BRAF35(K116I). NE, nuclear extract.
We then asked whether the mutant BRAF35(K116I) displaying the antirepression activity in transcription has lost its ability to interact with the DNA in vivo with ChIP. Interestingly, although both Flag-BRAF35 and Flag-BRAF35(K116I) were expressed to the same level (Fig. 5b), Flag-BRAF35(K116I) did not associate with the promoter DNA (Fig. 5c). Because the mutant BRAF35(K116I) retains the ability to associate with the other subunits of BHC (Fig. 5d), the absence of BRAF35(K116I) at the promoter may result from a role of the HMG domain in the recruitment step of BHC.
Discussion
We have purified and characterized BHC, a six-subunit BRAF35-containing complex that exhibits histone deacetylation activity. The polypeptide composition of the BHC has been completely defined, and therefore its structure and mechanism of action are open to study. The subunit composition of BHC resembles that of the two chromatin-remodeling complexes SWI/SNF and NuRD. Both the BHC and NuRD complexes share the HDAC 1/2 subunits. In addition, whereas BHC80 and CoREST contain similar domains to the Mi2 and MTA1 proteins, BHC110 and BRAF35 share motifs in common with the BAF170 and BAF57 subunits of the SWI/SNF complex. This conservation of domain structure among different chromatin-modifying complexes may not only underlie similar protein–protein interaction within the subunits of each complex but also represent a common mechanism by which they interact with the chromatin structure. The association of HDAC1/2 with CoREST (35–37) and BHC110 (35, 36) was reported recently.
The presence of the CoREST protein in BHC prompted us to ask whether BHC mediates repression by the transcriptional repressor REST. This question was explored by using multiple approaches. We first show that the endogenous RE1-responsive genes were sensitive to HDAC inhibitors, suggesting the contribution of HDACs in REST-mediated repression. We then generated a point mutant of BRAF35 by changing a conserved lysine residue within the HMG domain to isoleucine (K116I). Consistent with previous work with identical point mutations in SRY and BAF57 HMG-containing proteins (33, 34), BRAF35(K116I) displayed decreased binding activity to synapsin promoter in vivo. Moreover, increasing concentrations of BRAF35(K116I) abrogated the REST-mediated transcriptional repression. These results not only reveal a role for the BRAF35-containing complex in mediating REST-dependent repression but also implicate a critical role for the HMG domain of BRAF35 in transcriptional repression. Finally, we show that the overexpression of the BHC80 gene reverses the REST-dependent repression, and this effect can be rescued by the addition of REST, BRAF35, or HDAC1. We attribute this effect of BHC80 to competition of the exogenously overexpressed protein for the component of REST-repressive complexes and consequently interference with the transcriptional repression.
Both sequence-specific and nonspecific DNA architectural proteins have been identified. We have shown previously that BRAF35 is an architectural DNA-binding protein with a preference for four-way DNA junctions or cruciform DNA (25). The emerging theme for the function of such architectural DNA-binding proteins is their ability to increase the cooperative binding of sequence-specific activators to their regulatory sites (38–40). Therefore, BRAF35 by virtue of its HMG domain may stabilize REST DNA binding, which may account for the requirement of the HMG domain in transcriptional repression.
Repression of neuronal-specific genes is of fundamental importance in the development of both neuronal and nonneuronal tissues. Therefore, by establishing the BHC as a complex mediating the REST-dependent transcriptional repression we have identified one of the mechanisms underlying the neuron-specific gene repression.
Acknowledgments
Thanks to Dr. David Picketts for SNF2h constructs. We thank the National Cell Culture Center (Minneapolis, MN) for propagation of HeLa cells. M-.A.H. was supported by a postdoctoral fellowship from Association pour la Recherche sur le Cancer (Paris). G.M. was supported by National Institutes of Health Grant NS22518 and the McKnight Foundation for the Neurosciences. R.S. was supported by grants from the National Institutes of Health (GM61204), V Foundation, and American Cancer Society.
Abbreviations
- NuRD
nucleosome remodeling and deacetylating
- HDAC
histone deacetylase
- BHC
BRAF–HDAC complex
- P11
phosphocellulose
- ChIP
chromatin immunoprecipitation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY090777).
References
- 1.Kornberg R D, Lorch Y. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Laurent B C, Yang X, Carlson M. Mol Cell Biol. 1992;12:1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Côté J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 5.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 6.Imbalzano A N, Kwon H, Green M R, Kingston R E. Nature (London) 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 7.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nature (London) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Côté J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 9.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 10.Bochar D A, Wang L, Beniya H, Kinev A, Xue Y, Lane W S, Wang W, Kashanchi F, Shiekhattar R. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 12.Xue Y, Wong J, Moreno G T, Young M K, Côté J, Wang W. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 13.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Nature (London) 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 14.Hebbes T R, Thorne A W, Crane-Robinson C. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1997;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 16.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 17.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 19.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 20.Grimes J A, Nielsen S J, Battaglioli E, Miska E A, Speh J C, Berry D L, Atouf F, Holdener B C, Mandel G, Kouzarides T. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 21.Naruse Y, Aoki T, Kojima T, Mori N. Proc Natl Acad Sci USA. 1999;96:13691–13696. doi: 10.1073/pnas.96.24.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roopra A, Sharling L, Wood I C, Briggs T, Bachfischer U, Paquette A J, Buckley N J. Mol Cell Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong J A, Tapia-Ramirez J, Kim S, Toledo-Aral J J, Zheng Y, Boutros M C, Altshuller Y M, Frohman M A, Kraner S D, Mandel G. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 24.Schoenherr C J, Anderson D J. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 25.Marmorstein L Y, Kinev A V, Chan G K, Bochar D A, Beniya H, Epstein J A, Yen T J, Shiekhattar R. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 26.Andres M E, Burger C, Peral-Rubio M J, Battaglioli E, Anderson M E, Grimes J, Dallman J, Ballas N, Mandel G. Proc Natl Acad Sci USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 28.Sif S, Stukenberg P T, Kirschner M W, Kingston R E. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraner S D, Chong J A, Tsay H J, Mandel G. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 30.Mikaelian I, Sergeant A A. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battey J, Moulding C, Taub R, Murphy W, Stewart T, Potter H, Lenoir G, Leder P. Cell. 1983;34:779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree G R. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pontiggia A, Rimini R, Harley V R, Goodfellow P N, Lovell-Badge R, Bianchi M E. EMBO J. 1994;13:6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You A, Tong J K, Grozinger C M, Schreiber S L. Proc Natl Acad Sci USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphrey G W, Wang Y, Russanova V R, Hirai T, Qin J, Nakatani Y, Howard B H. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 37.Ballas N, Battaglioli E, Atouf F, Andres M E, Chenoweth J, Anderson M E, Burger C, Moniwa M, Davie J R, Bowers W J, et al. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 38.Ellwood K B, Yen Y M, Johnson R C, Carey M. Mol Cell Biol. 2000;20:4359–4370. doi: 10.1128/mcb.20.12.4359-4370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grosschedl R. Curr Opin Cell Biol. 1995;7:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 40.Paull T T, Haykinson M J, Johnson R C. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]