Abstract
Integrin activation states determine the ability of these receptors to mediate cell–matrix and cell–cell interactions. The prototypic example of this phenomenon is the platelet integrin, αIIbβ3. In unstimulated platelets, αIIbβ3 is inactive, whereas exposing platelets to an agonist such as ADP or thrombin enables αIIbβ3 to bind ligands such as fibrinogen and von Willebrand factor. To study the regulation of integrin activation states at the level of single molecules, we developed a model system based on laser tweezers, enabling us to determine the rupture forces required to separate single ligand-receptor pairs by using either purified proteins or intact living cells. Here, we show that rupture forces of individual fibrinogen molecules and either purified αIIbβ3 or αIIbβ3 on the surface of living platelets were 60 to 150 pN with a peak yield strength of 80–100 pN. Platelet stimulation using either ADP or the thrombin receptor-activating peptide enhanced the accessibility but not the adhesion strength of single αIIbβ3 molecules, indicating that there are only two states of αIIbβ3 activation. Thus, we found it possible to use laser tweezers to measure the regulation of forces between individual ligand-receptor pairs on living cells. This methodology can be applied to the study of other regulated cell membrane receptors using the ligand-receptor yield strength as a direct measure of receptor activation/inactivation state.
The detailed biochemistry of receptor–ligand interactions can be determined from solution and/or surface studies, but these results do not take into account the response of receptor-mediated cell functions to externally applied forces encountered in vivo. This consideration is particularly relevant for integrins because of the cellular capacity to regulate the state of integrin activation. The extent to which cells regulate integrin function is highly variable. In some cellular environments, the ability of a specific integrin to support cellular adhesion may be constitutive, whereas in others, the integrin may be inactive or only partially active (1). Thus, it has been concluded that integrins exist in a variety of activation states, although the basis for this conclusion has generally been indirect, based on whole-cell adhesion assays and the interaction of integrins with specific monoclonal antibodies. Accordingly, direct studies of the activation states of individual receptors are important. In addition, such studies can reveal the relative contribution to integrin regulation of changes in receptor conformation (affinity modulation) vs. receptor clustering (avidity; ref. 2).
The platelet integrin αIIbβ3 (GPIIb/IIIa), which is inactive on resting platelets but is activated by agonists such as ADP and thrombin, is the prototypic example of adhesion receptor modulation. This tight regulation of αIIbβ3 activity is imperative to prevent the spontaneous formation of thrombi. In this paper, we demonstrate a model employing laser tweezers to determine the force between single ligand-receptor bonds either in a purified protein system or on the surface of living, agonist-reactive cells. Laser tweezers are sensitive and accurate at the lower end of the force spectrum (0–150 pN; ref. 3); thus, they are a suitable system with which to study integrin–ligand interactions, because the binding forces involved have previously been reported to be in this range (4, 5). By using this methodology, we have demonstrated that there is only one αIIbβ3 activation state that is relevant to the biological endpoint, platelet aggregation or adhesion, and that cellular stimulation increases the probability, but not the strength, of ligand binding.
Materials and Methods
Description of the Laser Tweezers-Based Model System to Study Receptor–Ligand Interactions.
Laser tweezers are an optical system that use laser light to trap and manipulate dielectric particles such as small beads or cells (6–8). External forces applied to the trapped particle can be accurately measured because the angular deflection of the laser beam is directly proportional to the lateral force applied to the particle (9–11). To measure the bond strength between purified αIIbβ3 and fibrinogen, we covalently attached αIIbβ3 molecules to polyacrylamide-coated, spherical, silica pedestals anchored to a glass surface and covalently linked fibrinogen to smaller latex beads, one of which was trapped and moved toward and away from the receptor-coated pedestal, contacting it repeatedly (Fig. 1). The position of the optical trap was oscillated in a triangular waveform at 50 Hz with a peak-to-peak amplitude of 0.8 μm. The separation of the pedestal and the bead then was reduced in 10-nm steps with the piezostage until repeated contacts were observed. The distance between the bottom of the trapped bead and the coverslip was controlled so that bead-surface (as opposed to bead-pedestal) touching and linkages were excluded. Laser beam deflection, sensed by a photodetector, was displayed as a voltage signal, which was converted to a force value by using a Stokes' law calibration (11) with a calibration coefficient (pN/V) determined for every experiment. The measurements were arranged to test bond strength at a high repetition rate, so that reasonable statistics could be obtained. Rupture forces after contact were collected and displayed as normalized force histograms for each experimental condition. The results of many experiments under similar conditions were averaged so that each histogram represented from 104 to >2 × 105 contacts. The experimental protocol and subsequent analysis were validated with streptavidin and biotin because the interaction of these molecules has been studied by other methods. The rupture force distribution for streptavidin and biotin measured with laser tweezers ranged between 10 pN and >150 pN, which is consistent with the results of similar experiments using a biomembrane force probe (12); this result is in agreement with atomic force microscopy data that the rupture forces for this ligand-receptor pair may be beyond 150 pN (13, 14).
Figure 1.
Data trace for a typical interaction of fibrinogen and purified αIIbβ3 as measured with laser tweezers. The force that the laser trap exerted on the latex bead can be partitioned into four parts (A–D). The latex bead was trapped near the center of the laser beam while moving toward (Upper A) or away (Upper D) from the silica pedestal. In the absence of contact between the bead and pedestal the measured force was small. (Upper B) At the moment of contact the pedestal stopped the motion of the latex bead while the laser beam continued to move in the same direction (left). The laser trap exerted a positive, compressive force on the pedestal and latex bead. The trap motion then reversed, and the compressive force declined to zero. Peak B (Lower) represents contact duration time between the surfaces. (Upper C) When the pedestal and latex bead bind, either specifically or nonspecifically, the bead position remained nearly constant as the laser trap continued to move to the right. The force on the bead increased in the negative direction almost linearly until the pedestal-bead bond was ruptured and the force rapidly returned to nearly zero. If no attachment occurred, there was no negative force. (Lower E) Scanning electron microscope image of the fibrinogen-coated latex bead (right) attached to the integrin-coated pedestal.
Preparing αIIbβ3- and Fibrinogen-Coated Surfaces.
Both integrin and fibrinogen were covalently attached to surfaces, so that we would not have to worry about the possibility of pulling them off the surfaces with the laser tweezers. Integrin-coated pedestals were built by using plain silica microspheres 1.4 or 2.2 μm in diameter. A working suspension of the microspheres (5% vol/vol) was made in 30% (wt/vol) acrylamide/bis-acrylamide containing 2% (vol/vol) N,N,N',N'-tetramethylethylenediamine and 2% (vol/vol) amyl acetate. The microspheres were uniformly spread on a clean, dry microscope glass coverslip and air dried. Then, a freshly prepared saturated solution of ammonium persulfate (2 μl) was smeared uniformly over the surface. The polyacrylamide-coated surface was activated by a 10% (vol/vol) solution of glutaraldehyde (4 h at room temperature), followed by washing with 20 volumes of 0.055 M borate (“binding”) buffer, pH 8.5. A solution of purified human αIIbβ3 (1 mg ml−1), which had been dialyzed against the binding buffer containing 0.15 M NaCl and 1 mM CaCl2, was then applied for 12–16 h at 4°C. Human fibrinogen was covalently bound to 0.93 μm carboxylate-modified latex beads with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide as a crosslinking agent in a two-step procedure (based on TechNote no. 205, Bangs Laboratories, Carmel, IN) with minor modifications. The surface density of fibrinogen, determined by using 125I-labeled protein, was near the saturation point of (11 ± 2) × 10−9 μg/μm2; nonetheless, the fraction of reactive molecules with a conformation and orientation compatible with binding was indeterminate. Fibrinogen-coated beads were used at a final concentration of about 107 ml−1.
Laser Tweezers Setup.
The optical tweezers used for these experiments were assembled from a Nikon Diaphot 300 inverted microscope, 100 × 1.3 N.A. Fluor lens and a Spectra-Physics FCBar Nd:YAG laser. The tilt of the incoming laser beam at the back focal plane of the microscope objective, and consequently the trap position, was manually changed or computer controlled through a two-dimensional acousto-optical deflector (Brimrose, Baltimore, MD). The lateral forces that the trap exerted on a bead were measured with a quadrant detector conjugate to the back focal plane of the condenser (9) and were calibrated from the low-frequency component of the Brownian motion. Manipulations were visualized by a video charge-coupled device camera. All experiments were conducted at a trap stiffness of 0.2 pN/nm as computed from the bandwidth of the Brownian motion. Both the force calibration and trap stiffness were routinely confirmed by the Stokes' force method (11). LABVIEW software (National Instruments, Austin, TX) was used to control and record laser beam deflection, to move the piezoelectric stage (Queensgate, Berkshire, U.K.), and to analyze data subsequently off-line.
Results
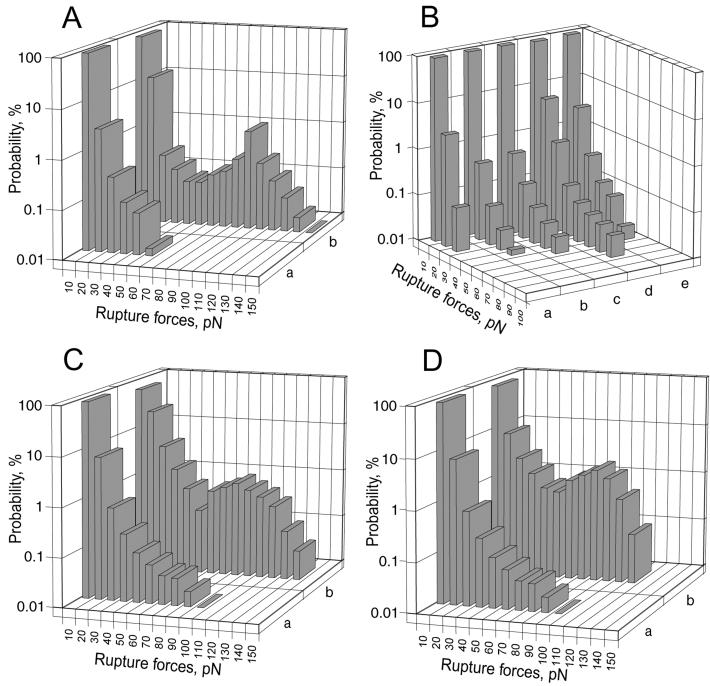
Observed signals using purified proteins could be experimentally partitioned into three categories: ligand-receptor binding and unbinding, nonspecific interactions, and small optical artifacts. The force distribution histogram of the interactions between intact purified αIIbβ3 and fibrinogen (Fig. 2Ab) revealed a major peak at 60 to 150 pN with a maximum bin at 90–100 pN, the latter defined as the yield strength, i.e., the average force required to rupture an αIIbβ3-fibrinogen bond. The fraction of contacts with rupture forces >60 pN, assumed to be the fraction of specific integrin–fibrinogen interactions, was as large as 3% for freshly purified intact αIIbβ3. In control experiments, αIIbβ3 was pretreated with 1 mM EDTA to prevent its specific association with fibrinogen (15). The force distribution histogram under this condition revealed no rupture forces >60 pN (Fig. 2Aa), indicating that high-affinity interactions did not occur.
Figure 2.
Comparative force histograms of various surface–surface interactions measured by laser tweezers. The individual forces measured during each contact-detachment cycle under similar experimental conditions were collected into 10-pN wide bins. The number of events in each bin was plotted against the average force for that bin after normalizing by the total number of interaction cycles. The percentage of events in a particular force range (bin) represented the probability of rupture events at that tension. (A) Comparison of the force distributions of the interactions between fibrinogen-coated beads and EDTA-pretreated αIIbβ3 molecules (a) or native immobilized αIIbβ3 (b). (B) Force distribution histograms of the nonspecific, control surface–surface interactions. (a) BSA-coated beads with untreated polyacrylamide-coated pedestals. (b) BSA-coated beads with BSA-coated pedestals. (c) Fibrinogen-coated beads with BSA-coated pedestals. (d) BSA-coated beads with αIIbβ3-coated pedestals. (e) Fibrinogen-coated beads with EDTA-pretreated αIIbβ3 bound to pedestals. (C) Comparison of the force distributions of the interactions between fibrinogen-coated beads and αIIbβ3 on the surface of resting platelets (a) or platelets stimulated with 10 μM ADP (b). (D) Comparison of the force distributions of the interactions between fibrinogen-coated beads and αIIbβ3 on the surface of resting platelets (a) or platelets stimulated with 50 μM TRAP (b).
Interactions between control beads and pedestals not coated with receptor-ligand pairs resulted in lower rupture forces that depended on the specific surface treatment. Optical artifacts observed with or without trapped latex beads produced signals that appeared as forces below 10 pN. The weakest interactions with rupture forces below 30 pN were observed for untreated polyacrylamide-coated pedestals in contact with BSA-coated beads (Fig. 2Ba). Pedestal surfaces were slightly more reactive after covalent coupling with BSA (Fig. 2Bb) and more reactive when coupled with fibrinogen (Fig. 2Bc) or αIIbβ3 (Fig. 2Bd). For comparison, beads coated with αIIbβ3 in the presence of EDTA again led to low-rupture forces (Fig. 2Be). The overall probability of weak nonspecific forces was higher than for specific interactions, but the probability of nonspecific forces of more than 60 pN varied from zero to only 0.1%. Thus, there was a clear quantitative difference between specific and nonspecific interactions, such that rupture forces for specific interactions between αIIbβ3 and fibrinogen were consistently >60 pN.
This conclusion was confirmed independently by probing the specificity of the forces with well characterized inhibitors of the interaction between fibrinogen and αIIbβ3 (16). The appearance of specific rupture forces >60 pN was prevented (Fig. 3A) by the presence of tirofiban, a tyrosine-based molecule that mimics the three-dimensional structure of the integrin inhibitor RGD (17), abciximab, a murine-human chimeric Fab fragment of the monoclonal antibody 7E3 that inhibits platelet aggregation (18), the H12 peptide from the carboxyl terminus of the fibrinogen γ-chain (19), or the αIIbβ3-specific monoclonal antibody A2A9 (20).
Figure 3.
Effect of specific αIIbβ3 antagonists on the rupture forces between fibrinogen and αIIbβ3. (A) Interactions between purified αIIbβ3 and fibrinogen in the presence or absence (positive control) of inhibitors: tirofiban (40 μM), abciximab (100 μg/ml), H12–fibrinogen-binding inhibitor dodecapeptide (1 mM), and mAb A2A9–αIIbβ3-binding specific “antiaggregant” monoclonal antibody (100 μg/ml). (B) Force distribution of the interactions between fibrinogen and platelets stimulated by 10 μM ADP in the presence or absence of the following inhibitors: abciximab (20 μg/ml), tirofiban (40 μM), cRGD–cyclic RGD peptide (80 μM), eptifibatide (100 μg/ml), EDTA-Na2–ethylendiaminetetraacetic acid (1 mM), PGE1–prostaglandin E1 (2 μM), and H12–fibrinogen-binding inhibitor dodecapeptide (1 mM). Because positive control data were collected separately for each experimental series with inhibitors, the probabilities displayed here were different from the data collected from the entire sum total of experimental data shown in Fig. 2.
Fibrinogen binding to αIIbβ3 on the platelet surface is absolutely regulated by platelet agonists (1). Therefore, we next studied interactions between a fibrinogen-coated bead and αIIbβ3 on the surface of living platelets. A platelet was trapped from a suspension of gel-filtered cells and attached to a 5-μm diameter silica pedestal coated with polylysine (Fig. 4). A fibrinogen-coated latex bead was then trapped and brought into intermittent contact with the platelet in the same way as for the experiments with purified proteins.
Figure 4.
Schematic representation (A) and scanning electron microscope image (B) of the interaction between an immobilized platelet and a fibrinogen-coated bead.
Force distribution histograms for the interactions between quiescent platelets and surface-bound fibrinogen were similar to those for the control histograms obtained between fibrinogen and BSA or BSA and purified αIIbβ3, except for rare events for which the magnitude of the recorded rupture forces reached a maximum of 100 pN (Fig. 2Ca).
On the other hand, platelet stimulation with ADP substantially affected their interaction with the fibrinogen-coupled beads, producing a major peak of rupture forces at 60 to 140 pN with a yield strength of 80–90 pN, without substantial change in lower nonspecific forces (Fig. 2Cb). Moreover, there was an ADP concentration-dependent increase in the probability of specific interactions having rupture forces >60 pN, with the greatest fraction observed after platelet stimulation with 10 μM ADP (Fig. 5A). At the same time, the average yield strength was constant at all ADP concentrations, with a value of about 80 pN (Fig. 5A). To confirm that the ADP-stimulated interactions were specific for αIIbβ3 and fibrinogen, two types of platelet inhibitors were used: competitive inhibitors of fibrinogen binding described above [abciximab, tirofiban, eptifibatide, A2A9, cyclic RGD-peptide (cRGD), and H12] and inhibitors of platelet metabolism (EDTA and PGE1). As shown in Fig. 3B, each of the inhibitors substantially decreased the frequency of attachments between platelets stimulated by 10 μM ADP and fibrinogen-coated beads, with more than a 10-fold reduction in the probability of high-yield force interactions.
Figure 5.
Probability and average yield strength of specific (>60 pN) fibrinogen–αIIbβ3 interactions plotted against the concentration of either ADP (A) or TRAP (B).
ADP is a “weak” platelet agonist. To determine whether stimulating platelets with a “strong” agonist results in similar fibrinogen-binding characteristics, we used the thrombin receptor-activating peptide (TRAP) as the platelet agonist. Like ADP, TRAP induced fibrinogen binding to αIIbβ3 with an average yield strength of 80–90 pN (Fig. 2Db). Moreover, increasing the TRAP concentration increased the probability, but not the average yield strength, of fibrinogen binding (Fig. 5B), implying that affinity of αIIbβ3 for ligands is independent of the nature of the platelet stimulus. Nevertheless, TRAP, at a concentration of 50 μM, induced significantly higher maximal probability of platelet interaction with fibrinogen consistent with its ability to induce a redistribution of αIIbβ3 molecules from an intracellular pool to the platelet membrane (21–23).
Discussion
By using laser tweezers, we found that αIIbβ3 resides on the platelet surface in either of two activation states, depending on the state of platelet stimulation. Nonetheless, because platelets express 80,000 or more copies of αIIbβ3 on their surface (24), it is possible that the rupture forces we measured represented the breaking of multiple rather than individual αIIbβ3-fibrinogen bonds. Two arguments support the view that our measurements represent individual bimolecular ligand-receptor binding events. First, the histograms of the distribution of rupture forces of multiple interactions should appear as a series of quantized peaks that are multiples of a single value of force and have probabilities inversely proportional to the number of bonds (25). However, apart from nonspecific interactions, we observed only a single well defined peak in the force histograms, suggesting that this peak represents individual ligand-receptor bonds. In quantitative terms, if we were to assume that the 100 pN peak we observed with a 3% probability represents the interactions of two ligand-receptor pairs, then the presumed bimolecular interactions at 50 pN would have a hypothetical square root probability of about 17%, whereas the observed probability was about 0.1% (Fig. 2Ab), indicating that the peak does indeed represent individual ligand–receptor interactions. Second, if multiple bonds were formed during a contact, it is unlikely that all would be ruptured simultaneously; rather, it is more probable that they would be ruptured sequentially so that multiple steps should have been observed during bond breakage. However, the rupture events we observed all occurred as a single step, at least with the time resolution of 0.5 ms.
Some of the events we measured may have resulted from pulling receptors out of the cell membrane, protein uprooting, or from disrupting integrin-receptor binding to the cytoskeleton (26), but such occurrences are unlikely for the following reasons. If receptor molecules are not linked to the cytoskeleton, tethers form because of reservoir buffering membrane tension rather than protein uprooting (27). Consistent with this report, we frequently observed tethers in resting or exhausted platelets after pulling the attached fibrinogen-coated beads apart from platelets. In activated platelets, although we occasionally observed tethers, the frequency was substantially lower compared with nonactivated cells. In addition, for most individual bead-platelet pairs, we observed multiple repeated binding and rupture signals, suggesting that the plasma membrane of the platelet was not drastically modified by either protein uprooting or cytoskeletal disruption.
Direct quantification of bimolecular interactions between fibrinogen and αIIbβ3 has not been reported previously, but two papers are relevant to our data. Goldsmith et al. (28) used external hydrodynamic force to break pairs of αIIbβ3-coated latex beads formed in the presence of free fibrinogen. They found that a substantial fraction of the fibrinogen-bridged beads separated at forces in the range of 70–150 pN, similar to the rupture forces we measured. However, a direct comparison between our work and theirs should be avoided because of substantial differences in the experimental models. Lee and Marchant used atomic force microscopy to measure the force required to rupture bonds between an immobilized RGD-containing peptide (GSSSGRGDSPA) and αIIbβ3 on the surface of adherent platelets (29). They reported that a value of 90 ± 45 pN represented the single molecular interaction between the RGD-ligand and the integrin at a loading rate of 18,000 pN/s, a value essentially identical to that used in our experiments (20,000 pN/s). The results reported in both of these papers are consistent with our data and support the view that the forces we measured are characteristic of bimolecular ligand–integrin interactions. Although not comparable in quantitative terms, our data are also in good qualitative agreement with recent work (30) in which the adhesion of ADP-activated vs. resting platelets to fibrinogen-coated latex beads was studied at different shear rates.
Our results have a number of physiological implications. First, the portion of contacts that lead to specific interactions between fibrinogen and αIIbβ3 is directly related to but not identical to the fraction of αIIbβ3 molecules with accessible exposed ligand-binding sites. On the other hand, the yield strength or, in terms of cell biology, the adhesion strength of activated receptor is an intrinsic property of the activated conformation of αIIbβ3 and is manifest as a distinctive range of forces with a particular set of loading rates (31). Therefore, the laser tweezers experiments allow a clear discrimination between accessibility and affinity of receptor-ligand binding. The probability of a specific interaction between fibrinogen and inactivated αIIbβ3 is low because the conformation of αIIbβ3 does not permit fibrinogen binding. Platelet stimulation by agonists increases the number of αIIbβ3 molecules with accessible fibrinogen-binding sites, but has no effect on the bond strength between fibrinogen and the activated form of the integrin. Thus, our results demonstrate that αIIbβ3 activation is an all-or-none phenomenon; each αIIbβ3 molecule resides on the platelet in either a completely on or a completely off conformation, which is consistent with structural data (32–34).
Second, in experiments with purified αIIbβ3, the fraction of specific interactions is related to the percentage of αIIbβ3 complexes in an active conformation and with the proper orientation on the pedestal surface. Because many experiments in the literature were performed with purified αIIbβ3, it is important to know whether the results are relevant to those obtained under physiologic conditions in live cells. The force histograms of fibrinogen binding to purified αIIbβ3 and to αIIbβ3 on the platelet surface were similar with regard to both the fraction of specific interactions and the average adhesion strength. It is important to mention that the force loading rate used in this work (20,000 pN/s) has the same order of magnitude that is imposed on platelets under physiologic shear rates (several hundred s−1), which is still low enough for αIIbβ3 to interact with surface-bound fibrinogen (35).
Third, the results from these experiments lead to an important addition to the cell adhesion paradigm. The remarkable number of weak interactions produced by quiescent platelets suggests that αIIbβ3-mediated platelet adhesion may be the result of the combination of numerous low-affinity attractive forces of nonspecific origin, together with forces arising from specific bond formation between ligand and receptor. The nonspecific protein–protein interactions may bring a ligand toward αIIbβ3 molecule and provide the spatial congruence required to initiate and/or reinforce specific bonding. Moreover, they may account for the ability of resting platelets to adhere spontaneously to fibrinogen-coated surfaces (1).
Our results indicate that at least for the integrin αIIbβ3, cells regulate the number of accessible binding sites, not the affinity of ligand binding. Because we have found that it is possible to measure and compare forces between individual ligand-receptor pairs on living cells, it will now be possible to determine whether this observation applies to the other members of the integrin superfamily.
Acknowledgments
We thank Yasuharu Takagi, Gaston Vilaire, and Chandrasekaran Nagaswami for their valuable methodological assistance. This work was supported by National Institutes of Health Grants HLBI 57407 and HLBI 30954.
Abbreviation
- TRAP
thrombin receptor-activating peptide
References
- 1.Bennett J S. Trends Cardiovasc Med. 1996;16:31–36. doi: 10.1016/1050-1738(95)00126-3. [DOI] [PubMed] [Google Scholar]
- 2.Hato T, Pampori N, Shattil S J. J Cell Biol. 1998;141:1685–1695. doi: 10.1083/jcb.141.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visscher K, Block S M. Methods Enzymol. 1998;298:460–489. doi: 10.1016/s0076-6879(98)98040-5. [DOI] [PubMed] [Google Scholar]
- 4.Lehenkari P P, Horton M A. Biochim Biophys Res Commun. 1999;259:645–650. doi: 10.1006/bbrc.1999.0827. [DOI] [PubMed] [Google Scholar]
- 5.Stout A L. Biophys J. 2001;80:2976–2986. doi: 10.1016/S0006-3495(01)76263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashkin A. Biophys J. 1992;61:569–582. doi: 10.1016/S0006-3495(92)81860-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashkin A. Proc Natl Acad Sci USA. 1997;94:4852–4860. [Google Scholar]
- 8.Svoboda K, Block S M. Annu Rev Biomol Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 9.Smith S B, Cui Y, Bustamante C. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 10.Allersma M W, Gittes F, deCastro M J, Stewart R J, Schmidt C F. Biophys J. 1998;74:1074–1085. doi: 10.1016/S0006-3495(98)74031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visscher K, Gross S P, Block S M. IEEE J Select Topics Quant Elec. 1996;2:1066–1076. [Google Scholar]
- 12.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Nature (London) 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 13.Florin E-L, Moy V T, Gaub H E. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 14.Moy V T, Florin E-L, Gaub H E. Science. 1994;266:257–259. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 15.Brass L F, Shattil S J, Kunicki T J, Bennett J S. J Biol Chem. 1985;260:7875–7881. [PubMed] [Google Scholar]
- 16.Bennett J S. Annu Rev Med. 2001;52:161–184. doi: 10.1146/annurev.med.52.1.161. [DOI] [PubMed] [Google Scholar]
- 17.Cook J J, Bednar B, Lynch J L, Gould R J, Egberson M S, Halczenko W, Duggan M E, Hartman G D, Lo M-W, Murphy G M, et al. Cardiovasc Drug Rev. 1999;17:199–224. [Google Scholar]
- 18.Coller B S. J Clin Invest. 1985;76:101–108. doi: 10.1172/JCI111931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kloczwiak M, Timmons S, Lukas T J, Hawiger J. Biochemistry. 1984;23:1767–1774. doi: 10.1021/bi00303a028. [DOI] [PubMed] [Google Scholar]
- 20.Bennett J S, Hoxie J A, Leitman S F, Vilaire G, Cines D B. Proc Natl Acad Sci USA. 1983;80:2417–2421. doi: 10.1073/pnas.80.9.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woods V L, Jr, Wolf L E, Keller D M. J Biol Chem. 1986;261:15242–15251. [PubMed] [Google Scholar]
- 22.Niiya K, Hodson E, Bader R, Byers-Ward V, Koziol J A, Plow E P, Ruggeri Z M. Blood. 1987;70:475–483. [PubMed] [Google Scholar]
- 23.Suzuki H, Kaneko T, Sakamoto T, Nakagawa M, Miyamoto T, Yamada M, Tanoue K. J Electron Microsc. 1994;43:282–289. [PubMed] [Google Scholar]
- 24.Wagner C L, Mascelli M A, Neblock D S, Weisman H F, Coller B S, Jordan R E. Blood. 1996;88:907–914. [PubMed] [Google Scholar]
- 25.Pierres A, Benoleil A-M, Bongrand P. Cell Adhes Commun. 1998;5:375–395. doi: 10.3109/15419069809010783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao J Y, Hochmuth R M. Biophys J. 1999;77:587–596. doi: 10.1016/S0006-3495(99)76915-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raucher D, Sheetz M P. Biophys J. 1999;77:1992–2002. doi: 10.1016/S0006-3495(99)77040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldsmith H L, McIntosh F A, Shahin J, Frojmovic M M. Biophys J. 2000;78:1195–1206. doi: 10.1016/S0006-3495(00)76677-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee I, Marchant R E. Microsc Microanal. 2000;2, Suppl. 6:974–975. [Google Scholar]
- 30.Bonnefoy A, Liu Q, Legrand C, Frojmovic M M. Biophys J. 2000;78:2834–2843. doi: 10.1016/S0006-3495(00)76826-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans E. Annu Rev Biophys Biomol Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 32.Hantgan R R, Paumi C, Rocco M, Weisel J W. Biochemistry. 1999;38:14461–14474. doi: 10.1021/bi9907680. [DOI] [PubMed] [Google Scholar]
- 33.Hantgan R R, Rocco M, Nagaswami C, Weisel J W. Protein Sci. 2001;10:1614–1626. doi: 10.1110/ps.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinogradova O, Haas T, Plow E F, Qin J. Proc Natl Acad Sci USA. 2000;97:1450–1455. doi: 10.1073/pnas.040548197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savage B, Saldivar E, Ruggeri Z M. Cell. 1996;84:289–297. doi: 10.1016/s0092-8674(00)80983-6. [DOI] [PubMed] [Google Scholar]