Abstract
We used a screening procedure to identify protein domains from phage-displayed cDNA libraries that bind both to bone marrow endothelial progenitor cells and tumor vasculature. Screening phage for binding of progenitor cell-enriched bone marrow cells in vitro, and for homing to HL-60 human leukemia cell xenograft tumors in vivo, yielded a cDNA fragment that encodes an N-terminal fragment of human high mobility group protein 2 (HMGN2, formerly HMG-17). Upon i.v. injection, phage displaying this HMGN2 fragment homed to HL-60 and MDA-MB-435 tumors. Testing of subfragments localized the full binding activity to a 31-aa peptide (F3) in the HMGN2 sequence. Fluorescein-labeled F3 peptide bound to and was internalized by HL-60 cells and human MDA-MB-435 breast cancer cells, appearing initially in the cytoplasm and then in the nuclei of these cells. Fluorescent F3 accumulated in HL-60 and MDA-MB-435 tumors after an i.v. injection, appearing in the nuclei of tumor endothelial cells and tumor cells. Thus, F3 can carry a payload (phage, fluorescein) to a tumor and into the cell nuclei in the tumor. This peptide may be suitable for targeting cytotoxic drugs and gene therapy vectors into tumors.
Keywords: angiogenesis‖bone marrow cells‖progenitor cells‖phage libraries‖ peptides
Tumors induce and sustain the growth of new blood vessels through angiogenesis (1–3). In the initial phases of tumor angiogenesis, new vessels are derived from existing capillaries by sprouting of the endothelial and mural cells (4, 5). Endothelial progenitor cells can be recruited to these new vessels from the bone marrow and from circulation (6–8). The recruitment of these progenitor cells and their proliferation are regulated by angiogenic factors such as vascular endothelial growth factor and angiopoietin-1 (9, 10), but the extent to which they contribute to angiogenesis in normal individuals is unclear. However, they appear to be of major importance in certain situations; Id-mutant mice are defective in angiogenic responses and do not support tumor growth (11), but transplanting wild-type bone marrow into these mice restores the angiogenic responses and allows for tumor growth (12).
The vasculature within tumors expresses distinct molecular markers that are characteristic of angiogenic vessels (13). These markers are also expressed in angiogenic vessels of nonmalignant tissues, such as wounds and inflammatory lesions. They include certain integrins (14, 15), receptors for angiogenic growth factors (16), proteases (17–19), and membrane proteins of unknown functions (20, 21). Pericytes of angiogenic vessels express a membrane proteoglycan known as NG2 or high molecular weight melanoma antigen (22, 23), and the extracellular matrix of angiogenic vessels contains an alternatively spliced form of fibronectin (24).
Several years ago, we developed an approach to study molecular specialization in the vasculature of tumors and normal tissues. The method is based on in vivo screening of large libraries of phage-displayed peptides to obtain peptides that have selective tissue affinities and direct the homing of phage to a specific target tissue in mice. When the libraries are injected into the circulation, the method primarily targets tissue-specific differences in endothelial cells. We have previously isolated peptides that home to the vasculature of individual normal tissues and tumors (25, 26).
Our previous tumor-homing peptides bind to molecules characteristic of angiogenic vessels (18, 25). In the present work, we modified the phage procedure to search for markers that would be shared by tumor vasculature and endothelial progenitor cells in the bone marrow. We screened cDNA libraries displayed on phage first on bone marrow cells ex vivo and then in tumor-bearing mice in vivo. We report a cDNA that encodes a fragment of the nuclear protein high mobility group (HMG) protein 2 (HMGN2). HMGN2 is a highly conserved nucleosomal protein thought to be involved in unfolding higher-order chromatin structure and facilitating the transcriptional activation of mammalian genes (reviewed in ref. 27). We derived a 31-aa synthetic peptide from this HMGN2 fragment, which, when injected i.v., accumulates in the nuclei of tumor endothelial cells and tumor cells. This peptide may find a use as a carrier of therapeutic molecules to the nuclei of tumor cells and tumor endothelial cells. The cell surface receptor for this peptide may be a useful molecular marker of endothelial progenitor cells.
Materials and Methods
Cell Lines and Experimental Animals.
Human myeloid leukemia HL-60 (American Type Culture Collection) and MDA-MB-435 human breast carcinoma cell lines were grown in RPMI 1640 media supplemented with 10% FBS (25). To establish xenograft tumors, 2- to 3-month old nude mice (Harlan Sprague–Dawley) were s.c. injected with 106 exponentially growing cells in 200 μl of culture media. The animals were used in experiments within 3–5 (HL-60) or 8–10 weeks (MDA-MB-435) of the injection.
Phage Libraries and Screening Protocol.
We constructed cDNA libraries by using mRNA purified from normal (human bone marrow, brain, and mouse embryo) and malignant (liver, lung, breast, and colon carcinoma) tissues (CLONTECH) and from mouse spleen and bone marrow (Oligotex Direct mRNA kit, Qiagen, Valencia, CA). The cDNA synthesis with random primers, cloning into the T7Select 10–3b vector, and phage packaging and amplification were done according to the manufacturer's instructions (Novagen). The cDNA libraries were pooled for the phage screening. The screening on isolated primary cells (ex vivo selection) and in vivo was performed as described (refs. 28 and 29; P.L., K.P., J.A.H., and E.R., unpublished work). In short, 109 plaque-forming units (pfu) of the phage library were incubated on target cells overnight at 4°C. Unbound phage were removed by extensive washing, and the phage bound to cells were rescued, amplified, and used for the subsequent round of selection. After two rounds of ex vivo selection, the phage pool was subjected to in vivo selection by injecting the pool (109 pfu) into the tail vein of a nude mouse bearing a HL-60 xenograft tumor. After two rounds of in vivo selection, 96 clones were selected from this tumor-homing pool and their protein-encoding inserts were sequenced.
Isolation of Murine Bone Marrow Progenitor Cells.
Mouse bone marrow was obtained by flushing both femoral and tibial bones with 3 ml of cold media (DMEM supplemented with 10% FBS). The bone marrow was then depleted of cells expressing common lineage-specific markers by using antibodies coupled to paramagnetic beads (StemSep Murine Kit, StemCell Technologies, Vancouver). The antibodies used were against mouse CD5 (clone Ly-1), myeloid differentiation antigen (Gr-1), CD45R (B220), erythroid cells (TER119), CD11b (Mac-1), and neutrophils (7–4) (StemCell Technologies). The remaining megakaryocytes were removed by filtering through a 30-μm nylon mesh filter (Miltenyi Biotec, Auburn, CA).
Flow Cytometry.
Human bone marrow specimens analyzed in this study represented excess material from samples collected for diagnostic purposes from adults with hematological malignancies. Informed consent was obtained before sample collection. A total of 2 ml of bone marrow was aspirated from the posterior iliac crest and stored in a citrate anticoagulant. Mononuclear cells were isolated by gradient centrifugation (Ficoll-Paque PLUS, Amersham Pharmacia) and incubated in RPMI 1640 media supplemented with 10% FBS for 2 h at 37°C. The cells were then transferred to 4°C and incubated with 1–2 μM fluorescein-conjugated peptides for 45 min and subsequently stained with PerCP- or phycoerythrin-conjugated CD34 and CD45 antibodies (Becton Dickinson) for 30 min. The sample was analyzed with either a FACSCalibur or a LSR flow cytometer (Becton Dickinson), and 100,000 events were collected.
Anti-T7 Antibodies.
New Zealand White rabbits were immunized with 1010 pfu of T7 nonrecombinant phage. The initial immunization was done in complete Freund's adjuvant and boosters were in incomplete Freund's adjuvant. The antiserum was absorbed on BLT5615 bacterial and mouse liver lysates, and the antibody titer was estimated by ELISA.
Peptides.
Peptides were synthesized with an automated peptide synthesizer by using standard solid-phase fluorenylmethoxycarbonyl chemistry (30). During synthesis, the peptides were labeled with fluorescein with an amino-hexanoic acid spacer as described (31). The concentration of unlabeled peptides was determined by weighing and from absorbance at 230 nm (32).
Histology.
Tissue distribution of homing ligands was examined by i.v. injection of fluorescein-coupled peptides (100 μl of a 1 mM solution) into the tail vein of anesthesized mice bearing xenografts. Blood vessels were visualized by i.v. injecting 200 μl of 0.5 μg/μl biotin-conjugated tomato lectin (Vector Laboratories). The injected materials (first peptide, then lectin) were allowed to circulate for 15 min, and the mouse, which remained anesthesized throughout the experiment, was perfused through the heart with 4% paraformaldehyde. Organs were removed and frozen in OCT embedding medium (Tissue-Tek, Elkhart, IN). The biotin-conjugated lectin was detected with streptavidin-Alexa 594 (Molecular Probes), mounted with Vectashield- 4′,6-diamidino-2-phenylindole (Vector Laboratories), and examined under an inverted fluorescent microscope.
Results
Tumor-Homing HMGN2 Fragment.
Hematopoietic and endothelial precursors have a common cell of origin (hemangioblast) and share several phenotypic characteristics. We devised a phage screening procedure that would capitalize on this shared phenotype and select clones that bind to both primitive bone marrow cells and angiogenic endothelial cells. The screening procedure included a preselection on lineage-depleted mouse bone marrow cells (putative progenitor cells) ex vivo and further selection for homing to HL-60 xenograft tumors in vivo.
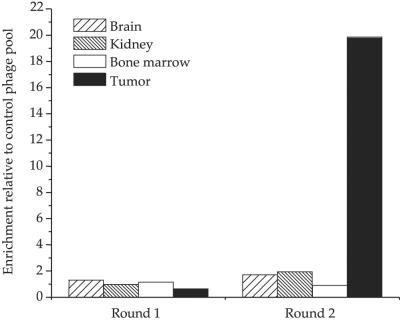
After two rounds of preselection on lineage-depleted mouse bone marrow cells, the resulting phage pool was injected in nude mice bearing HL-60 tumors. The selected phage pool showed a 20-fold enrichment for tumor homing relative to the unselected library after two rounds of in vivo selection (Fig. 1). Sequencing showed that the predominant cDNA in this pool was a 270-bp clone that contained an ORF encoding the first 73 N-terminal amino acids of the human HMGN2 protein; the clone also contained 51 bp from the 5′ noncoding sequence. We designated this clone HMGN2-N. Additional HMGN2 clones were also isolated from the phage pool; they all shared a common sequence corresponding to exons 3 and 4 in the HMGN2 sequence. The same HMGN2-N cDNA clone was also isolated in another independent screening.
Figure 1.
Phage enrichment in vivo. A cDNA phage pool that was preselected for ex vivo binding to bone marrow cells was injected into the tail vein of mice. After 10 min of circulation, the mice were perfused through the heart, and phage was rescued from various organs, amplified, and used for subsequent rounds of selection. Fold enrichment of selected phage pool relative to the unselected cDNA phage library pool is shown.
The HMGN2-N phage homed to the HL-60 tumors to about the same extent as the selected pool (data not shown). The injected phage also accumulated in the kidneys, but only when the number of phage injected was small, 109 pfu). Tumor homing was obtained both at this input level, and when 1012 pfu of phage were injected. We do not know the reason for the dose effect. About 1,000 times more HMGN2-N phage than nonrecombinant T7 phage bound to cultured HL-60 and MDA-MB-435 cells. Cell suspensions made from HL-60 tumors also bound the HMGN2-N phage with a 1,000-fold greater specificity relative to the control phage. Phage clones encoding fragments of some other proteins were identified in the screenings, but these fragments homed less well to tumors than HMGN2-N and were not studied further.
HMGN2-N Binding Domain and Specificity.
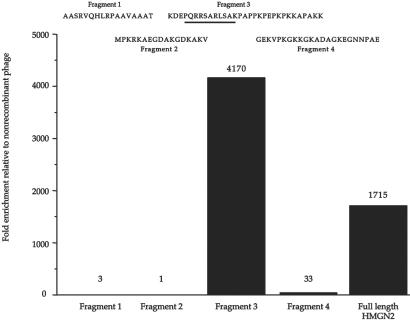
To identify the HMGN2-N domain responsible for the cell binding and in vivo homing, we constructed phage that display a set of fragments from HMGN2-N. The fragments were designed to include the complete exon sequences of the HMGN2 gene. A 31-aa fragment encoded by exons 3 and 4 (F3 peptide) showed substantial tumor cell binding (Fig. 2); F3 corresponds to the nucleosomal binding domain of HMGN2. The binding of the F3-displaying phage to tumor cells was inhibited by free F3 peptide in a dose-dependent fashion; complete inhibition was achieved with 100 μM peptide. The background binding of nonrecombinant phage was unaffected by this peptide. We further divided the F3 peptide into two peptides. Phage displaying exon 3, PQRRSARLSA, also bound to tumor cells but less well than F3. The specificity of the cell binding was further confirmed by comparing HL-60 cell binding of HMGN2 exon 3-displaying phage (insert sequence, PQRRSARLSA) and the homologous HMGN1 exon 3-displaying phage (insert sequence, PKRRSARLSA). The HMGN1 phage bound 90% less than the HMGN2 phage, indicating that the single amino acid change from glutamine to lysine substantially changes the cell binding specificity of the fragment. Database searches showed that the PQRRSARLSA sequence is present in a number of expressed sequence tags, the sequence of which is otherwise different from that of HMGN2. Thus, proteins other than HMGN2 may possess the same cell binding activity.
Figure 2.
Localization of the HMGN2-N cell binding site. The sequence and cell binding activity of individual HMGN2-N fragments. Inserts encoding for the various fragments were cloned into T7 phage, and the phage preparations were tested for binding to primary cells from HL-60 xenograft tumors. After a 1-h incubation at 4°C, the cells were washed and phage-quantified (the pfu are indicated above the columns). The enrichment is shown relative to a control phage (nonrecombinant T7 phage). One experiment representative of four is shown. The sequence encoded by exon 3 of HMGN2 is underlined.
Tissue and Cellular Localization of F3 Peptide.
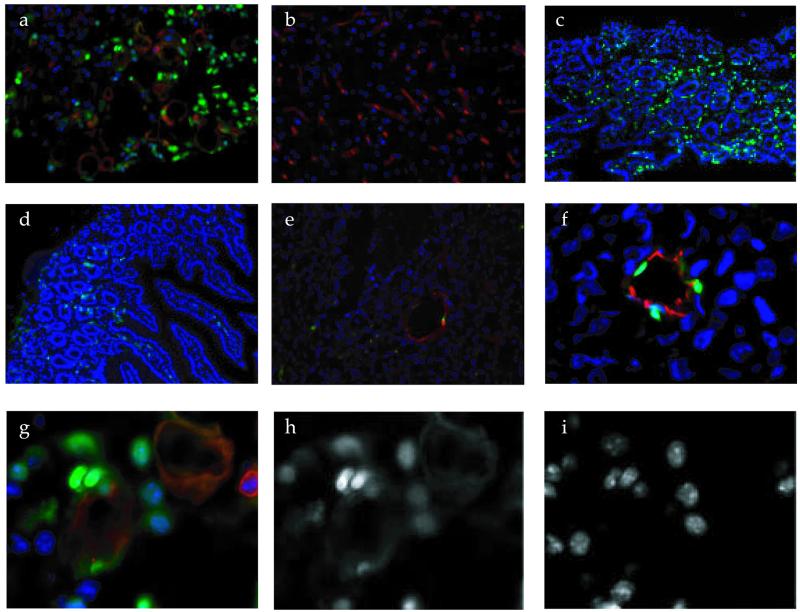
To be able to study the localization of the F3 peptide in tissues and cells, we synthesized this peptide as a conjugate with fluorescein. The labeled peptide was injected i.v. into mice bearing HL-60 or MDA-MB-435 tumors, and the localization of fluorescence was studied in tissues collected 10–15 min later. Strong fluorescence was seen in the tumor tissue (Fig. 3a), whereas little or no specific fluorescence was detected in the brain, liver, and spleen; the result for the brain is shown in Fig. 3b. F3 was present in a small population of cells in the skin and gut (Fig. 3 c and d). Diffuse fluorescence accumulated in proximal tubules of kidneys. Fluorescein-labeled scrambled exon 3 peptide (ARALPSQRSR) produced a similar fluorescence pattern in the kidney, but was essentially undetectable in other tissues, including the HL-60 tumors (Fig. 3e). Thus, the fluorescence in the kidneys is presumably caused by nonspecific uptake of peptides, or fluorescein, from the glomerular filtrate.
Figure 3.
Tissue localization of i.v.-injected F3 peptide. HMGN2-N F3 peptide and ARALPSQRSR control peptide were conjugated to fluorescein. Each labeled peptide was injected into the tail vein of mice bearing HL-60 or MDA-MB-435 xenografts. The peptide injection was followed 10 min later by an injection of biotinylated tomato lectin. After another 5 min, the mice were perfused through the heart with a fixative solution, and the organs were dissected, sectioned, and stained with streptavidin-Alexa 594. The slides were counterstained with 4′,6-diamidino-2-phenylindole and examined under an inverted fluorescent microscope. (e) An HL-60 tumor section from a mouse injected with fluorescein-labeled ARALPSQRSR control peptide, all other panels are from mice injected with fluorescein-labeled F3. (a) HL-60 tumor; (b) brain; (c) skin; (d) gut; (e) ARALPSQRSR control peptide in HL-60 tumor; (f) fluorescein-labeled F3 in an MDA-MB-435 tumor; (g) a higher magnification view from a showing the localization of F3 (green), lectin-stained vasculature (red), and 4′,6-diamidino-2-phenylindole-stained nuclei (blue). The green and blue images are shown individually in h and i. (Magnifications: a, b, and e, ×200; c and d, ×100; and f–i, ×400.)
In the HL-60 leukemia tumor tissue, F3 localized in tumor cells and cells lining tumor blood vessels, presumably the endothelial cells. The fluorescence was predominantly located in the nuclei (Fig. 3 a and g–i). Most of the tumor cells containing fluorescence were around blood vessels. In MDA-MB-435 breast cancer tumors, F3 also accumulated in the endothelial cells and tumor cells. In some microscopic fields, the F3 fluorescence was essentially limited to the endothelial cells, providing a particularly clear view of F3 within endothelial cells and their nuclei (Fig. 3f). Similar tumor localization was obtained with F3 coupled to another fluorescent molecule, rhodamine (not shown).
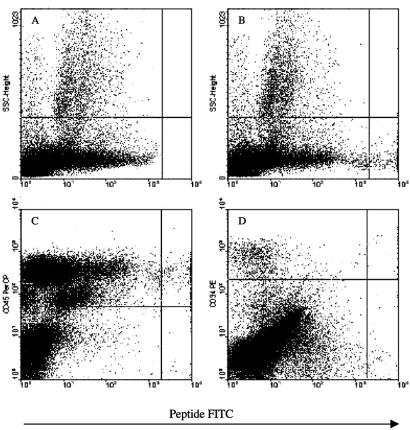
F3 also specifically bound to a small (0.3–0.8% of mononuclear cells) cell population in human bone marrow (Fig. 4). The F3-positive cells were lymphocyte-sized, agranular, CD45-positive, and mostly CD34-negative. This marker profile is similar to that of the lineage-depleted mouse bone marrow cell population we used in the phage preselection.
Figure 4.
FACS profile of bone marrow cells labeled with fluorescent F3 peptide and antibodies against cell differentiation markers. Fluorescein-labeled peptides, F3 and ARALPSQRSR control peptide at 2 μM, were incubated with gradient-depleted bone marrow cells and analyzed in a flow cytometer. (A) Control peptide (number/percentage of cells in lower right quadrant: 1/0.0); (B) F3 peptide (308/0.88); (C) F3 vs. CD45 (77% CD45-positive); (D) F3 vs. CD34 (75% CD34-negative).
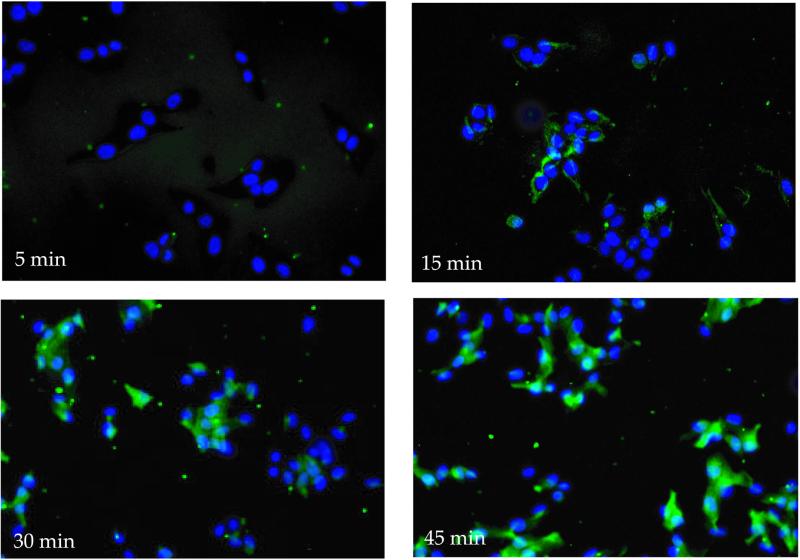
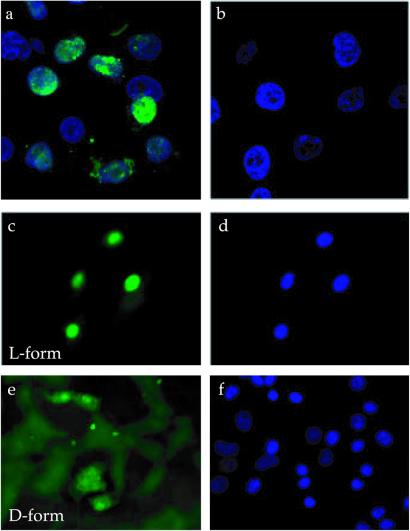
Cellular uptake and nuclear translocation of the F3 peptide were also observed in cultured HL-60 cells and MDA-MB-435 cells in vitro. The fluorescein-labeled peptide appeared in the cytoplasm of the cells where it was diffusely distributed (Fig. 5). Nuclear fluorescence started appearing at 30 min, and after 1 h most of the fluorescence was in the nuclei. The internalization process appeared to be slower in vitro than in vivo, possibly because of a more avid binding of the peptide to the target cells in vivo. The cellular uptake did not depend on the nature of the fluorescent dye, as rhodamine-labeled F3 also accumulated in the nuclei of the MDA-MB-435 cells (not shown). The D-amino acid form of F3 was also internalized by the MDA-MB-435 cells, but more slowly than the L-form, and it did not accumulate in the nucleus (Fig. 6). The uptake was energy dependent, as it was slower at 4°C than at 37°C (not shown).
Figure 5.
Kinetics of uptake and nuclear translocation of F3 peptide in cultured MDA-MB-435 cells. The cells were incubated with fluorescein-labeled F3 (1 μM) for the indicated length of time at 37°C, washed, fixed, and examined under an inverted fluorescent microscope.
Figure 6.
Nuclear localization of the L- and D-forms of the F3 peptide. Nuclear localization in HL-60 cells of F3 (a) or ARALPSQRSR control peptide (b). (c–f) Uptake by MDA-MB-435 cells of F3 synthesized either from L or D amino acids. The cells were treated as in Fig. 5A and stained with 4′,6-diamidino-2-phenylindole before examination under a confocal (a and b) or inverted fluorescent microscope (c–f). (c and e) Peptide staining (green); (d and f) nuclear staining (blue). (Magnifications: a and b, ×400; c–f, ×200.)
Discussion
We describe a tumor-homing protein fragment and a peptide derived from it, both of which bind to tumor blood vessels and tumor cells. The peptide homes selectively into tumors and has the remarkable property of being able to carry a payload into the nuclei of the target cells.
In this work, we used phage-displayed cDNA libraries in vivo to search for phage capable of homing to tumors, especially to their vasculature. Our thinking was that any proteins or protein fragments identified from the cDNA libraries might have higher affinities for tumor vasculature than the relatively short peptides obtained from random peptide libraries in previous work (13). Another expectation was that we might identify proteins that are functionally linked to the growth of tumors or their blood vessels. The cDNA library screening revealed a remarkably potent homing peptide, F3, indicating that the first expectation may have been fulfilled. Whether HMGN2, the nuclear protein that F3 is derived from, might have an extracellular function remains to be seen. It is interesting to note that there are some similarities between HMGN2 and amphoterin (HMGB1, formerly HMG-1), an intracellular and nuclear protein that is secreted in a signal sequence-independent manner and regulates cell migration and invasion through binding to its cell surface receptor, RAGE (33, 34). Blockade of RAGE-amphoterin signaling decreased tumor growth and metastases in mice (35).
Another modification to the original in vivo phage screening protocol was to enrich the library by selecting on isolated bone marrow cells before the in vivo screening step for tumor homing. Our aim was to identify proteins or peptides that would recognize an epitope shared by bone marrow progenitor cells and tumor endothelial cells. The F3 peptide appears to fulfill this expectation. It recognizes a minor population of progenitor cell-like bone marrow cells, the closer identity of which remains to the established, and it also binds to endothelial cells in tumors. This “dual target” screening approach is likely to further expand the utility of in vivo phage screening.
The specificity of the F3 peptide is broader than binding of the bone marrow subpopulation and tumor endothelial cells; F3 binding is a shared property of tumor cells and tumor endothelial cells. F3 appears to recognize a variety of tumor types; all tumors we have tested, including the TRAMP mouse prostate carcinomas (36), have been positive for F3 binding. In addition to the tumors and bone marrow, systemically injected F3 recognizes a minor population in both the normal skin and the gut. These cells are not associated with the vasculature, but their tissue localization did not suggest any obvious identity for these cells. The F3 binding specificity in the bone marrow and in tumors suggests that the positive cells in the skin and gut may represent some kind of a progenitor cell.
The strong accumulation of fluorescein-labeled F3 in tumor vasculature and tumor cells suggests that this peptide may be useful in targeting therapeutic agents into tumors. Other homing peptides isolated by in vivo phage display have given promising results in this regard (25, 37, 38). As fluorescein and rhodamine conjugated to F3 accumulate in tumor tissue, one can expect a drug conjugate to do the same.
A feature of F3 that makes it uniquely promising for drug targeting applications is its uptake by cells and its ability to carry a payload into the cell nucleus. We saw intracellular accumulation of fluorescence from fluorescein-labeled F3 in the cytoplasm of the target cells both in vitro and in vivo. The fluorescence accumulation was particularly striking in the nucleus. The F3 peptide is highly basic and is apparently recognized by the nuclear import machinery. The D-form of F3 also enters the cells, although it is not efficiently translocated into the nucleus. These properties are reminiscent of the cellular uptake and nuclear translocation of highly basic peptides from Tat protein and certain homeobox proteins (39–41). Such peptides are commonly used to introduce proteins and genes into cells. The mechanisms whereby these peptides enter cells are poorly understood, but their internalization is known not to require cellular energy. We find that the internalization of fluorescein-labeled F3 by tumor cells does not take place at 4°C, indicating energy dependence. A more important difference is that F3 only enters certain types of cells, whereas no cell type preference has been reported for other internalizing peptides. The cell type specificity of F3 can offer unique advantages. The numerous anticancer drugs that act in the nucleus (42) could particularly benefit from the nuclear-targeting capability provided by the F3 peptide.
Acknowledgments
We thank Dr. Edward Monosov for microscopy, Dr. Satu Mustjoki for help with flow cytometry, Dr. Fernando Ferrer for peptide synthesis, and Dr. Eva Engvall for comments on the manuscript. This study was supported by Grants CA74238, CA82713, and CA 30199 (Cancer Center Support Grant) from the National Cancer Institute and Grant 99-3339 from the Komen Foundation. K.P. is supported by the Finnish Medical Foundation, P.L. by fellowships from the Academy of Finland and the Finnish Cultural Foundation, J.A.H. by a National Cancer Institute Training Grant, and M.B. by a Swiss National Science Foundation Fellowship.
Abbreviations
- HMG
high-mobility group
- pfu
plaque-forming units
References
- 1.Hanahan D, Folkman J. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 2.Jain R K. Adv Drug Delivery Rev. 1997;26:71–90. doi: 10.1016/s0169-409x(97)00027-6. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Holash J, Maisonpierre P C, Compton D, Boland P, Alexander C R, Zagzag D, Yancopoulos G D, Wiegand S J. Science. 1999;284:1994–1998. doi: 10.1126/science.284.5422.1994. [DOI] [PubMed] [Google Scholar]
- 5.Yancopoulos G D, Davis S, Gale N W, Rudge J S, Wiegand S J, Holash J. Nature (London) 2000;407:242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 6.Davidoff A M, Ng C Y, Brown P, Leary M A, Spurbeck W W, Zhou J, Horwitz E, Vanin E F, Nienhuis A W. Clin Cancer Res. 2001;7:2870–2879. [PubMed] [Google Scholar]
- 7.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner J M. Science. 1997;275:964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 8.Shi Q, Rafii S, Wu M H, Wijelath E S, Yu C, Ishida A, Fujita Y, Kothari S, Mohle R, Sauvage L R, et al. Blood. 1998;92:362–367. [PubMed] [Google Scholar]
- 9.Hattori K, Dias S, Heissig B, Hackett N R, Lyden D, Tateno M, Hicklin D J, Zhu Z, Witte L, Crystal R G, et al. J Exp Med. 2001;193:1005–1014. doi: 10.1084/jem.193.9.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asahara T, Takahashi T, Masuda H, Kalka C, Chen D, Iwaguro H, Inai Y, Silver M, Isner J M. EMBO J. 1999;18:3964–3972. doi: 10.1093/emboj/18.14.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyden D, Young A Z, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader B L, Hynes R O, Zhuang Y, Manova K, et al. Nature (London) 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 12.Lyden D, Hattori K, Dias S, Costa C, Blaikie P, Butros L, Chadburn A, Heissig B, Marks W, Witte L, et al. Nat Med. 2001;7:1194–1201. doi: 10.1038/nm1101-1194. [DOI] [PubMed] [Google Scholar]
- 13.Ruoslahti E. Semin Cancer Biol. 2000;10:435–442. doi: 10.1006/scbi.2000.0334. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Bell K, Mousa S A, Varner J A. Am J Pathol. 2000;156:1345–1362. doi: 10.1016/s0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks P C, Montgomery A M, Rosenfeld M, Reisfeld R A, Hu T, Klier G, Cheresh D A. Cell. 1994;79:1157–1164. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 16.Korpelainen E I, Alitalo K. Curr Opin Cell Biol. 1998;10:159–164. doi: 10.1016/s0955-0674(98)80137-3. [DOI] [PubMed] [Google Scholar]
- 17.Bergers G, Brekken R, McMahon G, Vu T H, Itoh T, Tamaki K, Tanzawa K, Thorpe P, Itohara S, Werb Z, Hanahan D. Nat Cell Biol. 2000;2:737–744. doi: 10.1038/35036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun R A, Shapiro L H, Arap W, Ruoslahti E. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- 19.Silletti S, Kessler T, Goldberg J, Boger D L, Cheresh D A. Proc Natl Acad Sci USA. 2001;98:119–124. doi: 10.1073/pnas.011343298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.St Croix B, Rago C, Velculescu V, Traverso G, Romans K E, Montgomery E, Lal A, Riggins G J, Lengauer C, Vogelstein B, Kinzler K W. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 21.Carson-Walter E B, Watkins D N, Nanda A, Vogelstein B, Kinzler K W, St Croix B. Cancer Res. 2001;61:6649–6655. [PubMed] [Google Scholar]
- 22.Schlingemann R O, Rietveld F J, de Waal R M, Ferrone S, Ruiter D J. Am J Pathol. 1990;136:1393–1405. [PMC free article] [PubMed] [Google Scholar]
- 23.Ozerdem U, Grako K A, Dahlin-Huppe K, Monosov E, Stallcup W B. Dev Dyn. 2001;222:218–227. doi: 10.1002/dvdy.1200. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson F, Kosmehl H, Zardi L, Neri D. Cancer Res. 2001;61:711–716. [PubMed] [Google Scholar]
- 25.Arap W, Pasqualini R, Ruoslahti E. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 26.Ruoslahti E, Rajotte D. Annu Rev Immunol. 2000;18:813–827. doi: 10.1146/annurev.immunol.18.1.813. [DOI] [PubMed] [Google Scholar]
- 27.Bustin M. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. J Clin Invest. 1998;102:430–437. doi: 10.1172/JCI3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman J, Laakkonen P, Porkka K, Bernasconi M, Ruoslahti E. In: Phage Display: A Practical Approach. Clarkson T, Lowman H, editors. Oxford: Oxford Univ. Press; 2002. , in press. . [Google Scholar]
- 30.Atherton E, Sheppard R. Solid-Phase Peptide Synthesis. Oxford: IRL; 1989. [DOI] [PubMed] [Google Scholar]
- 31.Wender P A, Mitchell D J, Pattabiraman K, Pelkey E T, Steinman L, Rothbard J B. Proc Natl Acad Sci USA. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehresmann B, Imbault P, Weil J H. Anal Biochem. 1973;54:454–463. doi: 10.1016/0003-2697(73)90374-6. [DOI] [PubMed] [Google Scholar]
- 33.Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen J X, Nagashima M, Lundh E R, Vijay S, Nitecki D, et al. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- 34.Fages C, Nolo R, Huttunen H J, Eskelinen E, Rauvala H. J Cell Sci. 2000;113:611–620. doi: 10.1242/jcs.113.4.611. [DOI] [PubMed] [Google Scholar]
- 35.Taguchi A, Blood D C, del Toro G, Canet A, Lee D C, Qu W, Tanji N, Lu Y, Lalla E, Fu C, et al. Nature (London) 2000;405:354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 36.Gingrich J R, Barrios R J, Morton R A, Boyce B F, DeMayo F J, Finegold M J, Angelopoulou R, Rosen J M, Greenberg N M. Cancer Res. 1996;56:4096–4102. [PubMed] [Google Scholar]
- 37.Arap W, Haedicke W, Bernasconi M, Kain R, Rajotte D, Krajewski S, Ellerby M H, Bredesen D E, Pasqualini R, Ruoslahti E. Proc Nat Acad Sci USA. 2002;99:1527–1531. doi: 10.1073/pnas.241655998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curnis F, Sacchi A, Borgna L, Magni F, Gasparri A, Corti A. Nat Biotechnol. 2000;18:1185–1190. doi: 10.1038/81183. [DOI] [PubMed] [Google Scholar]
- 39.Lindgren M, Hallbrink M, Prochiantz A, Langel U. Trends Pharmacol Sci. 2000;21:99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 40.Schwarze S R, Ho A, Vocero-Akbani A, Dowdy S F. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 41.Gallouzi I E, Steitz J A. Science. 2001;294:1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- 42.Hurley L H. Nat Rev Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]