Abstract
TIP47 (tail-interacting protein of 47 kDa) binds to the cytoplasmic domains of the cation-dependent and cation-independent mannose 6-phosphate receptors (MPRs) and is required for their transport from endosomes to the trans-Golgi network in vitro and in living cells. TIP47 recognizes distinct determinants in the cytoplasmic domains of these two receptors, and its ability to bind to the cation-independent MPR is enhanced by the concomitant binding of the Rab9 GTPase. We show here that TIP47 residues 161–169 are essential, but likely not sufficient, for Rab9 binding. Mutation of these residues led to a significant decrease in Rab9 binding, but did not alter the global folding of the protein. The most impaired mutant was indistinguishable from wild-type TIP47 in its circular dichroism spectrum, and mutant proteins that showed decreased Rab9 binding retained full capacity to bind to MPR cytoplasmic domains. Closely related sequences in a related protein, adipophilin, did not confer Rab9 binding capacity to that protein. Partial proteolysis of TIP47 and TIP47 mutant proteins revealed subtle conformational differences, suggesting that residues 161–169 reside in a portion of TIP47 that is important for its conformation. These experiments reveal distinct binding domains for the Rab9 GTPase and MPR cytoplasmic domains in the cargo selection protein TIP47.
Mannose 6-phosphate receptors (MPRs) carry newly synthesized lysosomal enzymes from the Golgi to endosomes, where the enzymes are released. The receptors then return to the Golgi to pick up additional hydrolases for delivery to prelysosomes (1, 2). TIP47 (tail interacting protein of 47 kDa) binds to the cytoplasmic domains of MPRs and is required for their transport from endosomes to the trans Golgi, both in vitro and in living cells (3–5). TIP47 binds to distinct determinants on the cation-independent (CI-) and cation-dependent (CD-) MPRs: it recognizes a Phe-Trp in the CD-MPR (3, 6) and a proline-rich region in the CI-MPR (7). TIP47 does not bind to other proteins that cycle between endosomes and the Golgi, including TGN38, furin, and carboxypeptidase D (6).
We have recently shown that the ability of TIP47 to bind to the CI-MPR is enhanced in the presence of the Rab9 GTPase (4). Rab9 is a Ras-like GTPase that is located on late endosomes and is also required for the transport of MPRs from endosomes to the Golgi in vitro (8) and in living cells (9). Truncation analysis of TIP47 revealed that TIP47 requires the presence of residues 152–187 for Rab9 binding (4). However, removal of these residues had no influence on the affinity of the interaction of TIP47 with MPR cytoplasmic domains, consistent with a model in which the binding of Rab9 induces the formation of a TIP47 conformation that displays tighter binding to MPRs. The finding that MPR cytoplasmic domains also enhance TIP47 binding to Rab9 further supports the conclusion that TIP47 exists in at least two conformations, binary complex formation between either TIP47 and Rab9, or TIP47 and MPR, enhances ternary complex formation.
We present here a scanning mutagenesis of TIP47 to define more precisely the amino acid residues important for Rab9 binding. We have identified a stretch of nine residues that when mutated to alanine decrease the binding of TIP47 to Rab9 significantly. Although these residues are important for the interaction of TIP47 with Rab9 and for TIP47 function in living cells, our experiments suggest that they are not sufficient to comprise key determinants within an independent Rab9 binding pocket. Rather, these residues seem important for subtle conformational differences that influence the ability of TIP47 to bind Rab9.
Methods
Expression and Purification of Recombinant Proteins.
6xHis-TIP47 wild-type and mutant proteins, 6xHis–green fluorescent protein (GFP).
TIP47 Rab9-binding domain mutants were generated by using a QuikChange Site-Directed Mutagenesis kit (Stratagene). Plasmids containing the appropriate construct were transformed into Escherichia coli XL1-Blue cells and cultured at room temperature to an OD600 of ≈0.6. Cells were induced with 0.1 mM isopropyl β-d-thiogalactoside at room temperature for a maximum of 16 h. Cells were pelleted and resuspended in buffer A (50 mM Hepes-KOH, pH 7.5/150 mM KCl) with 3 mM PMSF, DNase I, and a protease inhibitor mixture (10 μg/ml leupeptin, 40 μg/ml aprotinin, and 1 μM pepstatin A). Cells were broken by two passes through a French press and centrifuged for 17 min at 17,000 rpm. Supernatants were collected and run three times through an 18-gauge needle, followed by three times through a 21-gauge needle to shear any remaining DNA. His-tagged proteins were then purified by using a 1-ml HiTrap nickel column (Amersham Pharmacia) according to the manufacturer's instructions. Eluates were dialyzed overnight to buffer B (50 mM Hepes-KOH, pH 7.5/150 mM KCl/1 mM MgCl2/10% glycerol). Proteins were analyzed for concentration and purity by BioRad protein assay using BSA as standard and 12.5% SDS/PAGE followed by densitometric analysis (National Institutes of Health image).
Glutathione S-transferase (GST)–CI-MPR (Δ12).
Expression and purification were as described (7). A truncation of the CI-MPR cytoplasmic domain, lacking the first 12 residues at the N terminus of the cytoplasmic domain (CI-MPRΔ12), was used because of its enhanced stability upon expression in E. coli; CI-MPRΔ12 retains wild-type binding capacity for TIP47 (7).
Rab9CLLL.
Expression and purification were as described (10). Rab9CLLL, a wild-type protein with an altered COOH terminus, was used because of its enhanced stability upon expression in E. coli. Rab9CLLL can be mono-geranylgeranylated in vitro and is functional in that it can support in vitro transport of CI-MPR from late endosomes to the trans-Golgi network (10). The preparation used was 83% active in terms of its capacity to bind GTP.
Adipophilin.
The adipophilin gene was cloned by using PCR from a skeletal muscle cDNA preparation. The resulting PCR product was cloned into the pTrcHis TOPO vector (Invitrogen) and sequenced to confirm the identity of the adipophilin gene. Adipophilin was expressed in XL-1 blue cells for 4 h at 37°C. Cells from a 2-liter culture were pelleted, resuspended in 25 mM Tris⋅HCl (pH 8.0) with protease inhibitors, and then broken as described above. The resulting mixture was centrifuged at 20,000 rpm in a JA-20 centrifuge for 1 h. The cell lysate was passed over a 5-ml Fast flow Q column (Amersham Pharmacia) and eluted with a linear gradient of 0–1.0 M NaCl. Fractions containing adipophilin were pooled, diluted to a lower salt concentration, and loaded onto a 1-ml Mono Q column. Proteins were eluted by using a linear gradient of 0–0.6 M NaCl. Adipophilin-containing fractions were concentrated by using a YM-10 Centriprep filter and loaded onto a 100-ml Superose 12 column in 50 mM Hepes (pH 7.5), 150 mM KCl, and 10% glycerol. Purification of adipophilin was monitored by immunoblot analysis using anti-HisG antibody (Invitrogen). The final concentration of adipophilin was determined as before.
In Vitro Binding Assays.
TIP47/Rab9 binding studies.
In vitro binding studies were carried out with a modification of Krise et al. (6). 6xHisTIP47 and Rab9CLLL at the indicated concentrations were incubated at 37°C for 1 h in binding buffer (50 mM Hepes-KOH, pH 7.5/150 mM KCl/8 mM imidazole/1 mM MgCl2) with 0.1% BSA and 50 μM GTP in 450 μl. Where indicated, 6xHis-GFP was used instead of TIP47 as a negative control. Fifty microliters of Ni-NTA (Qiagen, Chatsworth, CA) beads (50% slurry), prewashed three times in binding buffer plus 0.005% Triton X-100, were then added to each reaction, and samples were rotated at room temperature for 20 min. Samples were centrifuged at 4,000 rpm for 1 min, resuspended, and transferred to 1-ml syringe columns. Samples were washed three times with 0.75 ml binding buffer, and then eluted with 0.25 ml of binding buffer with 250 mM imidazole for 15 min at 20°C. One-tenth of the eluate was resolved by 12.5% SDS/PAGE and then transferred to nitrocellulose for Western blotting. The resulting immunoblot was scanned on a Typhoon imager and the amount of Rab9CLLL binding was quantitated with image quant software. Eluates were also analyzed by SDS/PAGE and Coomassie blue staining to verify that all eluates contained equal amounts of TIP47 protein.
TIP47/CI-MPR binding studies.
TIP47 (200 nM) and GST–CI-MPRΔ12 (200 nM) were incubated in binding buffer as above with 0.1% BSA in a final volume of 450 μl for 1 h at room temperature. Fifty microliters of Ni-NTA beads (50% slurry) was then added to each sample at room temperature for 20 min. Samples were centrifuged, loaded onto columns, washed, and eluted as above. A portion (8%) of the eluate was analyzed by immunoblot, which was probed with rabbit anti-GST antibody (1:1,000) for 2 h.
Adipophilin/Rab9 binding studies.
Binding of Rab9CLLL to His-tagged TIP47, His-tagged adipophilin, or the control protein, His-tagged GFP, was performed as follows: 2 μM Rab9CLLL was combined with 0.3 μM TIP47, adipophilin, or GFP. In addition, reactions contained 0.6 mg/ml BSA, 100 μM GTP, 50 μl nickel agarose (added as a 50% slurry) in 50 mM Hepes, pH 7.5/150 mM KCl/1 mM MgCl2/10% glycerol. The nickel agarose was prepared by washing in this buffer plus 0.005% Triton X-100. The above components were incubated at room temperature for 1 h with rotation. The beads were then spun down for 2 min at 4,000 rpm, resuspended, and poured into 1-ml columns. The columns were washed twice with 1 ml buffer and protein complexes were then eluted as before. Reactions were performed in duplicate, and 50% of each eluate was loaded onto a 12% gel. The gel was blotted for Rab9CLLL by using a Rab9 culture supernatant antibody and quantified as before. Note that these experiments contained higher levels of Rab9 to maximize potential interaction with adipophilin.
Nucleotide binding studies.
Rab9 nucleotide binding studies were carried out as described (10).
Proteolytic Footprinting.
Three micrograms of purified TIP47 wild-type or TIP47 SVV167–169AAA was incubated with protease as indicated in buffer (25 mM Hepes-KOH, pH 8.0/150 mM KCl) in 40 μl for 30 min at 37°C. Reactions containing bromelain were supplemented with 1 mM EDTA; reactions containing proteinase K were supplemented with 10 mM CaCl2. Reactions containing bromelain were stopped by the addition of 5 μl of 300 μM tosyl-lysine chloromethylketone; reactions containing elastase and proteinase K were stopped by addition of 5 μl of 100 mM PMSF. Control reactions containing no protease were carried out in the presence of tosyl-lysine chloromethyl ketone and PMSF as above. Reactions were analyzed by 12.5% SDS/PAGE followed by Coomassie blue staining.
Circular Dichroism.
CD spectra were measured by using an Aviv model 62 ADS spectropolarimeter (Aviv Associates, Lakewood, NJ) with a 1-mm pathlength quartz cuvette. Protein was analyzed at 0.2 mg/ml in 10 mM Tris⋅HCl, pH 7.4. Measurements were conducted at 4°C, 1.0-nm resolution, at a scan rate of 6 nm⋅min−1.
Results
Previous work has shown that deletion of TIP47 residues 1–152 yields a protein that can bind both MPR cytoplasmic domains and Rab9 (4). In contrast, a larger deletion of TIP47 residues 1–187 yields a protein that can bind MPR cytoplasmic domains but not Rab9 (4). Thus, residues 152–187 seem important for Rab9 binding. To explore the role of these residues in greater detail, we scanned across this region by mutating amino acids to alanine, three residues at a time; we then determined the consequences of the mutations on the biochemical properties of TIP47. Each of the mutant proteins was expressed in E. coli and purified by virtue of an N-terminal His tag (Fig. 1A).
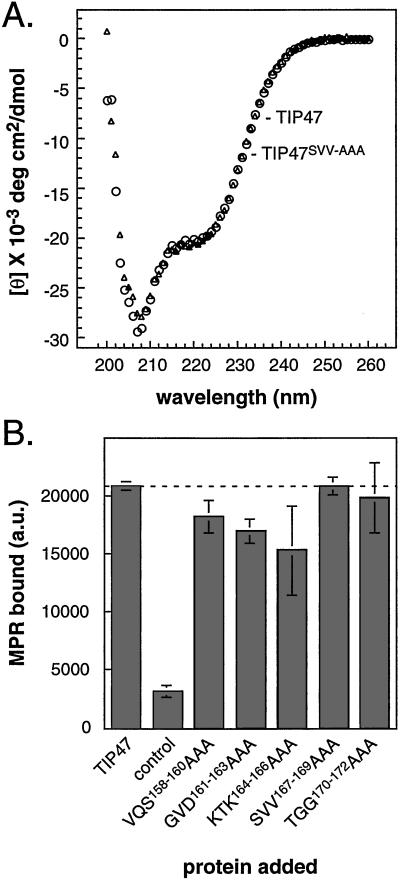
Figure 1.
(A) Purified TIP47 mutant proteins. Twelve mutants were generated that contain three alanines in place of residues 152–187 as described in Methods. Proteins were expressed in E. coli and purified. Shown is a 12.5% SDS-polyacrylamide gel stained with Coomassie blue. Lane 1, TIP47 wild type. Lanes 2–12, TIP47 mutants 1–10 and 12. TIP47 mutant 11 is not shown. The specific three residues mutated in each mutant protein are listed in B. (B) Alanine scanning mutagenesis reveals specific residues in TIP47 important for Rab9 binding. His-tagged TIP47 mutants (200 nM) were assayed for Rab9 (200 nM) binding. The data were corrected for background binding obtained by using His-tagged GFP instead of His-TIP47 and are representative of at least two, and for most mutants three, independent experiments. The dashed line is included to aid comparison with the wild-type binding level observed. Shown below, anti-Rab9 immunoblot data used for this analysis.
To test the ability of the mutant proteins to bind Rab9, TIP47 mutants were incubated with purified Rab9 and the amount of bound GTPase was determined by immunoblot. As shown in Fig. 1B, most of the mutant proteins bound essentially the same amount of Rab9 as wild-type TIP47. However, mutant proteins bearing alanine triplets instead of the normal residues 161–169 showed significantly diminished Rab9 binding; TIP47SVV-AAA bound only 22% the amount of Rab9 as wild-type TIP47. In these experiments, binding reactions were analyzed in parallel by SDS/PAGE and Coomassie blue staining, which confirmed that each sample contained equal amounts of TIP47.
Loss of Rab9 binding capacity was not caused by global misfolding by several independent criteria. First, CD spectra of TIP47 and the TIP47SVV-AAA mutant proteins were indistinguishable (Fig. 2A). CD analysis of wild-type TIP47 revealed that the protein contains a significant proportion of alpha helices (from the minima at 222 nm and 208 nm). These data show that the TIP47SVV-AAA mutant retains the same overall secondary structure as wild-type TIP47 and is not globally unfolded.
Figure 2.
(A) CD analysis of TIP47 (circles) and TIP47SVV-AAA (triangles). Units for mean residue ellipticity [θ] are degrees·cm2⋅dmol−1. (B) TIP47 and TIP47 mutants are fully active in binding MPR cytoplasmic domains. His-tagged TIP47 (200 nM) was incubated with GST–CI-MPRΔ12. Complexes were recovered on nickel beads and analyzed for the presence of GST–CI-MPR by immunoblot. Backgrounds were determined as in Fig. 1B and were subtracted. Shown are means and standard deviations of duplicate samples. Results are representative of three independent experiments. The dashed line is included as in Fig. 1.
To verify further that the loss of binding capacity for the mutant proteins was not caused by misfolding, we assayed their capacity to interact with MPR cytoplasmic domains. As shown in Fig. 2B, all of the TIP47 mutant proteins were functional in terms of their capacity to bind MPRs. Notably, the mutant that was most severely inhibited in terms of its ability to bind Rab9 (TIP47SVV-AAA) displayed 100% binding to MPR cytoplasmic domains. Thus, loss of Rab9 binding capacity cannot be attributed to global misfolding of the TIP47 mutant proteins.
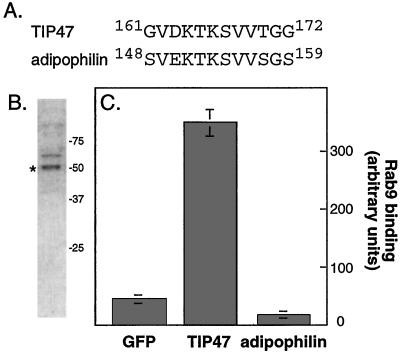
The residues 161GVD KTK SVV169 seemed most important for TIP47 binding. If these residues represent the core of an independent Rab9 binding motif, they may be unique to TIP47 and possibly other yet-to-be-identified Rab9 binding partners. TIP47 is related to a lipid droplet protein named adipophilin with ≈40% identity overall. Examination of the adipophilin sequence revealed the presence of sequences closely related to those important for Rab9 binding to TIP47, 148SVE KTK SVV156 (Fig. 3A). Thus it was essential to test whether adipophilin also bound to Rab9.
Figure 3.
(A) Sequence comparison of TIP47 and adipophilin. (B) Coomassie blue stained-SDS/PAGE analysis of purified adipophilin (*); adipophilin is the lower band in the doublet labeled, as determined by immunoblot. (C) Adipophilin does not bind Rab9. Binding was carried out as described in Methods; error bars represent the SE for duplicate measurements.
An adipophilin cDNA was amplified from a cDNA library and expressed in E. coli. The protein was purified from bacteria by ion exchange and gel filtration column chromatography (Fig. 3B). As shown in Fig. 3C, purified adipophilin failed to bind to Rab9 under conditions in which TIP47 showed significant binding. One model to explain these data would be that TIP47 residues 161–169 represent only part of a larger Rab9 binding site that is present in TIP47 and not in adipophilin. Alternatively, these residues may be important for the overall conformation of TIP47. In either case, they do not appear to represent an independent binding unit. The failure of adipophilin to bind Rab9 is consistent with adipophilin's distinct localization in relation to TIP47, in lipid droplets that lack MPRs and Rab9 (11). Thus, TIP47, but not adipophilin, binds Rab9.
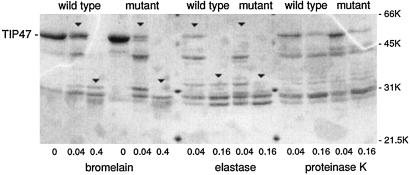
We also considered the possibility that residues key for Rab9 binding contributed in more subtle ways to the conformation of TIP47. In this scenario, such residues might or might not contribute to Rab9 binding directly. To test whether mutation of 167SVV169 subtly altered TIP47 conformation, we carried out partial proteolysis of wild-type TIP47 and TIP47SVV-AAA. As shown in Fig. 4, addition of increasing amounts of bromelain, elastase, and proteinase K generated a characteristic profile of cleavage products. In the case of bromelain and elastase, several cleavage products obtained with wild-type TIP47 were more resistant to cleavage at a given protease concentration than the TIP47SVV-AAA protein (compare downward arrowheads). Fewer differences were revealed by proteinase K cleavage.
Figure 4.
Proteolytic footprinting reveals minor conformational differences between TIP47 wild type and TIP47 SVV167–169AAA. TIP47 wild-type and mutant proteins (3 μg) were incubated with varying amounts of protease for 30 min at 37°C. Reactions were stopped by addition of protease inhibitors and analyzed by 12.5% SDS/PAGE. Results are representative of two, and for wild-type TIP47 three, independent experiments. Shown below the gel are the microgram amounts of protease used.
These minor differences in protease sensitivity are consistent with a slight alteration in the conformation of the TIP47SVV-AAA mutant protein. However, it was also possible that the differential cleavage was caused by the introduction (or removal) of a protease cleavage site upon mutation of residues SVV167–169 to AAA. This possibility seems unlikely for the following reason. A single cleavage at that site would yield two products, the larger of which would be 29.5 kDa, smaller than those observed. Even if the 29.5-kDa product migrated anomalously on this gel, the fact that bromelain and elastase yielded different products above 31 kDa demonstrates that at least one of these enzymes cleaves at a different site. Thus, mutant TIP47SVV-AAA differs slightly from the wild type in its overall protease sensitivity.
These experiments suggest that TIP47SVV-AAA is very subtly altered in its overall conformation; it is properly folded in that it can bind MPR cytoplasmic domains, but it is unable to bind Rab9. Further analyses will be needed to identify the precise Rab9 binding interface on TIP47.
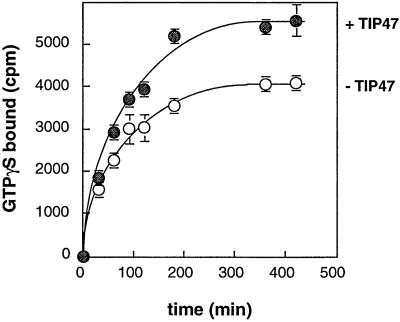
As an initial approach to exploring the extent of surface contacts between TIP47 and Rab9, we tested whether TIP47 altered the nucleotide binding properties of the Rab9 GTPase. When Rab9 was incubated in the presence of [35S]GTPγS, it exchanged bound GDP for the radiolabeled GTPγS over a period of approximately 3 h before reaching saturation (Fig. 5). In the presence of TIP47, the overall extent of nucleotide exchange was higher, although the rate of exchange was not enhanced. This increase was not caused by the binding of nucleotide to TIP47, as determined in control reactions (data not shown). These data confirm an interaction between TIP47 and Rab9 in solution and suggest that TIP47 is not a nucleotide exchange factor for Rab9. In support of this conclusion, the presence of TIP47 did not alter the off-rate for GDP from Rab9 (data not shown).
Figure 5.
TIP47 enhances Rab9 binding capacity, but is not a nucleotide exchange factor. Rab9 was incubated with 3 μM [35S]GTPγS at 37°C for varying times with or without TIP47 (100 nM) as indicated. Rab9-nucleotide complexes were isolated on nitrocellulose filters and analyzed by scintillation counting. Shown are means and standard deviations of duplicate samples. Results are representative of four independent experiments.
It is possible that the somewhat labile, purified Rab9 GTPase is stabilized in the presence of TIP47 and thus a larger proportion of the molecules can exchange nucleotide. The increase in nucleotide binding was not observed with two control proteins, α-SNAP or GFP (not shown). Thus, TIP47 binding to Rab9 GTPase appears to stabilize the GTPase from nonspecific, thermal denaturation, yielding more molecules that are competent to bind GTP, without influencing the rates of nucleotide binding or release.
Discussion
TIP47 is required in vivo for the transport of MPRs from late endosomes to the Golgi (3). In addition, the ability of TIP47 to bind Rab9 correlates strongly with TIP47's functionality in living cells (4). Expression of a mutant TIP47 that is impaired in Rab9 binding changes the morphological appearance of the Rab9-positive late endosome compartment (5) and blocks MPR transport back to the Golgi complex (4) and to the cell surface (5). Our current working model is that late endosome-localized Rab9 helps to recruit the predominantly cytosolic TIP47 onto MPRs present in late endosomes. We have recently shown that Rab9 is present on the vesicles that bud off of late endosomes and dock (and then fuse) with the trans Golgi (5); we presume that TIP47 is also present on these transport vesicles, at least when they are first formed.
We have defined a set of discrete residues that when mutated in TIP47 lead to a significant diminution in the ability of that protein to bind Rab9. Closely related residues in adipophilin are not sufficient to confer Rab9 binding capacity on that protein, thus the sequences alone do not appear to define an independent Rab9 binding site.
Mutant TIP47 proteins that displayed diminished Rab9 binding capacity were fully competent to bind MPR cytoplasmic domains. This observation showed that the mutants were not grossly misfolded and also confirmed the independence of Rab9 and MPR cytoplasmic domain interaction sites. Because the affinity of TIP47 for MPR cytoplasmic domains is enhanced in the presence of bound Rab9 and the affinity of TIP47 for Rab9 is enhanced by MPR cytoplasmic domains, the simplest model is that binding of either component to its respective binding site triggers a conformational change in TIP47. Partial proteolysis of a TIP47 Rab9-binding domain mutant suggested that loss of Rab9 binding capacity could be correlated with a subtle conformational alteration. Structure determination of TIP47 in the presence and absence of Rab9 will be needed to reveal the underlying structural changes involved.
Together our data suggest that TIP47 interacts with both MPRs and Rab9 on late endosome membranes and forms a microdomain on that compartment (3–5). This interaction is likely to occur via independent Rab9 and MPR binding sites on the TIP47 protein, as demonstrated here. The ability of TIP47 to interact with Rab9 helps TIP47 fulfill its role in transporting MPRs from late endosomes to the trans-Golgi complex. How this interaction is reversed to permit TIP47 release from transiting vesicles represents another important question for future investigation.
Acknowledgments
This research was supported by a research grant from the National Institutes of Health (DK37335).
Abbreviations
- TIP47
tail-interacting protein of 47 kDa
- MPR
mannose 6-phosphate receptor
- CI-MPR
cation-independent MPR
- GST
glutathione S-transferase
- GFP
green fluorescent protein
References
- 1.Kornfeld S, Mellman I. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- 2.Kornfeld S. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- 3.Diaz E, Pfeffer S R. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- 4.Carroll K S, Hanna J, Simon I, Krise J, Barbero P, Pfeffer S R. Science. 2001;292:1373–1376. doi: 10.1126/science.1056791. [DOI] [PubMed] [Google Scholar]
- 5.Barbero P, Bittova L, Pfeffer S R. J Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krise J P, Sincock P M, Orsel J G, Pfeffer S R. J Biol Chem. 2000;275:25188–25193. doi: 10.1074/jbc.M001138200. [DOI] [PubMed] [Google Scholar]
- 7.Orsel J G, Sincock P M, Krise J P, Pfeffer S R. Proc Natl Acad Sci USA. 2000;97:9047–9051. doi: 10.1073/pnas.160251397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombardi D, Soldati T, Riederer M A, Goda Y, Zerial M, Pfeffer S R. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riederer M A, Soldati T, Shapiro A D, Lin J, Pfeffer S R. J Cell Biol. 1994;125:573–582. doi: 10.1083/jcb.125.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro A D, Riederer M A, Pfeffer S R. J Biol Chem. 1993;268:6925–6931. [PubMed] [Google Scholar]
- 11.Barbero P, Buell E, Zulley S, Pfeffer S R. J Biol Chem. 2001;276:24348–24351. doi: 10.1074/jbc.M102468200. [DOI] [PubMed] [Google Scholar]