Abstract
The Ipl (Tssc3) gene lies in an extended imprinted region of distal mouse chromosome 7, which also contains the Igf2 gene. Expression of Ipl is highest in placenta and yolk sac, where its mRNA is derived almost entirely from the maternal allele. Ipl encodes a small cytoplasmic protein with a pleckstrin-homology (PH) domain. We constructed two lines of mice with germ-line deletions of this gene (Iplneo and IplloxP) and another line deleted for the similar but nonimprinted gene Tih1. All three lines were viable. There was consistent overgrowth of the Ipl-null placentas, with expansion of the spongiotrophoblast. These larger placentas did not confer a fetal growth advantage; fetal size was normal in Ipl nulls with the Iplneo allele and was decreased slightly in nulls with the IplloxP allele. When bred into an Igf2 mutant background, the Ipl deletion partially rescued the placental but not fetal growth deficiency. Neither fetal nor placental growth was affected by deletion of Tih1. These results show a nonredundant function for Ipl in restraining placental growth. The data further indicate that Ipl can act, at least in part, independently of insulin-like growth factor-2 signaling. Thus, genomic imprinting regulates multiple pathways to control placental size.
The Ipl gene, also known as Tssc3, lies on distal chromosome 7 of the mouse and human chromosome 11p15.5 (1, 2). This region contains multiple imprinted genes clustered in 1 Mb of DNA. Two of these, p57Kip2 (Cdkn1c) and Igf2, control fetal and placental growth in mice (3–7) and humans (8). Similar to these genes, Ipl is highly expressed in the extraembryonic tissues (1), but in contrast to these genes, Ipl is expressed only weakly in the embryo proper (1, 9). Ipl encodes a cytoplasmic protein with a pleckstrin-homology (PH) domain (9), thus by analogy with other PH-domain proteins it may modulate cell signaling, intracellular trafficking, or other processes that depend on phosphatidylinositol lipid second messengers. Ipl has two close relatives: TDAG51 and Tih1. Of these genes, Tih1 is most similar to Ipl (9). Tih1 is located in a nonimprinted region of mouse chromosome 1, and it is expressed biallelically (9). To determine the function of Ipl and to accumulate data relevant to the biological rationale for imprinting, we created mice with germ-line deletions of Ipl and Tih1.
Materials and Methods
Gene Deletions, Genotyping, and Gene Expression.
To make the Iplneo targeting vector, a 5-kb 5′ genomic NotI restriction fragment and a 6-kb 3′ EcoRI/XbaI fragment flanking Ipl were ligated upstream and downstream of Pgk-Neo in pPNT1, yielding pPNT-Ipl. To make the IplloxP targeting vector, the Pgk-Neo cassette of pPNT-ΔIpl was replaced by that from pLNL, which includes flanking loxP sites. To verify homologous recombination, we used genomic PCR products upstream and downstream of the 5′ and 3′ cassettes. Recombination of the loxP sites was induced by crossing to HSP-Cre transgenic mice (10). The Ipl deletions (encompassing nucleotides 77,444–78,990 of GenBank accession no. AP001294) eliminate both exons of the gene and delete the 5′ half of its upstream CpG island. To make the Tih1neo targeting vector, a 6-kb upstream KpnI fragment and a 6-kb downstream HindIII/NotI fragment were cloned into pPNT1. Homologous recombination was verified with flanking probes. The resulting deletion (nucleotides 579–2,096 of GenBank accession no. AF151099) eliminated all of exon 1 and part of exon 2, deleting the entire Tih1 coding region. The Igf2lacZ deletion was an insertion of lacZ-Neo in exon 4 of Igf2. Genotyping was performed by Southern blotting of EcoRI digests of genomic DNAs with a probe for Igf2 exon 1. Gene expression was measured by Northern blotting of total RNAs followed by PhosphorImaging (Molecular Dynamics).
Mouse Crosses and Weights.
Timed matings of heterozygotes were performed, and each conceptus was separated into yolk sac, placenta, and embryo. The placentas and embryos were fixed in 10% formalin and then weighed. The unfixed yolk sacs were divided in half for extraction of DNA and RNA and were used for genotyping by Southern blotting and detection of Ipl mRNA by Northern blotting. The crosses of female Iplneo heterozygotes with male Igf2lacZ heterozygotes were carried out similarly. t tests were two-sample, equal variance. The percentage differences in weights described in the text were calculated relative to the mean of the expression-positive conceptuses for that cross.
Immunohistochemistry and Morphometry.
Ipl protein was detected with affinity-purified polyclonal antiserum developed by immunoperoxidase (9). Antibodies against BrdUrd (Sigma) and Ki67 (NovoCastra, Newcastle, U.K.) were detected by the same method. BrdUrd was injected i.p. 40 min before killing the mice. Measurements of the placental labyrinth and spongiotrophoblast zone were made from digitized images of ×2 microscopic fields of midline sections stained with hematoxylin/eosin by using the lasso tool and histogram function of Adobe PHOTOSHOP.
Results
Mice with Deletions of Ipl Are Viable and Fertile.
We used gene targeting in embryonal stem cells to make two knockout (KO) alleles of Ipl. The first replaced the small Ipl gene by Pgk-Neo (Iplneo). The second (IplloxP) was made in an independent embryonal stem clone and led to replacement of Ipl by a recombined loxP site (Fig. 1 a and b). Both alleles were transmitted through the germ line. Breeding in the mixed C57BL/6;129/Sv background yielded progeny in Mendelian proportions (see Mendelian Proportions and Longitudinal Analyses of Adult Mice, which is published as supporting information on the PNAS web site, www.pnas.org). In placentas and yolk sacs there was an absence of Ipl mRNA in the homozygotes, nearly undetectable levels in the conceptuses that inherited the disrupted allele from the mother (+/−mat), and wild-type levels in the +/−pat conceptuses (Fig. 1c). This result highlighted the strong dosage regulation of Ipl by parental imprinting. Loss of Ipl protein in +/−mat and −/− placentas was verified by Western blotting (data not shown). Ipl-null heterozygotes and homozygotes appeared normal and showed normal activity and fecundity. Necropsies showed normal histology of the major organs, and we did not observe genotype-specific morbidity or neoplasia (see Mendelian Proportions and Longitudinal Analyses of Adult Mice).
Figure 1.
Germ-line deletions of the Ipl and Tih1 genes. (A) Replacement of Ipl by Pgk-Neo or loxP. Regions included in the targeting vector are indicated in bold. Recombined loxP sites are the triangle. Exons are rectangles. K, KpnI; B, BamHI; US, upstream probe; DS, downstream probe. (B) Southern blot of mouse DNAs with genotypes indicated. (C) Northern blot showing loss of Ipl mRNA in yolk sacs with +/− maternal (mat) and −/− genotypes and wild-type levels of Ipl mRNA in yolk sac with +/− paternal (pat) genotype. Similar results were obtained with placental RNAs. The arrow indicates the major Ipl transcript. Assignment of mat and pat alleles in this heterozygous cross is inferred from analyses of other crosses in which one parent was Ipl−/− and the other was Ipl+/+. (D) Northern blot characterizing the Tih1 deletion with RNA from embryo fibroblasts. EtBr, ethidium bromide.
Isolated Placental Overgrowth in Ipl-Null Conceptuses.
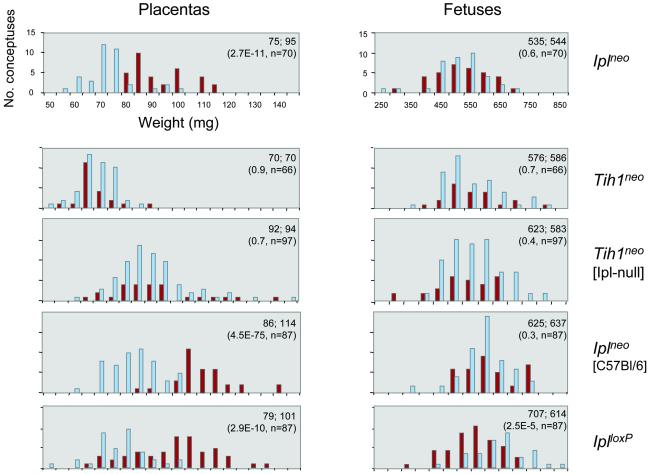
Expression of Ipl is highest in the placenta and yolk sac (1, 9). We therefore carried out timed matings of heterozygotes (het × het crosses) and weighed the placentas and embryos. This analysis revealed a consistent increase in placental weights of the Ipl-null conceptuses in both KO lines (Fig. 2). Because the imprint was robust, the Ipl-null data represent the combined Ipl−mat/+pat and Ipl−/− genotypes, and the Ipl-positive data represent the combined Ipl+mat/−pat and Ipl+/+ genotypes. For the Iplneo line in the mixed genetic background (C57BL/6 × 129/Sv), the placental overgrowth was significant by 12.5 dpc (22% increase in mean weight, t test 1.2E-05, which is 1.2 × 10−5, n = 59; data not shown) and was greater at 16.5 dpc (27% increase; Fig. 2). Crossing this allele into the pure C57BL/6 background accentuated the magnitude and significance of the placental overgrowth (33% increase; Fig. 2). Weighing desiccated placentas also revealed an increased mean weight among the Ipl nulls, excluding edema as an explanation for the placentomegaly (data not shown). The placental overgrowth was not accompanied by any change in weights of the corresponding fetuses in the Iplneo line. However, in the IplloxP line, we observed a small (13%) but significant decrease in the mean fetal weight among the Ipl nulls, an effect opposite in direction to the highly significant 28% increase in mean placental weight among these same conceptuses (Fig. 2). The basis for this subtle difference between Iplneo and IplloxP is not clear, but it may reflect cis effects of the Pgk-Neo insertion (see below).
Figure 2.
Isolated placental overgrowth in Ipl-null conceptuses. Data are shown for heterozygous crosses at 16.5 days postcoitum (dpc). Dark red bars denote conceptuses null for expression; light blue denotes positive for expression of the genes indicated on the right. Placental overgrowth is seen in the Ipl-null conceptuses with both the Iplneo and the IplloxP deletions and is accentuated after back-crossing to C57BL/6 for six generations (Iplneo [C57BL/6]). Fetal overgrowth is not observed. Deletion of Tih1 affects neither embryonic nor placental weight, but the placental weight distribution is shifted upward in the Ipl-null background (Tih1neo [Ipl-null]). Ipl-positive refers to combined +/+ and +mat/−pat genotypes, and Ipl-negative refers to combined −/− and −mat/+pat with assignment of paternal and maternal alleles inferred from expression in the yolk sacs. Tih1-positive refers to combined +/+ and +/− genotypes; Tih1-negative refers to the −/− genotype. Similar results were obtained when only Tih1+/+ and Tih1−/− genotypes were compared. Weights (mg) and t tests are shown as mean(expression-positive); mean(expression-negative); t value, number of conceptuses.
Pairwise analysis of fetal and placental weights showed positive but weak correlation coefficients (see Fig. 7, which is published as supporting information on the PNAS web site), which is consistent with previous findings in normal mice (11). In mouse development, placental weight reaches a maximum at 16.5 dpc and declines during the next 4 days before birth, but during this time the weight of the fetus more than doubles (4). In additional heterozygous crosses assessed at 18.5 dpc, the fetal weights doubled relative to 16.5 dpc, but there still was no evidence of a fetal growth advantage conferred by the larger Ipl-null placentas (see Fig. 6, which is published as supporting information on the PNAS web site).
Using BrdUrd incorporation, staining with anti-Ki67 antibody, and DNA end-labeling assays, we asked whether the increase in placental weights was caused by hyperproliferation of trophoblast cells and/or decreased apoptosis. At 12.5 dpc, apoptosis in the labyrinth and spongiotrophoblast was minimal even in wild-type placentas, although rare apoptotic cells were seen near the fetal/maternal interface. Counting of BrdUrd- and Ki67-positive nuclei at 12.5 and 16.5 dpc did not reveal significant differences in the Ipl-positive versus null placentas (data not shown). We also attempted to evaluate differences at earlier stages, but this failed because of very high baseline proliferation. Because Ipl is highly expressed from implantation onward (9), the net weight increase observed by midgestation in the Ipl-null placentas likely reflects the cumulative effect of a small net difference in trophoblast proliferation.
Ipl Protein Marks an Early Subpopulation of Trophoblast.
By midgestation, the placenta is composed of three zones (12). The labyrinth, containing trophoblast and fetal and maternal blood vessels, lies immediately under the chorionic plate. Trophoblast in this zone has a trilaminar arrangement, with type I cells adjacent to maternal vessels, proliferating type II cells centrally, and type III cells adjacent to the fetal vessels (13, 14). Deep to the labyrinth is the spongiotrophoblast followed by trophoblast giant cells abutting the maternal decidua. Ipl mRNA is abundant in extraembryonic ectoderm at the earliest stages postimplantation, with expression restricted to labyrinthine trophoblast by 10.5 dpc (9). Between 12.5 and 14.5 dpc, Ipl mRNA and protein in the placenta declines markedly (see Fig. 12, which is published as supporting information on the PNAS web site). This change does not reflect a decline in expression per cell, but instead is caused by a decrease in the number of Ipl-expressing cells (Fig. 3). On day 12 of gestation, the abundant Ipl-expressing cells are trophoblast at the chorionic plate and clustered type II trophoblast deeper in the labyrinth (Fig. 3). Terminally differentiated trophoblast expresses less Ipl protein, with faint staining in the cytoplasm of type III cells adjacent to the fetal blood vessels and no staining in the large type I cells adjacent to the maternal vessels (Fig. 3). As early as 14.5 dpc, Ipl protein becomes confined to a monolayer of cells at the chorionic plate and to rare cells in septa that protrude into the labyrinth (Fig. 3).
Figure 3.
Ipl protein marks a subpopulation of trophoblast. (A) Antipeptide antibody detects Ipl protein confined to the labyrinth. This layer has expanded by 14.5 dpc, but the number of Ipl-expressing cells is relatively reduced. The intensity of staining per expressing cell remains constant. sp, spongiotrophoblast; la, labyrinthine trophoblast. (B) Higher magnification shows that the Ipl-positive cells are clustered between blood vessels (small bold arrows; type II cells) and do not include the large type I trophoblast cells that are adjacent to maternal blood vessels (large bold arrows). The small plain arrows indicate type III cells that extend the Ipl-positive cytoplasmic process around fetal vessels. BrdUrd (BrdU) is incorporated in type II cell nuclei but not in differentiated type I cells. mv, maternal blood vessel; fv, fetal blood vessel.
Expansion of Spongiotrophoblast in Ipl-Null Placentas.
Despite the localized expression of Ipl, the placental overgrowth in the Ipl-null conceptuses was global, affecting both the labyrinth, which was slightly expanded laterally, and more dramatically the spongiotrophoblast, which was expanded laterally and in thickness (Fig. 4a). Microscopic examination did not show specific pathological abnormalities. However, the Ipl-null placentas had an increased number of glycogen-containing cells in the spongiotrophoblast zone (Fig. 4c). That the spongiotrophoblast was expanded disproportionately in the Ipl-null placentas was confirmed by morphometry, which showed that the ratio of cross-sectional area of the spongiotrophoblast to labyrinth was increased more than 1.5-fold (Fig. 4b; see Fig. 10, which is published as supporting information on the PNAS web site). This phenotype was reflected in a net overexpression of the spongiotrophoblast marker 4311 (15) and relative underexpression of the labyrinth marker Dlx3 (16) as measured by Northern blotting and microarray hybridization (see Figs. 11 and 13, which are published as supporting information on the PNAS web site). Relevant to the Ipl-null phenotype of placental overgrowth associated with either no change (Iplneo) or a slight reduction (IplloxP) in fetal growth, the abundance of glycogenated spongiotrophoblast was correlated previously with placental inefficiency, indicated by reduced fetal/placental weight ratios, in interspecific mouse crosses (17).
Figure 4.
Expansion of spongiotrophoblast in Ipl-null placentas. (A) Midline placental sections (hematoxylin/eosin) from the IplloxP line at 16.5 dpc. There is a global increase in placental size with deletion of Ipl, but the spongiotrophoblast (sp) is expanded disproportionately. la, labyrinthine trophoblast. (B) Ratios of the area of spongiotrophoblast to labyrinth (Spong/Lab) in midline sections of placentas from heterozygous crosses (16.5 dpc). Data from single crosses are shown. In data from four heterozygous crosses with the IplloxP line, combined after normalization to the mean value for each cross, the ratio was 1.22 ± 0.20 and 0.78 ± 0.13 for the Ipl-null (Neg) and Ipl-positive (Pos) placentas, respectively (n = 34). (C) High-power fields of Ipl-positive and Ipl-negative placentas. The panels on the left are from the Iplneo line at 12.5 dpc (PAS). Lines indicate the giant cell layer (gc) adjacent to maternal decidua (ma). The panels on the right are at 16.5 dpc (hematoxylin/eosin) centered on the spongiotrophoblast. There is expansion of this layer with an increase of glycogen-containing cells (arrow).
Deletion of Tih1 Does Not Affect Placental Growth.
Tih1 encodes a 125-aa protein that shares 53% identity with Ipl over its central 100-aa PH domain but which is expressed more broadly than Ipl (9). In the placenta, Tih1 mRNA can be detected in the labyrinth but at low levels (9). Because Tih1 is the closest relative of Ipl based on its sequence, we wished to test for overlap in the function of these two genes. Accordingly, we deleted the Tih1 gene by replacement with Pgk-Neo (Materials and Methods; Fig. 1d). Homozygotes lacking Tih1 were obtained in Mendelian proportions, were viable and fertile, lacked gross abnormalities, and did not show genotype-specific morbidity or neoplasia (see Mendelian Proportions and Longitudinal Analyses of Adult Mice). Northern blotting showed dose-dependent expression of Tih1 mRNA in these mice, which was independent of parental origin of the KO allele and consistent with a lack of imprinting (Fig. 1d). In contrast to the findings for Ipl, homozygotes lacking Tih1 showed normal placental and embryonic weights (t tests comparing wild-type, heterozygous, and homozygous conceptuses all were nonsignificant), and in crosses with Ipl KO mice, the absence of Tih1 did not increase the placental overgrowth (Fig. 2).
The Placental Overgrowth Is Not Caused by Cis Effects.
Ipl lies in an ≈1-Mb imprinted chromosomal domain, which contains several other genes that regulate growth. Placental size is affected by KOs of three of these genes: H19 (when deleted together with its 5′ insulator), Igf2 and p57Kip2, and a fourth imprinted gene in the region, Mash2, which controls placental development (4, 6, 7, 18). Instances in which the expression of one gene is affected by the deletion of an adjacent gene have been documented in imprinted regions, as exemplified by inverse regulation of Igf2 and H19 (19). We therefore wished to exclude cis effects of the Ipl deletions on neighboring genes. The Iplneo allele partially down-modulated the closest flanking gene, Impt1, (located 3 kb downstream of Ipl) but did not measurably affect the expression of several other genes in the domain. In particular, p57Kip2, which is located only 40 kb from Ipl, was expressed normally in yolk sacs, placentas, and fetuses, as was the Igf2 gene, located ≈1 Mb from Ipl (see Figs. 8, 9, and 14 and Table 1, which are published as supporting information on the PNAS web site). The IplloxP allele did not have detectable effects on expression of any of these flanking genes (see Figs. 8, 9, and 14 and Table 1).
Ipl Restrains Placental Growth in an Igf2-Null Background.
At least two imprinted genes, Igf2 and Igfr, affect the insulin-like growth factor-2 signaling pathway in the placenta (4, 20). In addition, certain members of the PH-domain superfamily are cytoplasmic components of this and related signaling pathways (21, 22). We therefore wished to determine whether the Ipl gene might restrain placental growth by antagonizing the growth-promoting effects of Igf2. To test for a genetic interaction, we crossed Ipl+/− females (Iplneo line) with Igf2+/− males (Igf2lacZ line) and measured fetal and placental weights at 16.5 dpc. Because the silenced allele of Ipl is paternal, whereas the silenced allele of Igf2 is maternal, all four possible combinations of Ipl and Igf2 mRNA expression (Ipl+ Igf2+, Ipl− Igf2+, Ipl+ Igf2−, and Ipl− Igf2−) were represented among the conceptuses. As shown in Fig. 5a, loss of Ipl mRNA conferred a growth advantage to the placentas in the Igf2-null background, which led to a partial rescue of the placental growth retardation caused by the deletion of Igf2. However, loss of Ipl mRNA did not rescue the fetal growth retardation (Fig. 5a). The ability of Ipl to restrain placental growth in an Igf2-null background indicates that the Ipl protein can act in a growth-regulating pathway that is distinct from Igf2 signaling. These crosses also substantiated the effect of Ipl deletion on the spongiotrophoblast/labyrinth ratio. Because the placentas in the Ipl− Igf2− group were reduced slightly in weight relative to those in the Ipl+ Igf2+ group, we could assess changes in this ratio independently of increases in placental size. Morphometry in Ipl− Igf2− and Ipl+ Igf2+ placentas showed that the spongiotrophoblast/labyrinth ratio was increased by 1.6-fold in the (Ipl− Igf2−) placentas (Fig. 5b), which indicates that the expansion of the spongiotrophoblast caused by deletion of Ipl is not a trivial consequence of the increased size of the Ipl-null placentas.
Figure 5.
Deletion of Ipl promotes placental but not fetal growth in an Igf2-null background. (A) Placental and fetal weights of conceptuses with the indicated Ipl and Igf2 expression status, measured at 16.5 dpc. Deletion of Ipl partially rescues the placental growth retardation but does not rescue fetal growth in the Igf2 nulls. The horizontal and vertical bars indicate means and standard deviations. t tests for equivalence of the weight distributions are indicated. (B) Ratios of the area of the spongiotrophoblast to labyrinth (Spong/Lab) in midline sections of all Ipl+Igf2+ and Ipl−Igf2− placentas obtained from pooling two crosses (16.5 dpc) carried out between the Iplneo and Igf2lacZ lines. Although the Ipl−Igf2− placentas are slightly smaller than the controls, they show an increased spongiotrophoblast/labyrinth ratio.
Discussion
Among vertebrates, genomic imprinting is restricted to placental mammals. Imprinted genes affecting the growth or development of the placenta include Mash2 (23), Igf2 (4, 6, 20, 24), Igf2r (20), p57Kip2 (6, 7), and Esx1 (25). With the addition of Ipl, the products of this group of genes span diverse biochemical functions. Mash2 and Esx1 encode transcription factors. Igf2 and Igf2r act in the Igf2 pathway, with Igf2 functioning as the agonist and Igf2r (because of its Igf2-clearance function) acting as an antagonist. The product of p57Kip2 acts as a cyclin/cyclin-dependent kinase inhibitor in the cell cycle, and this gene interacts genetically with Igf2 in controlling placental growth (6). The Ipl gene product represents yet another biochemical category, because it belongs to the class of cytoplasmic proteins that contain PH domains. The current data show that Ipl is an antagonist of placental growth, which can act via a mechanism that does not require inhibition of insulin-like growth factor-2 signaling. We conclude that genomic imprinting is an overarching mechanism that regulates multiple biochemical pathways to control placental growth.
A striking feature of the Ipl-null phenotype is the overgrowth of the spongiotrophoblast layer. Global placental overgrowth caused by deletion of a labyrinth-specific gene must reflect mechanisms that coordinate the growth of the labyrinth and spongiotrophoblast. A similar overgrowth of the spongiotrophoblast was reported after deletion of another imprinted gene, Esx1, the expression of which in midgestation is confined to the labyrinth (25). Precursor cells in the early extraembryonic ectoderm and the ectoplacental cone, tissues that strongly express Ipl (9), contribute to the definitive spongiotrophoblast layer (12), and thus it is possible that the overgrowth of spongiotrophoblast in the Ipl-null conceptuses results from an increased number of these cells. A non-cell-autonomous effect such as altered secretion of a trophic factor caused by deletion of Ipl, is an alternative. Another interesting point raised by the overgrowth of spongiotrophoblast in the Ipl nulls is the similarity of this phenotype to that observed in the placentas of cloned mice (26). The abnormalities seen in these conceptuses likely are accounted for by failure to establish appropriate epigenetic states at multiple loci (27), but an obvious question raised by our findings is whether the Ipl gene might be silenced in some clones.
Our findings also should be factored into attempts to evaluate the intergenomic conflict hypothesis for the evolution of imprinting (28, 29). This hypothesis, which is supported by data for a number of imprinted genes, invokes a selective advantage for maternal alleles if they carry an imprint that moderates the nutritional demands on the mother by the conceptuses. In contrast, paternal alleles are predicted to gain an advantage by having a pattern of imprinting that benefits the fetus or neonate at the expense of maternal resources. The phenotype described here suggests that expression of Ipl from the maternal allele conserves the mother's resources, because Ipl functions to restrain placental size, which is consistent with the conflict hypothesis from the maternal standpoint. But because the larger placentas of the Ipl mutants confer no fetal growth benefit, it is unclear how the epigenetic state of the paternal allele, which is decoded as gene silencing, could convey a selective advantage to the paternal genome. Placentomegaly without a fetal growth advantage is seen also in mice with deletions of the imprinted Esx1 gene and in mice with paternal uniparental disomy for chromosome 12 (25, 30). Additional examples will be needed to evaluate this issue further. It could be argued that the Ipl gene, which is under the control of cis-acting elements that establish the imprints of multiple genes in this chromosomal domain (31), might be an “innocent bystander” in the evolution of the domain. Nevertheless, the current data raise the possibility that maternal drive to restrain the size of the placenta may be a sufficient explanation for conservation of imprinting at some loci.
Supplementary Material
Acknowledgments
We thank G. Kelsey, M. Coustancia, and W. Dean for help with the Igf2 mutant mice and T. Bestor and A. Efstradiatis for comments. This work was supported by grants from the National Institutes of Health (to B.T. and D.F.) and the Human Frontiers Science Project (to B.T. and W.R.).
Abbreviations
- PH
pleckstrin homology
- KO
knockout
- mat
maternal
- pat
paternal
- dpc
days postcoitum
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Qian N, Frank D, O'Keefe D, Dao D, Zhao L, Yuan L, Wang Q, Keating M, Walsh C, Tycko B. Hum Mol Genet. 1997;6:2021–2029. doi: 10.1093/hmg/6.12.2021. [DOI] [PubMed] [Google Scholar]
- 2.Lee M P, Feinberg A P. Cancer Res. 1998;58:1052–1056. [PubMed] [Google Scholar]
- 3.DeChiara T M, Robertson E J, Efstratiadis A. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 4.Baker J, Liu J P, Robertson E J, Efstratiadis A. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 5.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Nature (London) 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 6.Caspary T, Cleary M A, Perlman E J, Zhang P, Elledge S J, Tilghman S M. Genes Dev. 1999;13:3115–3124. doi: 10.1101/gad.13.23.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang P, Wong C, DePinho R A, Harper J W, Elledge S J. Genes Dev. 1998;12:3162–3167. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maher E R, Reik W. J Clin Invest. 2000;105:247–252. doi: 10.1172/JCI9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank D, Mendelsohn C L, Ciccone E, Svensson K, Ohlsson R, Tycko B. Mamm Genome. 1999;10:1150–1159. doi: 10.1007/s003359901182. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich P, Dragatsis I, Xuan S, Zeitlin S, Efstratiadis A. Mamm Genome. 2000;11:196–205. doi: 10.1007/s003350010037. [DOI] [PubMed] [Google Scholar]
- 11.McLaren A. J Reprod Fertil. 1965;9:79–98. doi: 10.1530/jrf.0.0090079. [DOI] [PubMed] [Google Scholar]
- 12.Cross J C. Semin Cell Dev Biol. 2000;11:105–113. doi: 10.1006/scdb.2000.0156. [DOI] [PubMed] [Google Scholar]
- 13.Carpenter S J. Am J Anat. 1975;143:315–347. doi: 10.1002/aja.1001430305. [DOI] [PubMed] [Google Scholar]
- 14.Sapin V, Dolle P, Hindelang C, Kastner P, Chambon P. Dev Biol. 1997;191:29–41. doi: 10.1006/dbio.1997.8687. [DOI] [PubMed] [Google Scholar]
- 15.Lescisin K R, Varmuza S, Rossant J. Genes Dev. 1988;2:1639–1646. doi: 10.1101/gad.2.12a.1639. [DOI] [PubMed] [Google Scholar]
- 16.Morasso M I, Grinberg A, Robinson G, Sargent T D, Mahon K A. Proc Natl Acad Sci USA. 1999;96:162–167. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurz H, Zechner U, Orth A, Fundele R. Anat Embryol. 1999;200:335–343. doi: 10.1007/s004290050284. [DOI] [PubMed] [Google Scholar]
- 18.Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner A L. Nature (London) 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 19.Tilghman S M. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 20.Eggenschwiler J, Ludwig T, Fisher P, Leighton P A, Tilghman S M, Efstratiadis A. Genes Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yenush L, Zanella C, Uchida T, Bernal D, White M F. Mol Cell Biol. 1998;18:6784–6794. doi: 10.1128/mcb.18.11.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemmon M A, Ferguson K M. Biochem J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 23.Guillemot F, Caspary T, Tilghman S M, Copeland N G, Gilbert D J, Jenkins N A, Anderson D J, Joyner A L, Rossant J, Nagy A. Nat Genet. 1995;9:235–242. doi: 10.1038/ng0395-235. [DOI] [PubMed] [Google Scholar]
- 24.Sun F L, Dean W L, Kelsey G, Allen N D, Reik W. Nature (London) 1997;389:809–815. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Behringer R R. Nat Genet. 1998;20:309–311. doi: 10.1038/3129. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka S, Oda M, Toyoshima Y, Wakayama T, Tanaka M, Yoshida N, Hattori N, Ohgane J, Yanagimachi R, Shiota K. Biol Reprod. 2001;65:1813–1821. doi: 10.1095/biolreprod65.6.1813. [DOI] [PubMed] [Google Scholar]
- 27.Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout W M, 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 28.Haig D, Graham C. Cell. 1991;64:1045–1046. doi: 10.1016/0092-8674(91)90256-x. [DOI] [PubMed] [Google Scholar]
- 29.Moore T, Haig D. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 30.Georgiades P, Watkins M, Surani M A, Ferguson-Smith A C. Development (Cambridge, UK) 2000;127:4719–4728. doi: 10.1242/dev.127.21.4719. [DOI] [PubMed] [Google Scholar]
- 31.Cleary M A, van Raamsdonk C D, Levorse J, Zheng B, Bradley A, Tilghman S M. Nat Genet. 2001;29:78–82. doi: 10.1038/ng715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.