Abstract
In stimulated cells, the mitogen-activated protein kinase ERK2 (extracellular signal-regulated kinase 2) concentrates in the nucleus. Evidence exists for CRM1-dependent, mitogen-activated protein kinase kinase-mediated nuclear export of ERK2, but its mechanism of nuclear entry is not understood. To determine requirements for nuclear transport, we tagged ERK2 with green fluorescent protein (GFP) and examined its nuclear uptake by using an in vitro import assay. GFP-ERK2 entered the nucleus in a saturable, time- and temperature-dependent manner. Entry of GFP-ERK2, like that of ERK2, required neither energy nor transport factors and was visible within minutes. The nuclear uptake of GFP-ERK2 was inhibited by wheat germ agglutinin, which blocks nuclear entry by binding to carbohydrate moieties on nuclear pore complex proteins. The nuclear uptake of GFP-ERK2 also was reduced by excess amounts of recombinant transport factors. These findings suggest that ERK2 competes with transport factors for binding to nucleoporins, which mediate the entry and exit of transport factors. In support of this hypothesis, we showed that ERK2 binds directly to a purified nucleoporin. Our data suggest that GFP-ERK2 enters the nucleus by a saturable, facilitated mechanism, distinct from a carrier- and energy-dependent import mechanism and involves a direct interaction with nuclear pore complex proteins.
Keywords: mitogen-activated protein kinase‖import‖FXFG motif‖nucleoporins
Mitogen-activated protein (MAP) kinases are ubiquitous protein kinases that integrate cell surface signals by phosphorylating proteins throughout the cell. The MAP kinase ERK2 (extracellular signal-regulated kinase 2) targets proteins in multiple cell compartments after an activating stimulus. Stimuli induce the nuclear accumulation of ERK2 from its resting location in the cytoplasm of many types of cells, including fibroblasts, epithelial cells, and neuroendocrine cells (1, 2).
The location of ERK2 is a significant factor in determining its ability to phosphorylate key substrates and thereby influence cell behavior. Controlling the localization of ERK2 is a mechanism by which cell function may be influenced. For example, PEA15, which promotes the cytoplasmic retention of ERK2, is overexpressed in disease states, such as type II diabetes and breast cancer (3, 4). ERK2 is active and constitutively nuclear in certain breast cancer cells, but excluded from the nucleus in certain nonsmall cell lung cancers (ref. 5, S. Muneer, personal communication). In addition to extensive data suggesting a correlation between phenotypes and ERK2 localization, some cell behaviors have been demonstrated to occur only if ERK2 accumulates in the nucleus. For example, the nuclear localization of ERK2 is essential for morphological transformation of 3T3 fibroblasts and neurite extension in PC12 cells (6, 7). An understanding of how ERK2 localization is regulated will provide important insights into its normal function and disease mechanisms.
ERK2 subcellular localization is mediated by interactions with various proteins (8–11). Unphosphorylated ERK2 is anchored in the cytoplasm by its upstream activator MEK1 (mitogen-activated protein kinase kinase 1), which, like ERK2, is present at near micromolar concentrations. After stimulation, active ERK2 translocates to the nucleus, possibly in a dimeric form (10, 11). Phosphorylation is sufficient to cause the nuclear accumulation of ERK2, but its kinase activity is not required (10). Nuclear binding sites for ERK2 may retain it in the nucleus, and binding partners include topoisomerase II and kinetochores (12–15). Nuclear export of ERK2 also is mediated by MEK1, which has a nuclear export sequence (NES) that binds to the nuclear export receptor CRM1 (11). Dephosphorylated, nuclear ERK2 can bind MEK1 and be transported out of the nucleus, a process inhibited by leptomycin B, which disrupts CRM1-mediated export (9). Furthermore, fusion of ERK2 to MEK1 yields a protein that is excluded from the nucleus, unless the NES is inactivated by mutagenesis, demonstrating the dominance of the MEK1 NES (7). Interactions with several other proteins in addition to MEK1, including the phosphatases, PTP-SL and MKP-3, and the death effector domain-containing protein, PEA15, promote localization of ERK2 in the cytoplasm (16–18).
A complex series of events controls ERK2 cytoplasmic and nuclear localization, but the mechanism for nuclear entry of ERK2 is not known. To examine this mechanism, we investigated the entry of ERK2 into nuclei of permeabilized cells under conditions not supporting CRM1-mediated export. We conclude that ERK2 enters the nucleus through nuclear pore complexes (NPCs) by an energy- and carrier-independent, facilitated mechanism.
Experimental Procedures
Constructs and Recombinant Proteins.
The cDNA encoding rat ERK2 was subcloned into pRSETB-His-6-GFP (green fluorescent protein) by using KpnI and HindIII restriction sites. GFP-ERK2, ERK2, and thiophosphorylated ERK2 (ERK2-P2) were prepared as described by using the Escherichia coli strain BL21 (10). His-6-GFP, p38α, and c-Jun N-terminal kinase 2 (JNK2) were purified as described (10) for ERK2. Karyopherin-β1, karyopherin-α2, Ran, p10, and glutathione S-transferase (GST)-NUP153c were purified as described (19–21). To reconstitute classical import, rhodamine-labeled BSA was coupled to a peptide containing the nuclear localization sequence (NLS) of the simian virus T antigen (20).
Cell Culture.
BRL and REF52 cells were grown in DMEM supplemented with 10% FBS and 1% glutamine.
Import Assays.
Import assays were performed as described (19). Briefly, BRL (Fig. 1C) or REF52 cells were plated on coverslips 24 h before permeabilization. Cells were washed once with import buffer (20 mM Hepes-KOH, pH 7.3/110 mM potassium acetate/2 mM magnesium acetate/1 mM EGTA/2 mM DTT) and permeabilized with 70 μg/ml digitonin in transport buffer for 5 min on ice. Cells were washed and inverted over 40 μl of import mix, which contained 0.8 μM GFP-ERK2 (50 μg/ml), 10 μg/ml NLS-BSA-tetramethylrhodamine B isothiocyanate (TRITC) (≈0.14 μM), or indicated amounts of ERK2 or thiophosphorylated ERK2 (ERK-P2) (19). To determine requirements for import, the import mix also included cytosol from Xenopus oocytes (ref. 20, Fig. 1C) or 0.5 μM karyopherin-α2, 0.25 μM karyopherin-β1, 2 μM Ran, 0.4 μM p10/NTF2, plus 15 μM BSA (1 mg/ml), and as indicated, energy in the form of an ATP/GTP regenerating system (1 mM ATP, 1 mM GTP, 5 mM creatine phosphate, and 20 units/ml creatine kinase). Permeabilized cells were incubated for 15 min unless otherwise noted and were then washed and fixed in 3.7% formaldehyde for 15 min. Coverslips were mounted in 1 mg/ml phenylenediamine in 90% PBS plus 10% glycerol. GFP and TRITC were visualized by fluorescence microscopy with a Zeiss Axiocam microscope equipped with a Hamamatsu Orca II camera. Uptake was quantified by using Improvision open lab software. His-6-ERK2 was detected by immunofluorescence with an anti-His-6 antibody (CLONTECH) at 1:200 dilution and an Alexa anti-mouse secondary antibody (Molecular Probes) at 1:3,000 dilution (22).
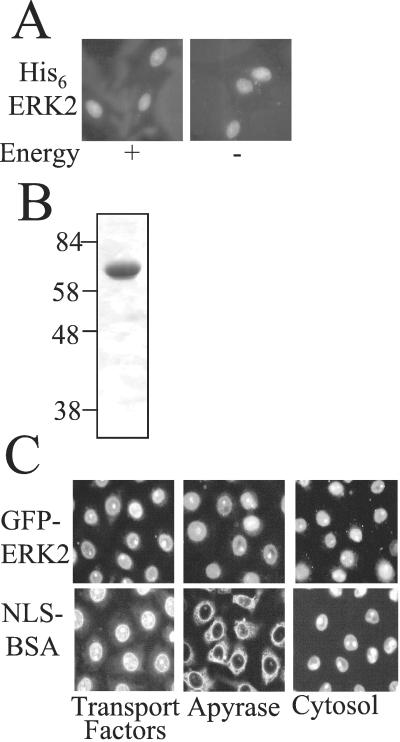
Figure 1.
Characterization of ERK2 import in permeabilized REF52 and BRL cells. (A) Nuclear import of His-6-ERK2 (0.8 μM, 50 μg/ml) in REF52 cells with or without an energy regenerating system (see Experimental Procedures) in the absence of transport factors was visualized by immunofluorescence. (B) Coomassie-stained polyacrylamide gel of purified GFP-ERK2. (C) Nuclear import of GFP-ERK2 (0.8 μM 50 μg/ml) and TRITC-NLS-BSA (1.4 μM) in BRL cells in the presence of an energy regenerating system plus the indicated factors. (Magnifications: ×63.)
Binding Experiments.
GST-Nup153c was incubated for 30 min with glutathione-agarose equilibrated in import buffer. His-6-ERK2 (200 nM) was added and mixed end over end at 23°C for 45 min. Beads were washed with 1 M NaCl, 1% Triton X-100, and 0.1% deoxcholate in 20 mM Hepes (pH 7.4) and analyzed on a 10% polyacrylamide gel in SDS. Proteins were transferred to nitrocellulose and immunoblotted with Y691 for 1 h at 1:2,500 dilution (7).
Results
ERK2 and GFP-ERK2 Accumulate in Nuclei by an Energy- and Carrier-Independent Mechanism.
To examine the mechanism of nuclear entry of ERK2, we used an import reconstitution assay in permeabilized cells. Permeabilized cells lose transport factors and thus a complementation assay with cytosol or recombinant transport factors and an ATP/GTP (energy) regenerating system can be used to investigate nuclear uptake of ERK2. ERK2, at 0.8 μM (50 μg/ml), close to its intracellular concentration, was added with or without energy or cytosol to permeabilized REF52 cells and its localization was detected by immunofluorescence (Fig. 1A and not shown). ERK2 accumulated in the nuclei of the permeabilized cells in an energy-independent manner, even in the absence of cytosol. Because ERK2 is only ≈41 kDa, it may have entered nuclei by diffusion. We increased the size of ERK2 to 68 kDa by incorporating a GFP tag; GFP-ERK2 was expressed and purified (Fig. 1B). Like ERK2, but unlike the control NLS-BSA-TRITC, GFP-ERK2 (0.8 μM) was taken up into nuclei of BRL and REF52 cells in the presence of apyrase, added to scavenge nucleotide (Fig. 1C and not shown). Over time, the concentration of GFP-ERK2 in the nucleus became greater than that in the import mixture (data not shown) and neither recombinant transport factors nor cytosol were required for its uptake (Fig. 1C). Quantification of the fluorescence intensity in the nuclei revealed no significant differences in GFP-ERK2 uptake in the presence or absence of these factors or cytosol in either cell type (data not shown). The control import substrate TRITC-NLS-BSA behaved in the expected manner, requiring energy and a source of transport factors for nuclear entry. We confirmed that the nuclear import of ERK2 was Ran independent, as the inclusion of Q69L Ran (defective in GTP hydrolysis) with cytosol inhibited import of NLS-BSA but not that of GFP-ERK2 (not shown) (23).
GFP-ERK2 Enters Nuclei by a Saturable, Facilitated Mechanism.
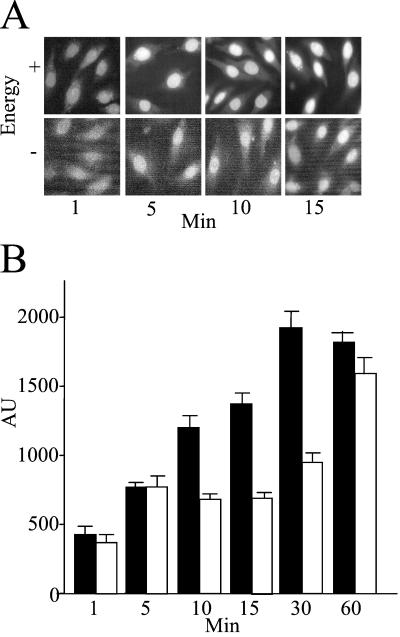
To determine whether the nuclear entry of GFP-ERK2 occurred by a facilitated mechanism, we examined the concentration dependence, time course, and temperature dependence of its nuclear uptake. The nuclear entry of GFP-ERK2 was time dependent (Fig. 2). To confirm that there was no energy requirement for GFP-ERK2 entry, a time course of its uptake in the presence and absence of an ATP/GTP regenerating system was examined at room temperature. No energy requirement was revealed at any time point (Fig. 2A). Furthermore, nonhydrolyzable analogs of ATP and GTP had no effect on ERK2 uptake (data not shown). Time courses of nuclear uptake were performed at both room temperature and 4°C. At the higher temperature, GFP-ERK2 uptake was readily detected after 10 min of addition to the permeabilized cells. The import of ERK2 was slowed but not abolished at 4°C (Fig. 2B), unlike that of NLS-BSA, which is completely inhibited at this temperature. This behavior of ERK2 is reminiscent of the movement of karyopherin-β in the absence of cargo and Ran (24). Using unfixed cells under these conditions, we could detect more ERK2 in the cytosol than in the bathing solution, possibly because of microtubule binding (25); but even so, some nuclear entry was detected within 1.5 min (not shown) (26, 27).
Figure 2.
GFP-ERK2 import is energy independent. (A) Import assay using GFP-ERK2 (0.8 μM, 50 μg/ml) was performed in REF52 cells with or without energy mix as indicated. Cells were fixed at indicated time points. (Magnification: ×63.) (B) Quantitation of REF52 nuclear fluorescence after import assay at room temperature (filled bars) or 4°C (open bars) in arbitrary units (AU) at indicated time points. Over a number of experiments, the relative rate of import at room temperature was approximately twice that at 4°C.
Nuclear uptake of GFP-ERK2 was concentration dependent (not shown). At 80 nM, rim staining of nuclei but little entry of GFP-ERK2 was observed. At 0.8 μM, GFP-ERK2 entry was readily observed. Pretreatment of ERK2 with an activated mutant of MEK1 under phosphorylating conditions to generate the phosphorylated form had no effect on its uptake, suggesting that phosphorylation of ERK2 does not influence its entry under these conditions (not shown).
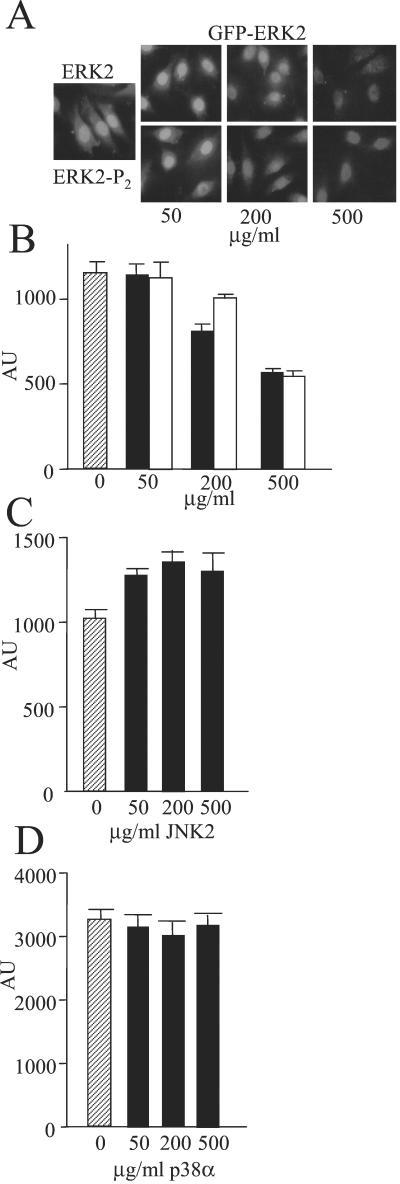
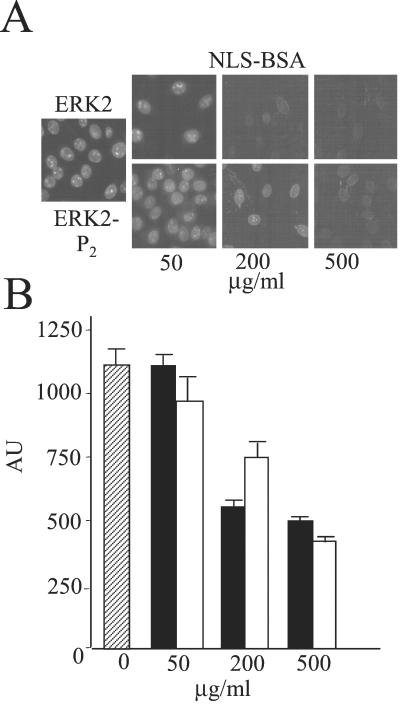
To examine whether the uptake system was saturable, increasing amounts of untagged ERK2 and ERK2-P2 were added along with a fixed amount of GFP-ERK2 (Fig. 3A). Both ERK2 and ERK2-P2 reduced the uptake of GFP-ERK2 in a concentration-dependent manner, indicating that the uptake system was saturable. These findings also support the above conclusion that ERK2 is equally effective in being taken up into the nucleus in the unphosphorylated and phosphorylated states. Because a 10-fold excess of the untagged proteins decreased GFP-ERK2 uptake by roughly half (Fig. 3B), the system is saturated at a concentration of ERK2 above that used for these assays. However, neither JNK2 nor p38α, two other MAP kinases, competed for ERK2 entry (Fig. 3 C and D).
Figure 3.
Effect of MAP kinases on nuclear uptake of GFP-ERK2. Import assay (A) and quantitation (B) using GFP-ERK2 (0.8 μM, 50 μg/ml) was performed in REF52 cells in the presence of increasing amounts of ERK2 (filled bars) and ERK2-P2 (open bars). Hatched bar is accumulation without excess ERK2. (C and D) Quantitation of nuclear fluorescence from import assay using GFP-ERK2 (0.8 μM, 50 μg/ml) in REF52 cells in the presence of increasing amounts of JNK2 (C) and p38α (D). Hatched bar is accumulation without excess MAP kinases. (Magnification: A, ×63.)
ERK2 Enters the Nucleus Through the NPC and Interacts Directly with Nucleoporins.
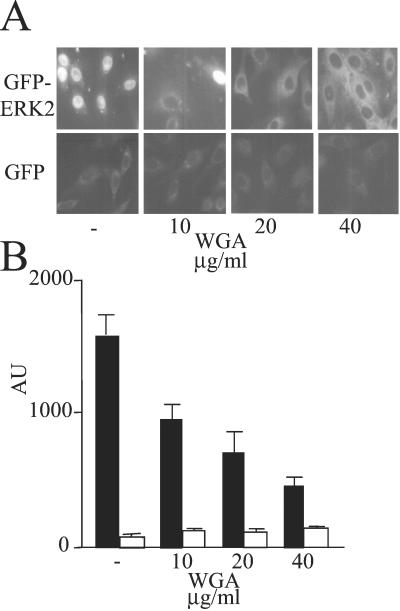
Some nucleoporins contain the covalently attached sugar moiety, N-acetyl glucosamine, which binds wheat germ agglutinin (WGA) with high affinity. Blockade of nuclear uptake by WGA is diagnostic for nuclear entry via the NPC (28). Therefore, we examined the effect of WGA on the nuclear entry of GFP-ERK2. The uptake of GFP-ERK2 was blocked by WGA in a concentration-dependent manner (Fig. 4), consistent with the conclusion that ERK2 enters the nucleus through the NPC. The nuclear accumulation of 27-kDa GFP, which enters the nucleus by diffusion, was unaffected by WGA (Fig. 4B).
Figure 4.
Effect of WGA on nuclear import of GFP-ERK2. Import assay (A) and quantitation (B) using GFP-ERK2 (0.8 μM, 50 μg/ml) (filled bars) and GFP (0.8 μM, 20 μg/ml) (open bars) was performed in REF52 cells in the presence of increasing amounts of WGA and energy mix. (Magnification: A, ×63.)
Our results indicate that ERK2 enters the nucleus through the NPC without a carrier and without the need for energy. Many nucleoporins contain FG repeats that often are found as part of a larger motif such as FXFG or GLFG. These repeats are recognized by nuclear carriers that transport their cargo across the NPC (29–35). MAP kinases have been shown to interact with many proteins through relatively well-defined targeting motifs. Interestingly, one of these is the FXF motif, which was identified by Kornfeld and colleagues as an ERK1/2-specific targeting domain (36). We considered the hypothesis that ERK2 can interact directly with nucleoporins.
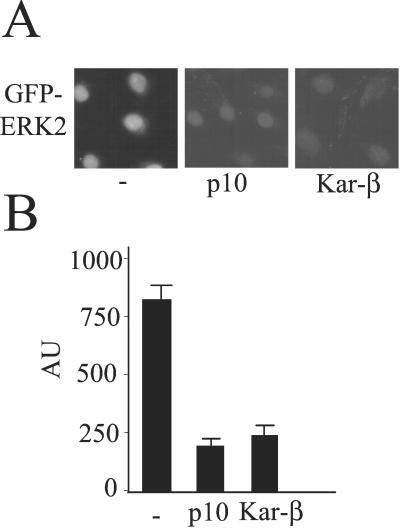
We tested this hypothesis by using the import assay. Carriers can compete with each other for shared docking sites (37). If ERK2 binds to FG repeat docking sites, it should compete with the import of NLS-BSA. Thus, we tested its capacity to interfere with import of NLS-BSA. Both ERK2 and ERK2-P2 reduced the uptake of NLS-BSA in a concentration-dependent manner (Fig. 5). As a further test, if ERK2 is binding directly to nucleoporins, the import receptors should compete for these binding sites and reduce ERK2 entry. We examined the effects of two carriers, karyopherin-β1 and p10/NTF2, on uptake of GFP-ERK2 (20). In support of the hypothesis, micromolar concentrations of both karyopherin-β1 and p10/NTF2 compete with GFP-ERK2 for nuclear entry (Fig. 6).
Figure 5.
Effect of ERK2 on import of NLS-BSA. Import assay (A) and quantitation (B) using TRITC-NLS-BSA (1.4 μM) was performed in REF52 cells in the presence of transport factors, energy mix, and increasing amounts of ERK2 (filled bars) and ERK2-P2 (open bars). Hatched bar is TRITC-NLS-BSA fluorescence in the absence of ERK2 or ERK-P2. (Magnification: A, ×63.)
Figure 6.
Effect of NPC binding proteins on import of GFP-ERK2. Import assay (A) and quantitation (B) using GFP-ERK2 (0.8 μM, 50 μg/ml) was performed in REF52 cells in the presence of energy mix and the indicated amounts of karyopherin-β1 or NTF2/p10. (Magnification: A, ×63.)
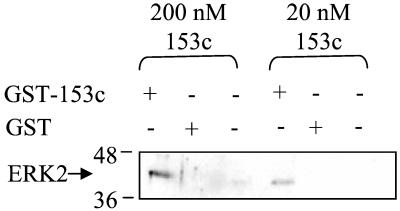
To determine whether ERK2 can interact directly with a repeat-containing nucleoporin, we expressed the 153c fragment of the nucleoporin 153 as a GST fusion in bacteria and tested its binding to ERK2 in vitro. ERK2 associated with GST-Nup153c but not with GST or empty beads (Fig. 7). Their binding can be detected with as little as 20 nM Nup153c and 200 nM ERK2. ERK2 phosphorylates Nup153c only weakly (data not shown), suggesting that the binding is not caused solely by an enyzme–substrate relationship. We hypothesize that the rapid entry of ERK2 into the nucleus is caused by facilitated entry via a direct interaction with nucleoporins.
Figure 7.
ERK2 binds to GST-Nup153c. GST-pulldown assay using GST-Nup153c at the indicated concentrations and 200 nM of ERK2 or buffer as indicated. Immunoblots were performed by using Y691 for ERK2.
Discussion
We find that ERK2 enters the nucleus by a saturable, facilitated process, distinct from the conventional import mechanism. Our data indicate that the rapid uptake of ERK2 into the nucleus, like that of nuclear carriers, depends on its direct interaction with FG repeat-containing nucleoporins, as demonstrated through competition studies and the finding that ERK2 binds directly to a repeat nucleoporin. This mechanism is attractive not only because ERK2 binds FXF motifs in other proteins, but also because ERK2 has been reported to affect the nuclear import of proteins in a manner consistent with interaction with the import machinery (36, 38). While our manuscript was in preparation, another group published a report concluding that ERK2 enters nuclei via an interaction with NPC proteins; ERK2 was found to bind to a different FG repeat-containing protein, Nup214, in addition to the binding to Nup153 that was shown here (39). These findings, together with ours, suggest that ERK2 binds to multiple NPC proteins, supporting the proposed mechanism.
These data in permeabilized cells are supported by previous findings in intact cells. We examined time courses of ERK2 localization after microinjection and found that the unphosphorylated protein rapidly entered the nucleus within 2 min, and then relocalized to the cytoplasm by 5–10 min (10). These previous experiments suggested that the localization of ERK2 is controlled by active export as well as import.
The facilitated import mechanism found here has not been previously reported for other protein kinases, but has been suggested for the transcriptional regulator β-catenin (40, 41). The major control of the nuclear function of β-catenin is its regulated association with the cytoskeleton. Once released, β-catenin binds nucleoporins without a requirement for carriers, competes with carriers for nuclear uptake, and enters the nucleus in an energy-independent manner, similar to the results of our studies of ERK2 import. In the case of ERK2, its nuclear accumulation appears to be regulated primarily by its export; it does not need an import carrier because it interacts directly with nuclear pore proteins. Thus, direct binding to nucleoporins may be a generally used mechanism for nuclear entry of proteins whose accumulation in the nucleus is controlled at other levels. This facilitated uptake mechanism is apparently not used by two other MAP kinases, JNK2 and p38α, based on their inability to compete for ERK2 entry. This result is consistent with the apparently selective interaction of FXF motifs with ERK2 among MAP kinases (36).
Phosphorylation of ERK2 promotes its nuclear localization in vivo. However, we were unable to find effects of phosphorylation on its uptake in permeabilized cells. This result is consistent with the observation that overexpressed ERK2 accumulates in the nucleus without being phosphorylated (42). Blockade of export, by favoring the dissociation of ERK2-MEK1 complexes, may be the primary event controlled by ERK2 phosphorylation. ERK2 forms dimers upon phosphorylation that promote its nuclear accumulation in vivo (10). An ERK2 dimer, as a consequence of having twice the sites to interact with FX motifs, might bind more tightly to NPCs and be imported more rapidly than a monomer. Alternatively, other import mechanisms for dimers may exist that could not be detected in this system. Finally, dimeric ERK2 may be further impaired in its interaction with proteins such as MEK1 that may promote its piggyback export.
In summary, multiple regulated events lead to the stimulus-dependent nuclear accumulation of ERK2. ERK2, phosphorylated or not, appears to enter the nucleus by a rapid, facilitated, energy- and carrier-independent mechanism that relies on its direct interaction with nucleoporins. A second entry mechanism also may exist for ERK2-P2, perhaps as a dimer, and/or by association with one or more other proteins that contain an NLS. ERK2 in the nucleus binds to proteins that retain it there even after it has been dephosphorylated. In stimulated cells, ERK2 that does not enter the nucleus is bound to cytosolic anchor proteins. Binding of ERK2 to proteins in the cytoplasm and nucleus may be as important as the transport mechanisms in determining its subcellular distribution. Unphosphorylated ERK2 is believed to be exported from the nucleus in a CRM-1-dependent manner primarily as a complex with MEK1 via its NLS. Other proteins also may mediate ERK2 export normally and under pathophysiological conditions. This model suggests strategies for therapeutic intervention in disease states in which the inappropriate localization of ERK2 is believed to contribute to symptoms.
Acknowledgments
We thank Tara Beers Gibson, Gray Pearson, Ewen Gallagher, and Bing-e Xu for critical suggestions and comments about the manuscript, Mike White for helpful discussions, Ian Mattaj, Gideon Dreyfuss, Natalie Ahn, and Rama Ranganathan for constructs, and Dionne Ware for administrative assistance. This work was supported by Grant DK34128 from the National Institutes of Health and Grant I1243 from the Welch Foundation (to M.H.C.). A.W.W. was supported by a National Institute of General Medical Sciences Pharmacological Sciences training grant. J.L.W. was supported by a fellowship from the Howard Hughes Medical Institute.
Abbreviations
- MAP
mitogen-activated protein
- MEK
mitogen-activated protein kinase kinase
- ERK
extracellular signal-regulated kinase
- JNK2
Jun N-terminal kinase 2
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- NPC
nuclear pore complex
- NLS
nuclear localization sequence
- TRITC
tetramethylrhodamine B isothiocyanate
- WGA
wheat germ agglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lewis T S, Shapiro P S, Ahn N G. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 2.Pearson G, Robinson F, Beers G T, Xu B E, Karandikar M, Berman K, Cobb M H. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 3.Condorelli G, Vigliotta G, Iavarone C, Caruso M, Tocchetti C G, Andreozzi F, Cafieri A, Tecce M F, Formisano P, Beguinot L, Beguinot F. EMBO J. 1998;17:3858–3866. doi: 10.1093/emboj/17.14.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukamoto T, Yoo J, Hwang S I, Guzman R C, Hirokawa Y, Chou Y C, Olatunde S, Huang T, Bera T K, Yang J, Nandi S. Cancer Lett. 2000;149:105–113. doi: 10.1016/s0304-3835(99)00350-x. [DOI] [PubMed] [Google Scholar]
- 5.Sivaraman V S, Wang H, Nuovo G J, Malbon C C. J Clin Invest. 1997;99:1478–1483. doi: 10.1172/JCI119309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowley S, Paterson H, Kemp P, Marshall C J. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 7.Robinson M J, Stippec S A, Goldsmith E, White M A, Cobb M H. Curr Biol. 1998;8:1141–1150. doi: 10.1016/s0960-9822(07)00485-x. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda M, Gotoh Y, Nishida E. EMBO J. 1997;16:1901–1908. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adachi M, Fukuda M, Nishida E. J Cell Biol. 2000;148:849–856. doi: 10.1083/jcb.148.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khokhlatchev A, Canagarajah B, Wilsbacher J L, Robinson M, Atkinson M, Goldsmith E, Cobb M H. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 11.Adachi M, Fukuda M, Nishida E. EMBO J. 1999;18:5347–5358. doi: 10.1093/emboj/18.19.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenormand P, Brondello J M, Brunet A, Pouyssegur J. J Cell Biol. 1998;142:625–633. doi: 10.1083/jcb.142.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro P S, Whalen A M, Tolwinski N S, Froelich-Ammon S J, Garcia M, Osheroff N, Ahn N G. Mol Cell Biol. 1999;19:3551–3560. doi: 10.1128/mcb.19.5.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro P S, Vaisberg E, Hunt A J, Tolwinski N S, Whalen A M, McIntosh J R, Ahn N G. J Cell Biol. 1998;142:1533–1545. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zecevic M, Catling A D, Eblen S T, Renzi L, Hittle J C, Yen T J, Gorbsky G J, Weber M J. J Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco-Aparicio C, Torres J, Pulido R. J Cell Biol. 1999;147:1129–1136. doi: 10.1083/jcb.147.6.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colucci-D'Amato G L, D'Alessio A, Califano D, Cali G, Rizzo C, Nitsch L, Santelli G, de Franciscis V. J Biol Chem. 2000;275:19306–19314. doi: 10.1074/jbc.275.25.19306. [DOI] [PubMed] [Google Scholar]
- 18.Formstecher E, Ramos J W, Fauquet M, Calderwood D A, Hsieh J-C, Canton B, Nguyen X-T, Barneir J-V, Camonis J, Ginsberg M H, Chneiweiss H. Dev Cell. 2001;1:239–250. doi: 10.1016/s1534-5807(01)00035-1. [DOI] [PubMed] [Google Scholar]
- 19.Schwoebel E D, Talcott B, Cushman I, Moore M S. J Biol Chem. 1998;273:35170–35175. doi: 10.1074/jbc.273.52.35170. [DOI] [PubMed] [Google Scholar]
- 20.Moore M S, Blobel G. Cell. 1992;69:939–950. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- 21.Nakielny S, Dreyfuss G. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 22.Christerson L B, Vanderbilt C A, Cobb M H. Cell Motil Cytoskeleton. 1999;43:186–198. doi: 10.1002/(SICI)1097-0169(1999)43:3<186::AID-CM2>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 23.Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reszka A A, Seger R, Diltz C D, Krebs E G, Fischer E H. Proc Natl Acad Sci USA. 1995;92:8881–8885. doi: 10.1073/pnas.92.19.8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribbeck K, Gorlich D. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Featherstone C, Darby M K, Gerace L. J Cell Biol. 1988;107:1289–1297. doi: 10.1083/jcb.107.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finlay D R, Newmeyer D D, Price T M, Forbes D J. J Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorlich D, Kutay U. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 30.Talcott B, Moore M S. Trends Cell Biol. 1999;9:312–318. doi: 10.1016/s0962-8924(99)01608-6. [DOI] [PubMed] [Google Scholar]
- 31.Strawn L A, Shen T, Wente S R. J Biol Chem. 2001;276:6445–6452. doi: 10.1074/jbc.M008311200. [DOI] [PubMed] [Google Scholar]
- 32.Wente S R. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- 33.Bayliss R, Littlewood T, Stewart M. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 34.Shah S, Forbes D J. Curr Biol. 1998;8:1376–1386. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- 35.Chook Y M, Blobel G. Nature (London) 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 36.Jacobs D, Glossip D, Xing H, Muslin A J, Kornfeld K. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 37.Lane C M, Cushman I, Moore M S. J Cell Biol. 2000;151:321–332. doi: 10.1083/jcb.151.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Czubryt M P, Austria J A, Pierce G N. J Cell Biol. 2000;148:7–16. doi: 10.1083/jcb.148.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsubayashi Y, Fukuda M, Nishida E. J Biol Chem. 2001;276:41755–41760. doi: 10.1074/jbc.M106012200. [DOI] [PubMed] [Google Scholar]
- 40.Fagotto F, Gluck U, Gumbiner B M. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 41.Yokoya F, Imamoto N, Tachibana T, Yoneda Y. Mol Biol Cell. 1999;10:1119–1131. doi: 10.1091/mbc.10.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubinfeld H, Hanoch T, Seger R. J Biol Chem. 1999;274:30349–30352. doi: 10.1074/jbc.274.43.30349. [DOI] [PubMed] [Google Scholar]