Abstract
Polydnaviruses are essential components mediating host–parasitoid relationships between some braconid wasps and their caterpillar hosts largely by suppressing or misdirecting the host immune systems. The polydnavirus–wasp relationship is an unusual apparent mutualism between viruses and eukaryotes and remarkably has evolved to the stage where the two entities no longer can be considered separate. Estimations of the age of the polydnavirus-bearing clade of braconid wasps based on separate calculations from the mitochondrial 16S rRNA and COI genes and the nuclear 28S rRNA gene, calibrated using fossil data, converge to indicate a date of origin of ≈73.7 ± 10 million years ago. This range provides an upper bound on the time during which these wasps and viruses have been functionally associated.
Some of the most complex and highly evolved interspecific interactions known among the insects occur among endoparasitoid Hymenoptera and their host insects (1–3). In these highly specialized systems, wasp larvae must survive and develop inside the bodies of other insects, typically caterpillars of Lepidoptera, despite the challenges such a habitat presents (4, 5). Among these challenges is the host cellular immune response (6, 7), which is designed to recognize, encase, and ultimately suffocate such macroscopic intruders in layers of melanized and fused hemocytes (8).
Several groups of endoparasitoid Hymenoptera have evolved tools to defeat these defense mechanisms by incorporating the services of symbiotic virus-like entities with which they coat their eggs (reviewed in refs. 9–11). These viruses deliver genes into host caterpillars that, when expressed, produce proteins implicated in immune and developmental alterations in the caterpillar (12–16). In addition, they also may wield proteins on their capsid surface that disguise the parasitoid egg as it enters the caterpillar's hemocoel (17). In some cases, the viral genomes seem to have incorporated genes of wasp origin, originally used as part of the wasp venom (18), and to have “engineered” themselves in a unique way to enhance gene expression in host caterpillars (19). The most fully understood systems of this sort are those of polydnaviruses (subclassified into bracoviruses and ichnoviruses; ref. 11), which are found in some ichneumonoid wasps as integrated proviruses. These proviruses are transmitted vertically within the wasp chromosomal DNA (20, 21), and their genes are exported into host caterpillars for expression.
Although all endoparasitoids are remarkable in some respects in their adaptations for manipulating host organisms, these virus-bearing wasps are unusual in having made use of one of the few known mutualisms between viruses and eukaryotes (10). What is even more remarkable is that this kind of association is now known to characterize several tens of thousands of species of parasitoid wasps that are virtually ubiquitous in terrestrial environments (11).
Currently, the source virus (or viruses) is not known for any of the wasp/virus associations. What does seem clear is that, at least in some cases, the virus associations are found only within certain phylogenetic lineages of the wasps and are pervasive within those lineages (11). This relationship is especially well established for the associations of polydnaviruses with braconid wasps (22). In this paper, the phylogenetic interrelationships between the symbiotic viruses and braconid wasps are reviewed, and estimates are made of the age of this remarkable association by using molecular clock-based extrapolations from DNA sequence data from three genes, calibrated with dates from wasp fossils. It is shown that the wasps and viruses are likely to have been associated with one another at least since the Cretaceous-Tertiary boundary and perhaps for even longer.
Monophyly of the Polydnavirus-Bearing Clade Within Braconid Wasps
The presence of a morphologically distinct class of polydnaviruses in braconid wasps (bracoviruses), different from those in ichneumonids (ichnoviruses), was recognized in the late 1970s (23). It was first suggested that both groups of wasps carrying polydnaviruses might themselves form evolutionary lineages. This prediction was made on the basis of a series of early experiments that appeared to show that viruses from more closely related wasps were more genetically similar in general than those from more distantly related wasps (24). It then was hypothesized that the braconid wasps carrying bracoviruses form a distinct evolutionary lineage (11), and this was supported by phylogenetic analysis of morphological and 16S rRNA data from the wasps (22, 25) and clinched by corroboration from analyses of 28S rRNA (26, 27). This limitation of the bracoviruses to a single lineage of wasps, although suggestive of phylogenetic coevolution between the two entities, was not fully demonstrative, because phylogenetic relationships among the viruses were not established yet.
Cophylogeny of Polydnaviruses and the Wasps That Carry Them
In 2000, parallel phylogenetic analyses of wasps in the genus Cotesia based on partial sequences from the mtDNA 16S and ND1 genes and of the bracoviruses they carry based on partial sequences of the glycosylating transmembrane protein gene CrV1 showed complete, statistically significant congruence between the histories of the two associated partners (28). Work is ongoing to expand this cophylogenetic analysis to include all major groups within the bracovirus-bearing clade of wasps. In the interim, the phylogenetic congruence between wasps and viruses within the genus Cotesia and the phylogenetic limitation of the viruses to a monophyletic clade of the Braconidae strongly suggest that there may have been a single origin of the association of bracoviruses. It is of considerable evolutionary and practical interest to know the age of the association between of wasps and viruses. That is, how long has it taken for the complex subtleties and interactive specializations to evolve? To estimate this age with any certainty, it is necessary to make use of information from available fossils and from comparative study of DNA sequences.
The Fossil Record of Braconidae
Fossils of braconid wasps are rare other than Miocene or more recent inclusions in amber, which mostly appear to represent genera still extant. This is especially true of braconid taxa associated with lepidopteran caterpillars. The oldest indisputable ichneumonoid fossils are Praeichneumon townesi Rasnitsyn and Eobracon inopinatus Rasnitsyn from the early Cretaceous of Mongolia, ≈140 million years ago (mya; ref. 29). The former fossil is not clearly assignable to either of the extant families Braconidae or Ichneumonidae, whereas the latter was assigned to the Braconidae. A variety of ichneumonoid fossils are known from slightly younger deposits in Mongolia, the Transbaikal, and Australia (29–32), and most of them have been assigned to the extinct family Eoichneumonidae (31, 32).
Within Braconidae, more modern looking taxa have been found from New Jersey and Baltic amber deposits. Protorhyssalus goldmani (33, 34), from ≈93 mya, was considered assignable to either the cyclostome (predominantly ectoparasitoid) lineage of braconid wasps or alternatively to the predominantly endoparasitoid helconoid assemblage. The more recent (≈45 mya) Baltic fossils all are referable to extant subfamilies, some of which possess polydnaviruses. Among these latter fossils are the Oligocene microgastrine representatives Eocardiochiles fritschii Brues (erroneously attributed in the original description to Cardiochilinae) and Snellenius succinalis Brues (35).
The current placement of these fossils, which provide a minimum age for the groups to which they belong, suggests that the microgastroid lineage carrying polydnaviruses must have originated and diversified between 93 and 45 mya. Estimation of a more precise date of origin in the absence of additional informative fossils requires the use of extrapolative methods using molecular clocks.
Using Molecular Clocks to Estimate Divergence Times
Because the original suggestion that DNA sequences may act in a more or less clock-like manner in recording substitutional changes (36, 37), it has been realized that “molecular clocks” require both careful calibration (not all genes evolve at the same rate; refs. 38 and 39) and testing to ensure that the assumptions of rate constancy are met. A variety of methods have been introduced to deal with various deviations from clock-like behavior.
That different genes evolve at different rates is not necessarily a problem for time estimation. After all, the existing range of variability allows for more efficient targeted analysis of phylogenetic relationships on different time scales by using different genes as markers (40). However, the use of different genes for estimating divergence times requires that each gene be calibrated independently. It is a more serious problem for analysis if evolutionary rates in the same gene are not similar among different lineages (41, 42) or if they are not similar within lineages over different time spans. To tackle these latter problems, methods have been developed to either prune out lineages exhibiting highly divergent rates (43, 44) or allow rates to vary among lineages by using rate-smoothing (45, 46) or Bayesian estimation techniques (47–49). Relative rates and divergence time estimates using any of these methods can then be “calibrated” (converted to absolute times of divergence) using fossils to establish limits on the age of one or more nodes in the phylogeny (50). A common type of calibration is to use relatively recent fossils to extrapolate backwards in time to estimate dates of earlier divergences, as in the “quartets method” of Rambaut and Bromham (51). In practice, any node or nodes in the phylogeny other than the terminal tips can serve for calibration, depending on the availability of fossils.
Unless rates of evolution among lineages vary strikingly, relative divergence time estimates from these various methods tend to be fairly similar as long as multiple substitutions superimposed at the same site are adequately taken into account. A more significant source of error in estimation of absolute divergence times are the uncertainties associated with accurately placing fossils relative to nodes in the phylogeny and with determining the maximum age of taxa, because the fossils can only represent minimum ages (52–54).
The assessment of potential error in estimation of divergence times for particular nodes in a phylogeny can be strengthened by employing multiple genes for estimating relative rates and multiple fossils for calibration. In this study, portions of three genes, two mitochondrial (16S rRNA and cytochrome oxidase I) and one nuclear (28S rRNA), are used comparatively to estimate rates of wasp gene evolution within the microgastroid lineage, and three different fossil calibration points are used for each gene. In addition, several different rate-estimation methods are used for each comparison to obtain a “cloud” of divergence time estimates for each node of interest. This cloud of more or less independent estimates is likely to provide a more realistic view of the accuracy of the divergence time estimates than would standard confidence intervals based only on the error associated with single extrapolations from the rate models.
Materials and Methods
Taxa Sampled.
Table 1 provides a list of the taxa used for the divergence time estimates with GenBank accession numbers for DNA sequence data from each gene. The taxon representation is essentially the same for all three genes for the microgastroid complex of subfamilies but differs for some outgroups. Each major internal node is represented by at least two (usually more) taxa per lineage for each gene, giving a large number of pairwise divergence estimates for each node of interest.
Table 1.
Source sequences for the calculations of divergence times in this study (with GenBank accession numbers)
| Taxon | 16S | COI | 28S |
|---|---|---|---|
| Ichneumonidae | |||
| Venturia canescens (Gravenhorst) | U06961 | U59221 | AJ245958 |
| Dusona egregia (Viereck) | — | AF146682 | AF146670 |
| Ichneumon promissorius Erichson | U06960 | — | — |
| Pimpla aequalis Provancher | — | AF146681 | AF146665 |
| Xorides sp. | AF003520 | — | Z83612 |
| Cyclostome Braconidae | |||
| Aphidius ervi Haliday | AF176067 | — | Z83582 |
| Dolopsidea sp. | — | AF379990 | — |
| Bracon sp. | Z93722 | — | AJ296037 |
| Jarra maculipennis Marsh & Austin | AF003485 | AF379991 | Z97970 |
| Helconoid Braconidae | |||
| Agathis sp. | — | AF078468 | — |
| Meteorus sp. | U68146 | — | Z97953 |
| Neoneurus mantis Shaw | U68147 | — | AF029133 |
| Peristenus pallipes (Curtis) | — | AF189242 | — |
| Microgastroid Braconidae | |||
| Adelius sp. | AF029111 | — | AF029117 |
| Chelonus (Microchelonus) sp. | U68150 | AF102723 | AF029123 |
| Mirax lithocolletidis Ashmead | AF102765 | AF102722 | AF102747 |
| Cardiochiles fuscipennis Szepligeti | AF029112 | AY044207 | AF029118 |
| Toxoneuron nigriceps (Viereck) | U68151 | AF102724 | AF029120 |
| Microgastrinae s. s. (10) | |||
| Apantelescanarsiae Ashmead | AF102750 | AF102703 | AF102728 |
| Diolcogaster schizurae (Muesebeck) | AF102759 | AF102716 | AF102741 |
| Dolichogenidea lacteicolor (Viereck) | AF102760 | AF102717 | AF102742 |
| Glyptapanteles indiensis (Marsh) | AF102757 | AF102713 | AF102738 |
| Hypomicrogaster ectdytolophae (Muesebeck) | AF102756 | AF102712 | AF102737 |
| Microgaster canadensis Muesebeck | U68154 | AF102708 | AF102733 |
| Microplitis maturus Weed | U68155 | AF102702 | AF102727 |
| Parapanteles paradoxus (Muesebeck) | AF102753 | AF102709 | AF102734 |
| Pholetesor ornigis (Weed) | AF102755 | AF102711 | AF102736 |
| Sathon falcatus (Nees) | AF102764 | AF102721 | AF102746 |
Obtaining Molecular Data.
Sequence data were used primarily from refs. 22, 55, and 56, but additional data from several other studies using the same gene regions for some of the nonmicrogastroids were drawn from GenBank. The data were aligned as described in detail in refs. 22 and 55, with the inclusion of the additional outgroup sequences. Briefly, 16S sequences were fit to secondary structure, whereas other sequences were aligned by using CLUSTAL X (57) with the parameters described in ref. 56. In summary, the data set comprised 438 aligned bp from the 16S gene, 432 aligned bp from the COI gene, and 466 bp from the 28S gene (incorporating portions of the D2 expansion loop).
Experimental Design for Comparisons.
Pairwise comparisons were made assuming the following basic relationships, which have been supported strongly by many morphological and molecular studies: Ichneumonidae + (cyclostome Braconidae + helconoid complex + microgastroid complex). A sister-group relationship between representatives of the helconoid and microgastroid complexes was not assumed a priori but resulted from analyses. The monophyly of the helconoid complex was not assumed, because it was not necessary for the present analysis and is not well established. However, monophyly of the microgastroid complex is well supported (22, 26, 27) and was assumed a priori. The taxon representation allowed comparisons that elucidate the divergence times between (i) Braconidae and Ichneumonidae (providing age estimates for the braconid lineage), (ii) microgastroid versus nonmicrogastroid lineages (providing age estimates for the bracovirus-bearing lineage), and (iii) Microgastrinae versus nonmicrogastrine microgastroids (providing age estimates for the Microgastrinae).
Testing for Constant Rate Across the Tree.
Relative rate tests were conducted on all possible triplets of taxa by using both the model-based method of Wu and Li (58) and the nonparametric (model-free) method of Tajima (59) as implemented in the program R8S (available at http://ginger.ucdavis.edu/r8s/, ref. 46).
Divergence Time Estimation.
Uncorrected distances (p distances or observed percentage differences between pairs of sequences) underrepresent the divergence of anciently diverged lineages because of the problem of saturation of substitutions at variable sites. Observed distances thus were corrected by using several models that take saturation and various substitution biases into account with PAUP* 4.0b4a (60). Those models used in divergence time estimation were the Kimura two-parameter (61) model for all genes, the HKY85 (62) model incorporating γ-distributed rates among sites (63) for the 28S gene, and the general time-reversible model (64) with γ-distributed rates among sites (for the two mtDNA genes to accommodate the AT-rich bias). The above estimates all assumed a molecular clock with no lineage-specific rate variation (following ref. 65). To accommodate lineage-specific rate differences, the latter two models also were used by using the nonparametric rate-smoothing method of Sanderson (45) as used in the program R8S (46).
Calibrating the Clocks with Fossils.
Three calibration points were used independently for each gene in each analysis. First, the age of the ichneumonoid lineage can be estimated minimally at 150 mya based on the ages of the Praeichneumon and Eoichneumonidae fossils. Second, the age of the Braconidae could be set minimally at 130 mya based on the fossil of Eobracon. Finally, the fossil Protorhyssalus indicates that the modern cyclostome lineage might extend back at least 93 mya (this fossil was not considered to be unambiguously cyclostome in oral morphology but has otherwise entirely cyclostome morphological characters). Each of these calibration points was used in the program R8S to fix the age of the appropriate node for estimation runs using each gene/model combination.
Results
Relative Rate Tests.
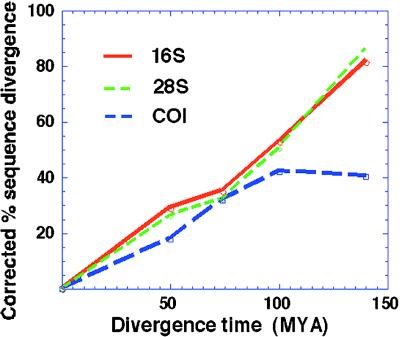
All tests indicated nonsignificant variation in rates among lineages except some combinations of exemplars of cyclostome/ichneumonid/microgastroid using the COI gene. In these cases, the COI gene showed larger divergence estimates between cyclostomes and microgastroids than between ichneumonids and braconids. Fig. 1 shows how the divergence estimates from COI, even when corrected for “multiple hits,” appear to saturate at higher levels of divergence. Thus, it would be expected that COI comparisons would overestimate divergence times for more recent divergences when older fossils are used for calibration and underestimate older divergence times when younger fossils are used.
Figure 1.
Plots of corrected (HKY85 + γ for 28S and GTR + γ for 16S and COI) sequence divergence in the three genes against estimated time. Note that COI loses its linear relationship at more ancient divergence times.
Divergence Time Estimates.
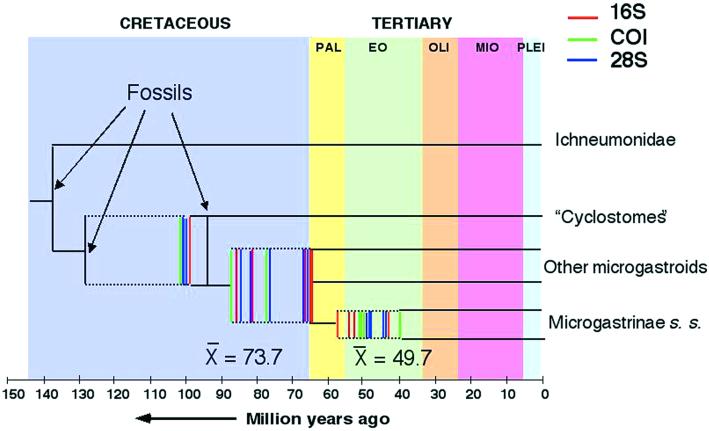
Fig. 2 plots the range of divergence time estimates for the three nodes of interest, including the principal focus, in the base of the bracovirus-bearing microgastroid lineage. Table 2 provides the actual divergence time estimates for each of the three genes by using each of the three fossil calibrations using the HKY85 + γ model for 28S and the GTR + γ model for 16S and COI, either assuming a molecular clock or compensating for rate differences by nonparametric rate smoothing.
Figure 2.
Plots of divergence time estimates for the age of the microgastroid and microgastrine lineages for different combinations of assumptions, fossil calibrations, and genes. PAL, Paleocene; EO, Eocene; OLI, Oligocene; MIO, Miocene; PLEI, Pleistocene.
Table 2.
Divergence time estimates based on various gene/model/fossil combinations
| 16S Clock, mya | 16S NPRS, mya | COI Clock, mya | COI NPRS, mya | 28S Clock, mya | 28S NPRS, mya | |
|---|---|---|---|---|---|---|
| Using Praeichneumon + Eoichneumonidae to calibrate (base 150 mya) | ||||||
| Base of tree | 150 | 150 | 150 | 150 | 150 | 150 |
| Braconidae | 130 | 97.1 | 130 | 130 | 150 | 150 |
| Microgastroid lineage | 84.5 | 64.6 | 141 | 104.1 | 76.3 | 85.9 |
| Microgastrinae | 55.8 | 53.4 | 49.5 | 87.6 | 48.6 | 41.7 |
| Using Eobracon to calibrate tree (Braconidae > 130 mya) | ||||||
| Base of tree | — | 200 | 150 | 150 | 150 | 150 |
| Braconidae | 130 | 130 | 130 | 130 | 130 | 150 |
| Microgastroid lineage | 84.5 | 86.6 | 141 | 104.1 | 66.1 | 85.9 |
| Microgastrinae | 55.8 | 71.5 | 49.5 | 87.6 | 42.1 | 41.7 |
| Using Protorhyssalus to calibrate (cyclostome/noncyclostome > 93 mya) | ||||||
| Base of tree | — | 153.6 | — | — | — | — |
| Braconidae | 99.4 | 99.4 | 98.5 | 98.5 | 97.9 | 97.9 |
| Microgastroid lineage | 64.4 | 66.2 | 106.8 | 78.8 | 49.8 | 64.7 |
| Microgastrinae | 42.5 | 54.7 | 38.9 | 52.8 | 31.7 | 31.4 |
| Means excluding outliers | ||||||
| Base of tree | 156 | |||||
| Braconidae | 130 | |||||
| Cyclo/noncyclo split | 102 | |||||
| Microgastroid lineage | 73.7 | |||||
| Microgastrinae | 49.7 | |||||
Considered broadly, the mean estimate of the age of the microgastroid lineage is 73.7 mya. The actual individual estimates ranged from a minimum of 49.8 mya (28S assuming a clock using Protorhyssalus to calibrate) to 141 mya (COI assuming a clock using Praeichneumon and Eoichneumonidae or Eobracon to calibrate). Excluding these obvious outliers (the exclusion of which does not affect the overall mean values), the estimates were bimodally distributed, either concentrated tightly around the Cretaceous–Tertiary boundary at 65 mya or spread over a broader range between 76 and 88 mya. The range of estimates for the age of the subfamily Microgastrinae is similarly broad with clusters of estimates and a mean of 49.7 mya (thus slightly older than the known Baltic amber fossils from this group). Estimates for the timing of the divergence between cyclostome and noncyclostome lineages (centered around 100 mya) fall far short of the oldest fossil (Eobracon) attributed to the Braconidae unless one uses Eobracon to calibrate the divergence times to begin with.
Discussion
Convergence of Estimates from Different Genes.
In general, the divergence time estimates from the three genes overlap and accord well with the exception of some point estimates from COI and to a lesser extent from 28S. The difficulty with COI in tracking the older divergences probably can be attributed to the apparent lineage-idiosyncratic saturation of sites free to vary (Fig. 1); thus in this gene rate constancy is violated most strongly, at least in observable differences (as detected by the relative rates tests). The remainder of the spread in estimates is less because of disagreement among genes than extrapolation from different fossil calibrations.
Uncertainties and Limitations of the Clock Assumptions.
In general, nonparametric rate smoothing (relaxing the assumption of a molecular clock) tended to reduce estimates of the age of the microgastroid lineage for the 16S and COI genes and to increase them for the 28S gene. The magnitude of these effects clearly depends on the fossil date used for calibration of the underlying clock (Table 2; Fig. 2). It is not clear that relaxing the assumption of a molecular clock led to any increase in accuracy overall. The use of the date from Protorhyssalus, the most recent calibration fossil and the one closest to the braconid/virus lineage age being estimated, seems to provide the strongest congruence among genes.
Uncertainties About Placement and Dating of Fossils.
One clear anomaly in this study is the discrepancy between the younger estimated age of the basal splits within the Braconidae using molecular data and the far older age of the Eobracon fossil attributed to Braconidae. One is forced to conclude that (i) Eobracon is attributed erroneously to Braconidae, (ii) its age is overestimated, or (iii) the Braconidae existed for a long time before its major extant lineages diverged. None of these scenarios can be ruled out at present, although scenario ii is less likely considering the apparent accuracy of the geological source. The other (older) fossils provide estimates from the three genes that are more in agreement with one another except COI, which tends to systematically overestimate the age of the microgastroid lineage.
It is possible that eventually the fossil calibration issue will be circumvented by more accurate rate estimation procedures based on many genes and using only 4-fold degenerate (silent) sites, which have been shown to behave in a remarkably clock-like manner (66). However, the current genetic database for parasitoids is not adequate for such an analysis yet.
Assumptions About Extent of Cophylogeny.
These estimates of the age of the microgastroid lineage (73.7 ± 10 million years), which should provide minimum estimates of the age of the braconid/bracovirus symbiosis, depend on the critical assumption that the two lineages have been associated at least since the origin of the wasp lineage that currently carries the viruses. This seems to be the case, and given the known biology and mode of inheritance of the viruses, it would be difficult to explain alternative routes to the current association. Nevertheless, it should be kept in mind that any data suggesting horizontal transmission of bracoviruses at any point in the evolutionary history of the wasps might cast doubt on these conclusions about age of the association.
The tight biological linkages we see today between wasp and virus are among the most remarkable interspecific integrations known in nature and have evolved into bewilderingly sophisticated functional complexes. However, it should be kept in mind that the association between bracoviruses and wasps, throughout its history, may not have played the important role in wasp–caterpillar interactions that it does today. Indeed, it is quite possible that the tight biological linkages we see between wasp and virus today were assembled over time from genes of both wasp and viral origin, the principal role of the viruses being a “delivery system” of functional genes into host caterpillars.
Acknowledgments
I would especially like to thank Mike Sanderson for help with using his program R8S, Mike Sanderson, Chris Simon, and John Huelsenbeck for discussions of divergence times methods in general, and Donald Quicke for information concerning the fossil Protorhyssalus. Sydney Cameron and three anonymous reviewers also provided valuable comments on the manuscript. This work was supported by National Science Foundation Grant DEB 02-96147.
Abbreviation
- mya
million years ago
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Askew R R. Parasitic Insects. London: Heinemann; 1971. [Google Scholar]
- 2.Godfray H C J. Parasitoids: Behavioral and Evolutionary Ecology. Princeton: Princeton Univ. Press; 1994. [Google Scholar]
- 3.Quicke D L J. Parasitic Wasps. London: Chapman and Hall; 1997. [Google Scholar]
- 4.Whitfield J B. J Hymenopt Res. 1992;1:3–14. [Google Scholar]
- 5.Whitfield J B. Annu Rev Entomol. 1998;43:129–151. doi: 10.1146/annurev.ento.43.1.129. [DOI] [PubMed] [Google Scholar]
- 6.Strand M R, Pech L L. Annu Rev Entomol. 1995;40:31–56. doi: 10.1146/annurev.en.40.010195.000335. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt O, Theopold U, Strand M R. BioEssays. 2001;23:344–351. doi: 10.1002/bies.1049. [DOI] [PubMed] [Google Scholar]
- 8.Gupta A P. Hemocytic and Humoral Immunity in Insects. New York: Wiley; 1986. [Google Scholar]
- 9.Fleming J G W. Annu Rev Entomol. 1992;37:401–425. doi: 10.1146/annurev.en.37.010192.002153. [DOI] [PubMed] [Google Scholar]
- 10.Whitfield J B. Parasitol Today. 1990;6:381–384. doi: 10.1016/0169-4758(90)90146-u. [DOI] [PubMed] [Google Scholar]
- 11.Stoltz D B, Whitfield J B. J Hymenopt Res. 1992;1:125–139. [Google Scholar]
- 12.Edson K M, Vinson S B, Stoltz D B, Summers M D. Science. 1981;211:582–583. doi: 10.1126/science.7455695. [DOI] [PubMed] [Google Scholar]
- 13.Beckage N E. In: Parasites and Pathogens of Insects, Volume1: Parasites. Beckage N E, Thompson S N, Federici B A, editors. San Diego: Academic; 1993. pp. 25–57. [Google Scholar]
- 14.Asgari S, Hellers M, Schmidt O. J Gen Virol. 1996;77:2653–2662. doi: 10.1099/0022-1317-77-10-2653. [DOI] [PubMed] [Google Scholar]
- 15.Asgari S, Schmidt O, Theopold U. J Gen Virol. 1997;78:3061–3070. doi: 10.1099/0022-1317-78-11-3061. [DOI] [PubMed] [Google Scholar]
- 16.Beckage N E. Sci Am. 1997;277(5):82–87. [Google Scholar]
- 17.Asgari S, Schmidt O. J Insect Physiol. 1994;40:789–795. [Google Scholar]
- 18.Webb B A, Summers M D. Proc Natl Acad Sci USA. 1990;87:4961–4965. doi: 10.1073/pnas.87.13.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb B A, Cui L. J Insect Physiol. 1998;44:785–793. doi: 10.1016/s0022-1910(98)00011-0. [DOI] [PubMed] [Google Scholar]
- 20.Stoltz D B. J Gen Virol. 1990;71:1051–1056. doi: 10.1099/0022-1317-71-5-1051. [DOI] [PubMed] [Google Scholar]
- 21.Fleming J G W, Summers M D. Proc Natl Acad Sci USA. 1991;88:9770–9774. doi: 10.1073/pnas.88.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitfield J B. Naturwissenschaften. 1997;84:502–507. [Google Scholar]
- 23.Stoltz D B, Vinson S B. Adv Virus Res. 1979;24:125–171. doi: 10.1016/s0065-3527(08)60393-0. [DOI] [PubMed] [Google Scholar]
- 24.Cook D, Stoltz D B. Virology. 1983;130:215–220. doi: 10.1016/0042-6822(83)90129-0. [DOI] [PubMed] [Google Scholar]
- 25.Whitfield J B, Mason W R M. Syst Entomol. 1994;19:61–76. [Google Scholar]
- 26.Belshaw R, Fitton M, Herniou E, Gimeno C, Quicke D L J. Syst Entomol. 1998;23:109–123. [Google Scholar]
- 27.Dowton M, Austin A D. Mol Phylogenet Evol. 1998;10:354–366. doi: 10.1006/mpev.1998.0533. [DOI] [PubMed] [Google Scholar]
- 28.Whitfield J B. In: The Hymenoptera: Evolution, Biodiversity, and Biological Control. Austin A D, Dowton M, editors. Melbourne: CSIRO; 2000. pp. 97–105. [Google Scholar]
- 29.Rasnitsyn A P. Contr Am Entomol Inst. 1983;20:259–265. [Google Scholar]
- 30.Rasnitsyn A P. Hymenoptera Apocrita of Mesozoic. Vol. 174. Moscow: Trans. Paleontol. Inst. Acad. Sci.; 1975. [Google Scholar]
- 31.Jell A P, Duncan P M. Mem Assoc Aust Palaeontol. 1986;3:111–205. [Google Scholar]
- 32.Rasnitsyn A P, Sharkey M J. Adv Parasitic Hymenopt Res. 1988;1988:169–197. [Google Scholar]
- 33.Basibuyuk H H, Rasnitsyn A P, van Achterberg K, Fitton M G, Quicke D L J. Zool Scripta. 1999;28:211–214. [Google Scholar]
- 34.Quicke D L J, Basibuyuk H H, Fitton M G, Rasnitsyn A P. Zool Scr. 1999;28:175–202. [Google Scholar]
- 35.Brues C T. Bernstein Forsch. 1933;3:4–178. [Google Scholar]
- 36.Zuckerkandl E, Pauling L. In: Horizons in Biochemistry. Kasha M, Pullman B, editors. New York: Academic; 1962. pp. 189–225. [Google Scholar]
- 37.Zuckerkandl E, Pauling L. In: Evolving Genes and Proteins. Bryson V, Vogel H J, editors. New York: Academic; 1965. pp. 97–166. [DOI] [PubMed] [Google Scholar]
- 38.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 39.Wilson A C, Ochman H, Prager E M. Trends Genet. 1987;3:241–247. [Google Scholar]
- 40.Avise J C. Molecular Markers, Natural History, and Evolution. New York: Chapman & Hall; 1994. [Google Scholar]
- 41.Ayala F J. J Hered. 1986;77:226–235. doi: 10.1093/oxfordjournals.jhered.a110227. [DOI] [PubMed] [Google Scholar]
- 42.Britten R J. Science. 1986;231:1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- 43.Li W-S, Tanimura M. Nature (London) 1987;326:93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- 44.Takezaki N, Rzhetsky A, Nei M. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 45.Sanderson M J. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- 46.Sanderson M J. R8S Analysis of Rates (r8s) of Evolution (and Other Stuff) Davis, CA: Univ. of California; 1997b. [Google Scholar]
- 47.Thorne J, Kishino H, Painter I S. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 48.Huelsenbeck J P, Larget B, Swofford D. Genetics. 2000;154:1879–1892. doi: 10.1093/genetics/154.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn B H, Wolinsky S, Battacharya T. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 50.Hillis D M, Mable B K, Moritz C. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 515–543. [Google Scholar]
- 51.Rambaut A, Bromham L. Mol Biol Evol. 1998;15:442–448. doi: 10.1093/oxfordjournals.molbev.a025940. [DOI] [PubMed] [Google Scholar]
- 52.Doyle J A, Donoghue M J. Paleobiology. 1993;19:141–167. [Google Scholar]
- 53.Springer M. J Mol Evol. 1995;41:531–538. [Google Scholar]
- 54.Sanderson M J. In: Molecular Systematics of Plants II: DNA Sequencing. Soltis D E, Soltis P S, Doyle J J, editors. Boston: Kluwer; 1998. pp. 242–264. [Google Scholar]
- 55.Mardulyn P, Whitfield J B. Mol Phylogenet Evol. 1999;12:282–294. doi: 10.1006/mpev.1999.0618. [DOI] [PubMed] [Google Scholar]
- 56. Whitfield, J. B., Mardulyn, P., Austin, A. D. & Dowton, M. (2002) Syst. Entomol., in press.
- 57.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 58.Wu C-I, Li W H. Proc Natl Acad Sci USA. 1985;82:1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tajima F. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swofford D L. PAUP*, Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer; 1998. , Version 4.0. [Google Scholar]
- 61.Kimura M. J Mol Evol. 1980;16:11–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 62.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;21:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 63.Yang Z. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 64.Rodriguez F, Oliver J L, Marin A, Medina J R. J Theor Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- 65.Langley C H, Fitch W M. J Mol Evol. 1974;3:161–177. doi: 10.1007/BF01797451. [DOI] [PubMed] [Google Scholar]
- 66.Kumar S, Subramanian S. Proc Natl Acad Sci USA. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]