Abstract
A self-encoding system designed to have strict “compartition” of the molecules, i.e., to contain only a single molecule of DNA in each compartment, was established, and its evolutionary fate was analyzed. The system comprised the Thermus thermophilus DNA polymerase gene as the informational molecule and its protein product replicating the gene as the functional molecule. Imposing strict compartition allows the self-encoding system to last up to at least the tenth generation, whereas the system ceased to work after the third generation when loose compartition initiated with 100 molecules was imposed. These results provide experimental evidence on the importance of compartition for the maintenance of a self-encoding system. In addition, the extent of diversity in self-replication activity of the compartments was found to be another vital difference in the evolutionary dynamics between the strict and loose compartitions. Although the system with strict compartition provides widely diversified activity of the compartments at each generation, the values of the activity diverge only within a small range in the system with loose compartition. When the variety in the activity of a compartment is small, functional selection becomes weak, and to conform Darwinian evolution may become unfeasible. Therefore, strict compartition is essential for the evolvability of a self-encoding system.
Researchers have dealt with the construction of informational replicating systems (1) based on oligonucleotides (2–4), peptides (5), artificial complementary molecules (6, 7), and RNA molecules with (8–10) or without (11–14) using the natural polymerases. Although conditions used in such in vitro informational replicating systems may not be very close to the supposed prebiotic environment (15), great contributions were made to the conceptual knowledge on the origin of life (1).
According to the RNA world theory (16, 17), evolution began with the appearance of a self-replicating RNA, catalyzing its own replication and function as both gene and enzyme, and finally constituted a mutually dependent system. To maintain and further evolve the newly emerged mutually dependent system, two organizational principles, “hypercycle” and “compartition,” complementing each other were proposed to be indispensable (18). Hypercycle is a superimposed high-order “closed” cyclical coupling of individual replication cycles organized in a feedback loop connecting the functional molecule (enzyme or protein) to its informational molecule (RNA). The second organizational principle, compartition, works to complement the hypercycle by encapsulating the informational and functional molecules in some kind of a compartment, as the feedback loop can come into effect only if the genotype and phenotype remain in each other's vicinity. The hypercycle can perform coupling, such as binding, between the genotype and phenotype, functional integration, and growth regulation (18). However, present organisms seem to have only weak binding between the gene and its products. Therefore, sometime along the line of evolution, the binding between the gene and its products might have weakened and may have caused the degeneration of the hypercycle. Without compartmentalization, the potential fitness benefits of other molecules encoded by the gene would be lost (18). Compartmentalization, then, might have provided a way of linking genotype and phenotype, of which the resultant is the evolution of a vast range of molecules with diverse activities.
In this report, we show an example of an in vitro self-encoding system with the efficient cycling of the DNA polymerase gene, an informational molecule, expressing its protein product, a functional molecule that duplicates the gene. The role of compartmentalization when mutations are accumulated in the system, i.e., when the binding between the genotype and phenotype weakens, is examined. We conclude that compartition is an essential factor for the maintenance and evolution of a self-encoding system.
Materials and Methods
Materials.
pLED-HB (DDBJ/EMBL/GenBank accession no. AB025788) and pLED-HBB are hybrid plasmids harboring the wild-type (19) gene of Thermus thermophilus DNA polymerase and a mutant (G1851T) gene, respectively. Purified wild-type DNA polymerase was kindly provided by Toyobo (Osaka).
Nested PCR.
Nested PCR was done according to ref. 20. To eliminate any contaminating DNA completely, the reaction mixture (45 μl) consisting of 30 mM N-tris(hydroxymethyl)glycine (Tricine)/NaOH (pH 8.4), 1 mM MgCl2, 0.2 mM each of the dNTPs, 0.4 unit of purified T. thermophilus DNA polymerase (wild type), and 50 pmol each of primers A3 (ACCTCCTGGACCCCTCCAACAC) and B2 (CTTCTCTCATCCGCCAAAACAGCC) was pretreated with HpaII (2 units) at 37°C for 2 h, and the enzyme was inactivated at 65°C for 20 min before the addition of 5 μl of the highly diluted DNA template solution. The positions of the primers used in this work are shown in Fig. 1A. The reaction mixture was preheated at 94°C for 5 min before being subjected to 45 cycles of amplification (94°C for 1 min, 55°C for 1.5 min, and 72°C for 2 min). After the first amplification, an aliquot (5 μl) was transferred to a fresh tube for the second round of the same 45 cycles of amplification, but with primers A4 (TTAGTAGCTCCGACCCCAAC) and B3 (CCAGGGGCACGGCGAGGG) instead. In addition, take note that the reaction mixture, although basically the same on the second cycle of amplification, used 0.8 mM MgCl2 and 1.2 unit of the polymerase instead. The resulting A4/B3 fragment was digested with BamHI, HindIII, and SacII, and the resultant BamHI/HindIII fragment was used hereafter. The restriction endonuclease SacII was used to facilitate the isolation of the BamHI/HindIII fragment.
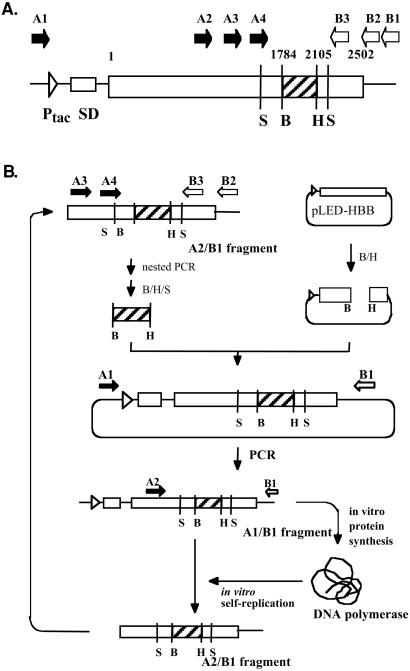
Figure 1.
Structure of T. thermophilus DNA polymerase gene (A) and schematic diagram of the life cycle of the self-encoding system (B). Restriction sites are abbreviated as follows: B, BamHI; H, HindIII; S, SacII. Other abbreviations are Ptac, tac promoter; SD, Shine–Dalgarno sequence.
Replacement of the BamHI/HindIII Region in the DNA Polymerase Gene.
The BamHI/HindIII fragment prepared above was substituted for the corresponding region of the mutant gene G1851T harbored in pLED-HBB by ligating the fragment with the plasmid that had been digested with BamHI and HindIII (Fig. 1B). The G1851T mutation generates a stop codon. The region including the polymerase gene in the ligated sample was amplified by using a commercially available DNA polymerase (0.25 μl/50 μl reaction, Extaq, Takara) with primers A1 (CCACCTCTGACTTGAGCGTCG) and B1 (GTTCTGATTTAATCTGTATCAGGCTG). After agarose gel electrophoresis, the amplified A1/B1 fragments were recovered with QIAquick (Qiagen), quantified based on absorbance at 260 nm, and used for in vitro protein synthesis and self-replication.
In Vitro Synthesis of DNA Polymerase from Its Gene.
In vitro protein synthesis from the DNA polymerase gene was carried out as described (21), except that Escherichia coli DPB267 (22) was used for the preparation of S-30 fraction. The reaction mixture (30 μl), including 2 μg of the A1/B1 fragment, was incubated at 37°C for 2 h, heat-treated at 75°C for 20 min, and centrifuged. An aliquot (2.5 μl) of the supernatant was used for in vitro self-replication.
In Vitro Self-Replication.
A reaction mixture (50 μl), consisting of the A1/B1 fragment, self-produced protein product of DNA polymerase, 30 mM Tricine/NaOH (pH 8.4), 1.0 mM MgCl2, 0.2 mM each of the dNTPs, and 25 pmol each of primers A2 (CTTCCTGGAGAGGCTGGAGTTC) and B1, was subjected to the following thermal cycle: preheat at 94°C for 5 min and 30 cycles of consecutive 94°C for 1 min, 55°C for 1 min, and 72°C for 10 min. The PCR product was analyzed by agarose gel electrophoresis of an aliquot (3 μl) of the mixture together with the internal markers and the standard DNAs. The amount of the amplified DNA was quantified by the intensity of the ethidium bromide fluorescence of each band scanned and analyzed by an Atto densitograph image analyzer AE-6900-F (Atto, Tokyo). The amount of amplified DNA was used as a measure of the activity of the self-encoding system. Equal volumes of the reaction mixture from each compartment were mixed, and the pooled A2/B1 fragments were recovered from a 1% agarose gel. DNA concentration of the recovered fragments determined by A260 was in the range of 10–100 ng/μl. The pooled fragment solution was then highly diluted (10−9 to 10−11) and an aliquot (5 μl) of the diluted solution was distributed to each of the compartments for the nested PCR of next generation. The dilution factor was determined such that DNA amplification by nested PCR will proceed in only a maximum of 30% of the total number of compartments. Among the compartments subjected to nested PCR, 10 of those positive in DNA amplification comprised the individual compartments of the next generation.
Results and Discussion
To analyze the effects of compartment formation on the evolutionary fate of a self-encoding system, we established a method that will afford DNA amplification from the least number of DNA molecules in each compartment, a single molecule. In our self-encoding system, the T. thermophilus DNA polymerase gene (19) serves as an informational molecule and the test tube takes the role of a compartment. The life cycle of the system in a compartment with strict compartition starts from a single molecule of a fraction of the gene, the A2/B1 fragment (Fig. 1). The gene fragment was amplified by nested PCR (20) using two pairs of primers, A3/B2 and A4/B3, and subjected to the in vitro self-replication reaction using the DNA polymerase transcribed from the gene containing the amplified fragment (see Materials and Methods). The DNA fragments amplified through self-replication in all of the compartments were mixed and appropriately diluted, and each aliquot, containing at most a single DNA molecule, was passed to the next cycle. The probability for genetic information in a compartment to recur in the next generation depends on the number of the DNA molecules after amplification.
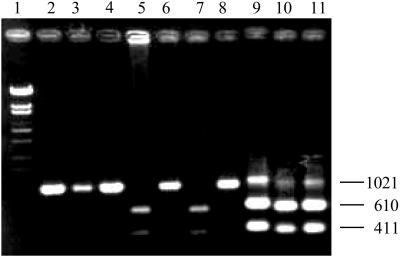
To start a life cycle of the system with strict compartition, the solution of the gene fragment was appropriately diluted such that only in a maximum of 30% of the total number of compartments did DNA amplification proceed successfully after the nested PCR. Assuming a Poisson distribution for the number of the DNA molecule in each compartment, the probability for an amplification to be prompted by a single DNA molecule is estimated to be at least 0.83. The reliability of DNA amplification from a single DNA molecule through nested PCR was confirmed by subjecting to the nested PCR a mixture containing equal amount of two types of A1/B1 fragments, one is wild-type and the other the mutant (G1851T) possessing an additional BglII site on the DNA polymerase gene. The nested PCR of the highly diluted DNA mixture giving about 0.3 molecule per compartment in average produced either of the sequences in most cases, as shown in Fig. 2 (lanes 2–8), with a low frequency of occurrence of the two sequences in a compartment (data not shown). Conversely, the nested PCR in each compartment initiated with a DNA mixture that is 10 times higher in concentration always yielded both sequences (Fig. 2, lanes 9–11). This manner of picking either of the two fragments during amplification shows the efficacy of the nested PCR from a single DNA molecule.
Figure 2.
Confirmation for DNA amplification from a single DNA molecule. Nested PCR was conducted on a mixture containing equal amounts [about 7.0 × 10−20 g/μl (lanes 2–8) or 7.0 × 10−19 g/μl (lanes 9–11)] of two A1/B1 fragments of the wild-type and a mutant (G1851T) DNA polymerase genes as described in Materials and Methods. As the two types of A1/B1 fragments are discriminated based on the additional BglII site on the DNA polymerase gene of the mutant G1851T, the amplified fragments were digested with BglII before being subjected to agarose gel electrophoresis. Lane 1, molecular weight marker.
In the life cycle of the self-encoding system, the A4/B3 fragments amplified by nested PCR were digested with BamHI and HindIII, and the BamHI/HindIII fragment was substituted for the corresponding region in the wild-type DNA polymerase gene to obtain the whole genetic information (Fig. 1). The newly constructed gene was subjected to the in vitro protein synthesis reaction, and the product DNA polymerase was used for the in vitro self-replication reaction, i.e., the PCR amplification of the A2/B1 region of its own gene (Fig. 1). This strategy ensures that of the spontaneous mutations on the amplification reactions including the nested PCR and self-replication, mutations only in the BamHI/HindIII region were accumulated and inherited by the next generation. The overall mutation rate for one life cycle was estimated to be 1.2 ± 0.2 nucleotide substitutions per gene based on the nucleotide sequences of 30 genes sampled from the gene libraries obtained after one life cycle of model experiments initiated from a single molecule of the wild-type and mutant genes with known sequences.
The schematic diagram of the life cycle of the self-encoding system, shown in Fig. 1B, consists in total three consecutive PCR steps: nested PCR starting from a single gene, amplification PCR of the DNA polymerase gene before in vitro protein synthesis, and self-replication PCR. The first two PCR steps, involving the commercially available DNA polymerases, were conducted to achieve a highly sensitive measurement of the activity of the self-encoding system, i.e., the level of the self-replication reaction. Agarose gel electrophoresis was then performed to purify the amplified DNA fragments so obtained and guarantee the complete elimination of the DNA polymerases used in the first two PCRs. Therefore, in the final self-replication PCR, the DNA polymerase was generated through in vitro protein synthesis with the use of E. coli S-30 extract.
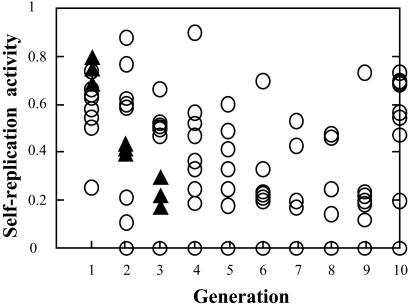
To observe the evolutionary dynamics, we monitored the self-encoding system with strict compartition in 10 compartments for 10 generations (Fig. 3, circles). The activity of each compartment was measured as the amount of DNA molecules amplified by the self-replication reaction in each generation, and the values were expressed relative to the wild-type gene. It should be noted that the mutational effect on the activity includes not only that of the activity of the enzyme transcribed from the gene, but also the properties of the gene as a template for the PCR and for protein synthesis. In addition, for every generation, mutations on the gene accumulate during PCR amplifications and accordingly, polymerase molecules prepared by in vitro protein synthesis vary in accord with the mutated genes in a compartment. Hence, the self-replication reaction in a compartment proceeds with heterogeneous polymerase molecules and heterogeneous templates. Consequently, the self-replication activity of a compartment is an actual summation of activities from all mutated polymerases and genes in the compartment.
Figure 3.
Molecular evolution of the self-encoding system. The A1/B1 fragment serving as the initial material for the first generation of the life cycle was prepared by PCR using ex-Taq DNA polymerase with pLED-HB (19) as a template and A1/B1 primers. From the second generation of the system, A2/B1 was used as the template as shown in Fig. 1B. For the system with strict compartition (○), the evolutionary dynamics were observed for 10 generations and each generation comprised 10 compartments. More than 83% of the compartments were estimated to contain a single molecule of the A1/B1 or A2/B1 fragment at the initial stage of the life cycle. For the system with loose compartition (▴), the dynamics were observed for three generations and each generation comprised three compartments, which had about 100 molecules of the A1/B1 or A2/B1 fragment at the start of the life cycle. The activity of the self-encoding system in a compartment was measured as the amount of DNA molecules amplified by the self-replication reaction in each generation, and the values were expressed relative to the self-replication activity of the wild-type gene. The accuracy of the activity assay was within 10% error for the system of the wild-type gene.
A distinct feature of the evolutionary dynamics observed in our system with strict compartition is that every generation has a population exhibiting a wide variety of activity (Fig. 3, circles). As all compartments possess lower self-replication activity than the wild-type gene, the mutations incurred seem to be deleterious or neutral. Because the system was carried out by mixing the genes in all of the compartments before their passage to next generation, the probability of the genes in a compartment with higher activity being inherited by the next generation is higher, whereas those with lower activity tend to become extinct. This selection pressure was clearly shown when the genetic code in the compartment with highest activity in the ninth generation and the genetic codes in each compartment of the tenth generation were determined. Indeed, nine of the ten compartments at the tenth generation had genes with the same or derived sequences as that found in the compartment with the highest activity in the ninth generation.
Theoretically, Eigen proposed that to maintain and evolve a self-replicating system organized in a feedback loop coupling a functional molecule and an informational molecule to each other leading to the generation of a hypercycle of replication, compartition of an information molecule is necessary (18). To assess the principle, we relaxed the criteria of compartition in our self-encoding system such that, instead of having a single DNA molecule to initiate each generation, we increased the number to 100 DNA molecules, and observed the self-encoding system with loose compartition in three compartments (Fig. 3, triangles). The results show that the activity of all of the compartments decreased sharply, and the self-encoding system ceased to work after three generations. In contrast, with the case of strict compartition initiated with a single molecule, the system was carried over even to the tenth generation of our evolutionary process (Fig. 3, circles). The activity of a compartment initiated with 100 molecules depicts an average activity over 100 compartments each originating from a single DNA molecule. In this case, the value of the activity averaged out the consequence of compartments with higher activity, especially when the effects of mutations are mostly deleterious or neutral as mentioned above. Eventually, the effects of disadvantageous mutations were augmented in the averaged activity and explain degeneration of the self-encoding system with loose compartition. On the contrary, with strict compartition, an enduring self-encoding system is obtained and catastrophic consequences rendered by deleterious mutation are neglected. The results shown in Fig. 3, hence, provide experimental evidence on the importance of compartition, in combination with the hypercycle, for the maintenance of a self-encoding system as proposed by Eigen (18), and recently by Kaneko and Yomo (23).
The extent of diversity in activity of the compartments comprises another vital difference in the evolutionary dynamics between the strict and loose compartitions. Applying strict compartition to the system resulted to widely diversified activity of the compartments at each generation (Fig. 3, circles), whereas the values of the activity fall only within a small range in the system with loose compartition (Fig. 3, triangles). This difference is attributed to the weak law of large numbers that the variance of a sample is larger than that of the average of samples. When the variety in the activity of a compartment is small, functional selection becomes weak or dormant, and Darwinian evolution may not be followed as stated. Given that the mutations produced higher- and lower-activity mutants distributed evenly in the population, the activity will improve recurrently in each generation for the self-encoding system with strict compartition, but will not vary much from the original level in the case of loose compartition. Even with a negative average effect of mutations, chances for an activity to improve in a system with strict compartition still prevail (see ref. 24 for statistical analysis). Therefore, we propose that compartition is essential for the evolvability of a self-encoding system, as it increases the variety of the competing population and makes the functional selection effective.
Here, we showed a sustainable in vitro self-encoding system, where the genotype–phenotype dichotomy gyrated in a cyclical feedback coupling of the translated product, the DNA polymerase, and the replicated gene of its own throughout the evolutionary process. In addition, the system was also proven effective in evolving molecules based on functional selection. From the fact that functional selection comes into effect through imposed strict compartition on a self-encoding system, we may assume that primitive life used occasional subsequent dilution and compartition for its evolution. When some of the replicators adopt strict compartition, their information will become digitalized and the system will become evolvable. Oberholzer et al. (25) had shown that the RNA molecules were amplified through the added RNA polymerase in the dividing lipid vesicles serving as a compartment or protocell. As evolution proceeded, the replicator may have taken a more sophisticated mechanism, such as dividing vesicles, leading to the development of a whole arsenal of integrated networks that render the present status of cells. Considering the robotics-supported technology of directed molecular evolution (26), it may not be too far to see the evolution of more sophisticated self-encoding system in the near future.
Acknowledgments
This work was supported in part by Ministry of Education, Science, Sports and Culture of Japan Grants 11CE2006 and 09555253.
References
- 1.Orgel L E. Nature (London) 1992;358:203–209. doi: 10.1038/358203a0. [DOI] [PubMed] [Google Scholar]
- 2.Inoue T, Orgel L E. Science. 1983;219:859–862. doi: 10.1126/science.6186026. [DOI] [PubMed] [Google Scholar]
- 3.Sievers D, von Kiedrowski G. Nature (London) 1994;369:221–224. doi: 10.1038/369221a0. [DOI] [PubMed] [Google Scholar]
- 4.Li T, Nicolaou K C. Nature (London) 1994;369:218–221. doi: 10.1038/369218a0. [DOI] [PubMed] [Google Scholar]
- 5.Lee D H, Granja J R, Martinez J A, Severin K, Ghadiri M R. Nature (London) 1996;382:525–528. doi: 10.1038/382525a0. [DOI] [PubMed] [Google Scholar]
- 6.Tjivikua T, Ballester P, Rebek J., Jr J Am Chem Soc. 1990;112:1249–1250. [Google Scholar]
- 7.Hong J-I, Feng Q, Rotello V, Rebek J., Jr Science. 1992;255:848–850. doi: 10.1126/science.255.5046.848. [DOI] [PubMed] [Google Scholar]
- 8.Mills D R, Peterson R L, Spiegelman S. Proc Natl Acad Sci USA. 1967;58:217–224. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer G J, McCaskill J S, Otten H. Proc Natl Acad Sci USA. 1989;86:7937–7941. doi: 10.1073/pnas.86.20.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breaker R R, Joyce G F. Proc Natl Acad Sci USA. 1994;91:6093–6097. doi: 10.1073/pnas.91.13.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel D P, Szostak J W. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 12.Bohler C, Nielsen P E, Orgel L E. Nature (London) 1995;376:578–581. doi: 10.1038/376578a0. [DOI] [PubMed] [Google Scholar]
- 13.Wright M C, Joyce G F. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 14.Ekland E H, Bartel D P. Nature (London) 1996;382:373–376. doi: 10.1038/382373a0. [DOI] [PubMed] [Google Scholar]
- 15.Joyce G F. Nature (London) 1989;338:217–224. doi: 10.1038/338217a0. [DOI] [PubMed] [Google Scholar]
- 16.Cech T R. Proc Natl Acad Sci USA. 1986;83:4360–4363. doi: 10.1073/pnas.83.12.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert W. Nature (London) 1986;319:618. [Google Scholar]
- 18.Eigen M. Steps Towards Life. New York: Oxford Univ. Press; 1992. [Google Scholar]
- 19.Asakura K, Komatsubara H, Soga S, Yomo T, Oka M, Emi S, Urabe I. J Ferment Bioeng. 1993;76:265–269. [Google Scholar]
- 20.Porter-J K, Rosenberg E I, Keiser J F, Gross J D, Ross A M, Nasim S, Garrett C T. J Med Virol. 1990;30:85–91. doi: 10.1002/jmv.1890300202. [DOI] [PubMed] [Google Scholar]
- 21.Yomo T, Habu T, Soga S, Matsuura T, Shima Y, Urabe I. In: Artificial Life. Langton C G, Shimohara K, editors. Vol. 5. Cambridge, MA: MIT Press; 1996. pp. 402–405. [Google Scholar]
- 22.Biek D P, Cohen S N. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko K, Yomo T. J Theor Biol. 2002;214:563–576. doi: 10.1006/jtbi.2001.2481. [DOI] [PubMed] [Google Scholar]
- 24.Matsuura T, Yomo T, Trakulnaleamsai S, Ohashi Y, Yamamoto K, Urabe I. Protein Eng. 1998;11:789–795. doi: 10.1093/protein/11.9.789. [DOI] [PubMed] [Google Scholar]
- 25.Oberholzer T, Wick R, Luisi P L, Biebricher C K. Biochem Biophys Res Commun. 1995;207:250–257. doi: 10.1006/bbrc.1995.1180. [DOI] [PubMed] [Google Scholar]
- 26.Christians F C, Scapozza L, Crameri A, Folkers G, Stemmer W P. Nat Biotechnol. 1999;17:259–264. doi: 10.1038/7003. [DOI] [PubMed] [Google Scholar]