Abstract
Noncoding RNA (ncRNA) genes that produce functional RNAs instead of encoding proteins seem to be somewhat more prevalent than previously thought. However, estimating their number and importance is difficult because systematic identification of ncRNA genes remains challenging. Here, we exploit a strong, surprising DNA composition bias in genomes of some hyperthermophilic organisms: simply screening for GC-rich regions in the AT-rich Methanococcus jannaschii and Pyrococcus furiosus genomes efficiently detects both known and new RNA genes with a high degree of secondary structure. A separate screen based on comparative analysis also successfully identifies noncoding RNA genes in P. furiosus. Nine of the 30 new candidate genes predicted by these screens have been verified to produce discrete, apparently noncoding transcripts with sizes ranging from 97 to 277 nucleotides.
Noncoding RNA (ncRNA) genes are genes for which RNA, rather than protein, is the functional end product. The number and diversity of ncRNA genes is a subject of active research (1). In principle, the availability of many genome sequences makes it possible to search computationally for novel ncRNA genes. Computational protein gene finders search for ORFs that have certain statistical biases in their nucleotide composition (2–4). Unfortunately, ncRNA genes have neither ORFs nor (generally speaking) nucleotide composition biases, making ncRNA gene-finding a more formidable problem.
Hyperthermophiles must stabilize double-stranded DNA and RNA against thermal denaturation (5). The simplest stabilization strategy is increased GC content. However, the GC content of hyperthermophile genomes does not correlate with optimal growth temperature (5–7). Hyperthermophiles use various other mechanisms to stabilize their DNA, including increased intracellular ionic concentrations, cationic proteins, and supercoiling (5, 7). Intramolecular RNA secondary structure, however, seems to be partially stabilized by increased hydrogen bonding, as the GC content of ribosomal RNA and transfer RNA genes in hyperthermophiles shows a strong correlation with optimal growth temperature (6). We reasoned that in an AT-rich extreme hyperthermophile, structural RNA genes (i.e., ncRNA genes with a high degree of secondary structure) might be found just by searching for regions of elevated GC content. Such a gene finder would not be able to be generalized. However, one might use novel ncRNAs identified in these unusual genomes to identify homologous RNAs in a variety of other genomes.
Several recent reports describe computationally aided screens for ncRNA genes in Escherichia coli. Argaman et al. (8) searched for strong promoter and terminator signals appropriately spaced over intergenic regions. This approach obviously requires the genome sequence of an organism for which transcriptional regulation is well understood. Carter et al. (9) used a neural network to classify genomic sequences based on several features, including GC composition. Two other approaches used a comparative genomics approach, requiring genomic sequence from related organisms as well as that of E. coli. Whereas Wassarman et al. (10) simply looked for conserved intergenic regions, Rivas et al. (11) further processed sequence alignments of the conserved intergenic regions to decide whether the pattern of mutation was most consistent with a protein-coding gene, an ncRNA gene with secondary structure, or simply random mutation. This latter approach has been converted into a general gene finder, QRNA, which can be used for any genome for which additional comparative genomic sequence is available (12).
To date, detailed analysis of the performance of QRNA has been performed only in E. coli. Furthermore, comparison of the performance of QRNA with that of an alternative gene finder would prove informative on the trustworthiness of both screens. That is, even though a GC-based screen may work only in unusual organisms, those organisms provide a test bed for further validation of QRNA as a general RNA gene finder. Therefore, we screened for novel ncRNAs by using both the GC content bias and QRNA to compare their performance and results. Here we identify novel ncRNAs in Methanococcus jannaschii by using the GC content bias alone and in Pyrococcus furiosus by using both the GC bias and QRNA-based comparative analysis. We find that the two screens performed in P. furiosus identified nearly exactly overlapping sets of ncRNA genes.
Methods
Genomes Used.
Fifty-one complete prokaryotic genome sequences in GenBank as of June 21, 2001, were downloaded from ftp://ftp.ncbi.nlm.nih.gov/genbank/genomes/Bacteria. The sequence of P. furiosus was downloaded from the Utah Genome Center (http://www.genome.utah.edu/sequence.html) on August 27, 2001. trnascan-se version 1.21 (13) was used to identify tRNA genes.
Computational Screens.
A hidden Markov model with two states (“RNA” and “background genome”) was used. The emission probabilities of the genome state were set to the low GC content of the overall genome, whereas the emission probabilities of the RNA state were set to the higher GC content of the tRNA and rRNA genes. Transition probabilities were set by assuming that the number of ncRNA genes in the genome was equal to the known ncRNA gene number, and that all ncRNA genes should be around 100 nucleotides long. Standard Viterbi and posterior decoding algorithms were used (14). In the Viterbi screen of M. jannaschii, only nine candidate RNA regions of at least 50 nucleotides were considered; one shorter region was discarded. For the posterior decoding screen, regions of at least 50 nucleotides were selected in which all bases had a posterior probability of the RNA state over a chosen cutoff. The cutoff probabilities were set so that all tRNAs were successfully proposed as GC-rich regions. These cutoffs were 0.130 for P. furiosus, 0.052 for Pyrococcus. abyssi, and 0.147 for Pyrococcus horikoshii. Conserved GC-rich P. furiosus candidate ncRNAs were then identified with wu-blast version 2.0 with W = 4 (ref. 15; http://blast.wustl.edu/) by requiring a P. furiosus GC-rich region to hit a GC-rich region from both P. abyssi and P. horikoshii with an E-value less than 10−5. The source code and parameters for the screening program are freely available from http://www.genetics.wustl.edu/eddy.
To perform the comparative analysis by examining the pattern of mutation in the alignments, we first used wu-blastn (version 2.0) with default parameters except hspmax = 100,000 to compare related genomes. We used the P. furiosus genome as a query against the P. abyssi and P. horikoshii genomes. We kept only alignments with E < 0.01, 65–85% identity, and at least 50 nucleotides long for further analysis. These alignments were then analyzed with QRNA 1.1 (12), and a list of candidate ncRNA genes was produced by merging all overlapping P. furiosus regions scoring at least 5 bits.
Northern Blotting and Rapid Amplification of cDNA Ends (RACE)-PCR Analysis.
M. jannaschii frozen cell paste was provided by J. Brown (North Carolina State University). These cells were grown in 12-liter batch fermentations in American Type Culture Collection (ATCC) media 2121 at 83°C with continuous sparging with 60% H2/40% CO2 (vol/vol) and daily replacement with Na2S. Cultures were harvested after 2–3 days, approximately during late logarithmic growth. RNA was prepared from cell paste by mortar and pestle lysis and phenol/chloroform extraction by modifying a DNA extraction protocol (16). P. furiosus was grown at 95°C in rich medium containing peptides and maltose, but without sulfur, as described (17). Cells were harvested in mid-log phase, and the RNA was extracted as described (18).
Northern blots were performed by running 10 μg of total RNA on a 6% denaturing polyacrylamide gel. Size standards were 5′ end-labeled 100- (New England Biolabs) and 25-bp (Promega) denatured DNA ladders. Gels were electroblotted to Zeta-Probe membrane (Bio-Rad), hybridized to 106 cpm of labeled oligonucleotide probe, and visualized on a Molecular Dynamics PhosphorImager system.
To perform 5′ and 3′ RACE, total RNA was purified further by treating with RNase-free DNase (Promega), polyadenylated with E. coli poly(A) polymerase (GIBCO/BRL), then reverse transcribed with the SMART RACE cDNA Amplification kit (CLONTECH). Specific 5′ and 3′ cDNA ends were amplified with a gene-specific primer and the UPM-Long primer in a PE GeneAmp System 9700 thermocycler (Perkin–Elmer), then reamplified with the same gene-specific primer and the UPM-Short primer with HotStar Taq (Qiagen, Chatsworth, CA). Products were cloned with the pCRII vector in the TA Cloning kit (Invitrogen). Five to 10 independent clones of each end were sequenced with M13 Reverse primer with the Applied Biosystems Big Dye Sequencing kit version 2. Some false 5′ ends were identified on the basis of an internal GGG in the ncRNA molecule and were not considered to be true 5′ ends.
Computational Analysis of the RNAs.
wu-blastn version 2.0 with W = 3 was used to search the NCBI nonredundant nucleotide database (version May 16, 2001) and a database of all of the available Archaeal genomes in GenBank as of June 21, 2001 (Aeropyrum pernix, Archaeoglobus fulgidus, Halobacterium sp. NRC-1, Methanobacterium thermoautotrophicum, M. jannaschii, P. abyssi, P. horikoshii, Sulfolobus solfataricus, Thermoplasma acidophilum, and Thermoplasma volcanium), as well as P. furiosus from August 27, 2001. Secondary structure prediction was assisted by mfold version 3.1 (19, 20).
Results
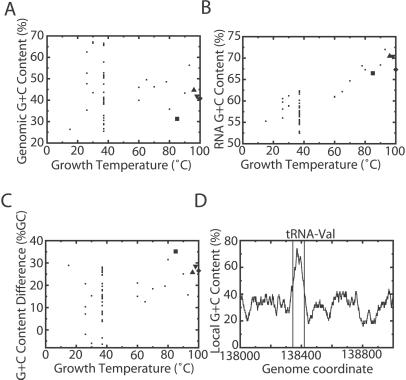
We first tested whether previous observations on the relationship between genomic and ncRNA GC content with optimal growth temperature held for 52 prokaryotic genomes available in the summer of 2001 (5–7). The overall GC content of each genomic sequence shows no correlation with optimal growth temperature (Fig. 1A). On the other hand, the transfer RNA GC content does clearly correlate with growth temperature (Fig. 1B). Surprisingly, a large GC content difference is not restricted to thermophiles, although it is more pronounced there (Fig. 1C), suggesting that a screen on the basis of GC bias may work in some mesophiles. In the organism with the largest GC content difference, M. jannaschii, tRNAs are readily apparent because of the GC content difference (Fig. 1D; ref. 21). We therefore decided to screen M. jannaschii for ncRNAs based solely on local GC content.
Figure 1.
GC content as a basis for finding ncRNA genes. (A) GC content of whole genomes vs. optimal growth temperature. In this and subsequent images, the large square represents M. jannaschii, the up triangle P. abyssi, the down triangle P. horikoshii, and the diamond P. furiosus. (B) GC content of tRNA genes vs. optimal growth temperature. (C) Difference in tRNA and genomic GC content vs. optimal growth temperature. (D) GC content of a 1-kb region of the M. jannaschii genome containing a tRNA gene calculated in a 100-bp sliding window.
To objectively define high-GC regions, we parameterized a two-state hidden Markov model and used the Viterbi algorithm to parse the M. jannaschii genome. This approach identified 43 different regions. Because Viterbi decoding produces the best (maximum likelihood) assignment of nucleotides to either “RNA” or “background genome,” there is no score associated with individual regions or any cutoff parameters to set. To evaluate sensitivity, we asked what percentage of tRNA genes were correctly identified by overlapping with predicted “RNA” regions; all 37 known M. jannaschii tRNAs were identified, as were ribosomal RNA genes, RNase P RNA, and 7S (signal-recognition particle) RNA. After accounting for regions that contain these genes, 9 regions at least 50 nucleotides in length remained as candidate ncRNA genes (Table 1). To evaluate specificity, we analyzed 1,000 random genome sequences with the same overall G+C composition and length and detected 33 GC-rich regions, indicating that the expected number of GC-rich regions detected by chance is about 0.03 per genome.
Table 1.
Candidate ncRNAs
| Candidate no. | Detected by | Predicted start | Predicted length | % G+C | Flanking % G+C | Northern + strand | Northern − strand | Genetic locus | Real length | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| Mj1* | V | 16816 | 84 | 64 | 21 | — | — | |||
| Mj2 | V | 118079 | 101 | 63 | 23 | — | 125 | hgcA | 129 | AF447575 |
| Mj3 | V | 325029 | 68 | 65 | 34 | — | 105, 130 | hgcB | 127 | AF447576 |
| Mj4 | V | 412582 | 54 | 72 | 31 | — | — | |||
| Mj5 | V | 774708 | 81 | 63 | 35 | — | — | |||
| Mj6 | V | 951852 | 117 | 64 | 29 | 110, 120 | — | hgcC | 129 | AF447577 |
| Mj7 | V | 1129126 | 69 | 70 | 20 | — | 105 | hgcD | 127 | AF447578 |
| Mj8 | V | 1553923 | 61 | 67 | 24 | — | — | |||
| Mj9 | V | 1659451 | 70 | 73 | 22 | — | — | |||
| Pf1 | P, Q | 163924 | 75 | 71 | 34 | — | 105 | sR9 | 128 | AF468960 |
| Pf2 | P | 505759 | 62 | 69 | 30 | — | — | |||
| Pf3 | P | 942541 | 170 | 73 | 34 | — | 160 | hgcE | (132) | AF468961 |
| Pf4 | P | 1226100 | 65 | 69 | 38 | — | — | |||
| Pf5 | P, Q | 1333169 | 147 | 62 | 37 | — | — | |||
| Pf6 | P, Q | 1666314 | 157 | 68 | 35 | 145, 155 | — | hgcF | 168 | AF468962 |
| Pf7 | P, Q | 1732711 | 215 | 68 | 36 | 200, 300 | — | hgcG | 277 | AF468963 |
| Pf9 | P | 1865084 | 83 | 71 | 38 | — | — | |||
| PfQ1 | Q | 15714 | 148 | 32 | 16 | — | — | |||
| PfQ2 | Q | 210249 | 96 | 23 | 11 | — | — | |||
| PfQ3 | Q | 272045 | 73 | 36 | 18 | — | — | |||
| PfQ4 | Q | 338679 | 94 | 39 | 20 | — | — | |||
| PfQ5 | Q | 647264 | 59 | 32 | 16 | — | — | |||
| PfQ6 | Q | 659448 | 240 | 38 | 19 | — | — | |||
| PfQ7 | Q | 661470 | 111 | 27 | 14 | — | — | |||
| PfQ8 | Q | 753505 | 51 | 43 | 22 | — | — | |||
| PfQ9 | Q | 856398 | 150 | 33 | 17 | — | — | |||
| PfQ10 | Q | 1016055 | 91 | 43 | 21 | — | — | |||
| PfQ11 | Q | 1289953 | 195 | 43 | 22 | 48, 98 | — | sscA | 97 | AF468964 |
| PfQ12 | Q | 1792629 | 124 | 52 | 26 | — | — | |||
| PfQ13 | Q | 1897919 | 105 | 45 | 22 | — | — | |||
| Mj6A | H | 1622879 | 117 | 56 | 33 | 100, 125, 200 | — | hhcA | 127 | AF468965 |
| Pf8 | H | 1768894 | 127 | 66 | 38 | — | 110 | hhcB | 127 | AF468966 |
The real size of the gene products given is the maximal size as determined by 5′ and 3′ RACE as shown in Fig. 3 and thus may be bigger than the bands visible on the Northern blots. V, Viterbi screen; P, posterior decoding with cutoff set to identify all tRNAs + conservation among GC-rich regions of all three Pyrococcus species; Q, QRNA screen; H, homologue of Mj6/hgcC.
Located on large extrachromosomal element (ECEL).
As we wished to compare the performance of the GC-content ncRNA gene finder with that of QRNA, we needed to consider a set of related organisms in which to do comparative analysis. Although no nearby relative of M. jannaschii has yet been completely sequenced, there are three genome sequences available of the AT-rich hyperthermophilic genus Pyrococcus—P. furiosus, P. abyssi, and P. horikoshii (ref. 22; http://www.genoscope.fr/Pab and http://www.genome.utah.edu/sequence.html). A GC-content screen with Viterbi parsing was less successful for these genomes; the highest sensitivity observed was in P. furiosus, where 67% of tRNAs were identified. Therefore, we decided to include simple comparative analysis in the GC screen. We used a hidden Markov model posterior-decoding algorithm to relax the specificity of the Viterbi screen and identify GC-rich regions with more sensitivity, and then considered only regions from P. furiosus that showed significant blastn similarity to regions in the other two Pyrococcus genomes. The threshold was set such that all 46 known P. furiosus tRNAs were found by definition. This screen initially identified 51 conserved GC-rich regions. All of the tRNAs, ribosomal RNAs, RNase P RNA, and 7S RNA were also identified. After accounting for these known ncRNAs, eight regions remained as putative ncRNA genes (Table 1). To test specificity, we made a data set for each genome consisting solely of regions identified as protein-coding ORFs at least 200 amino acids long with glimmer (4). We assumed that protein-coding sequences should contain few stable structural RNA regions. The posterior-decoding screen identified no regions in the ORF data set, suggesting that specificity is near 100%.
We next wished to see how these results in P. furiosus compared with a QRNA screen. We used QRNA to compare alignments between P. furiosus and each of the other two Pyrococcus genomes and identify putative ncRNA loci (12). This screen identified 73 candidate ncRNA regions. Among these, 45 of 46 tRNAs were found, as were the ribosomal RNAs and 7S RNA. After accounting for these known RNAs, 32 candidate regions remained. Many of these were either partially or completely overlapped by protein-coding gene annotation; therefore, areas of overlap were eliminated from further consideration. This approach left 17 intergenic regions at least 50 nucleotides in length for further consideration. Four of the 17 correspond to regions identified by the posterior-decoding GC screen (Table 1). To estimate specificity, we performed QRNA analysis as before but shuffled the individual columns of each alignment before scoring with QRNA. This analysis resulted in 41 loci being called as putative ncRNAs, predicting a specificity of about 44% [(73–41)/73]. Because 53% of the loci already contained at least one ncRNA characterized before this study, the number of novel ncRNAs was expected to be small (or zero).
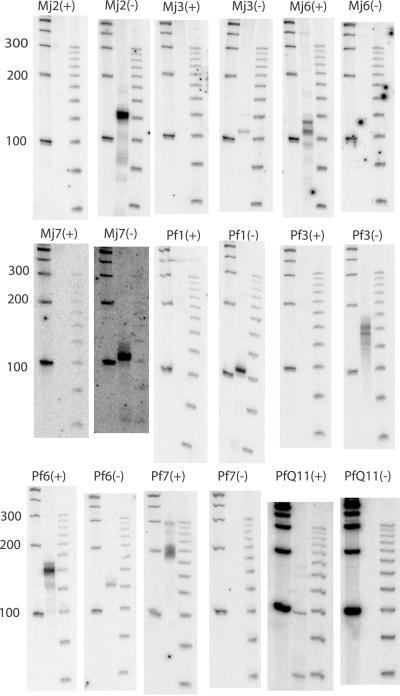
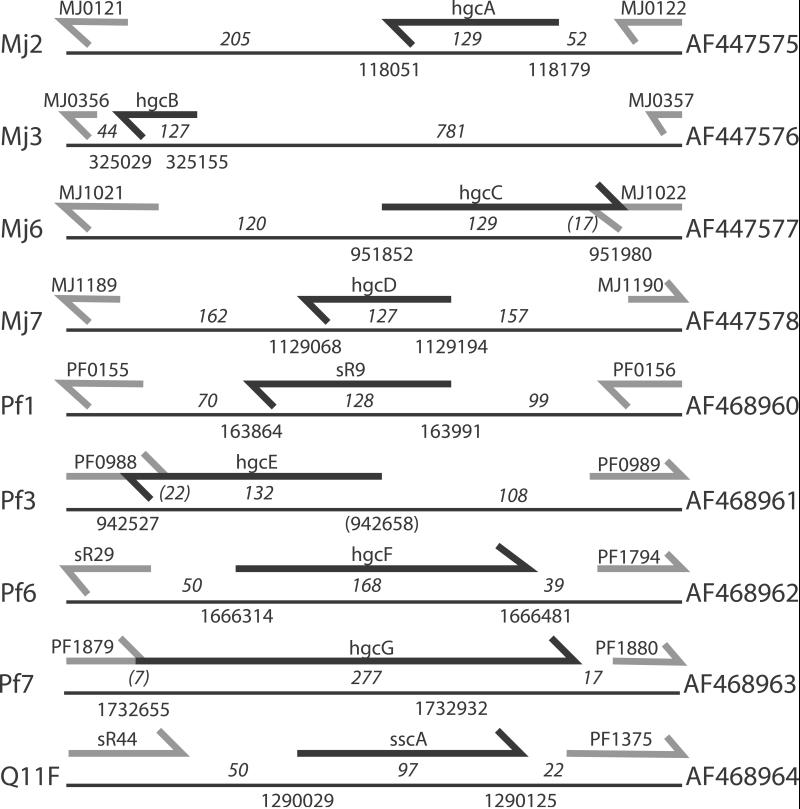
To test whether these candidate loci express a detectable RNA transcript, we used Northern blot analysis with strand-specific oligonucleotide probes and RNA from log-phase cultures grown in standard lab conditions. We detected small, stable RNA transcripts from nine candidate loci in total—four of the nine M. jannaschii candidates, four of the eight P. furiosus conserved GC-rich candidates, and four of the 17 P. furiosus QRNA candidates (Fig. 2). Three of the five expressed Pyrococcus candidates were found by both screens. Candidate PfQ11 was not found by the GC screen because its GC content is the same as background; candidate Pf3 was not found by QRNA because its identity was above 85% in all windows tested. (Although a higher percent identity cutoff would have allowed identification of Pf3, it would also have increased the expected false positive rate to unacceptable levels.) Overlapping 5′ and 3′ end fragments of these RNA transcripts were amplified by RACE, cloned, and sequenced, defining transcripts ranging in length from 55 to 277 nucleotides. At only one locus was the maximal length transcript significantly shorter than the major Northern band; we believe we could not find the true 5′ end of Pf3 because of an extremely abnormal GC composition (Table 1). None of these transcripts seem to have any significant coding potential. One corresponds to a known ncRNA, sR9 (see below). We named the other seven GC-rich loci hgcA through hgcG (“high GC”), whereas the RNA identified only through QRNA was named sscA (“secondary structure, conserved”; Fig. 3).
Figure 2.
Northern blots of novel ncRNAs. Each pair of blots represents probing with oligonucleotides for RNA on the + or − strand, respectively. On each blot, the leftmost lane is a 100-bp ladder, the center lane is the RNA sample, and the rightmost lane is a 25-bp ladder.
Figure 3.
The genomic context of each novel ncRNA gene. The left-hand column gives the candidate name from the screen. In the center is an independently scaled schematic of the genomic locus. The black arrow represents the longest copy we could find of the ncRNA gene, whereas the gray arrows represent 50 nucleotides of the flanking annotated genes. The numbers above the lines and below the arrows indicate the maximal length of the ncRNA gene as determined by 5′ and 3′ RACE. The other two numbers above the lines are the distances between the gene and its flanking genes, where a number in parentheses indicates the length of the overlap between the two genes. The numbers below the lines represent the start and stop coordinates of the gene. The right-hand column contains the GenBank accession no. for the cDNA sequence.
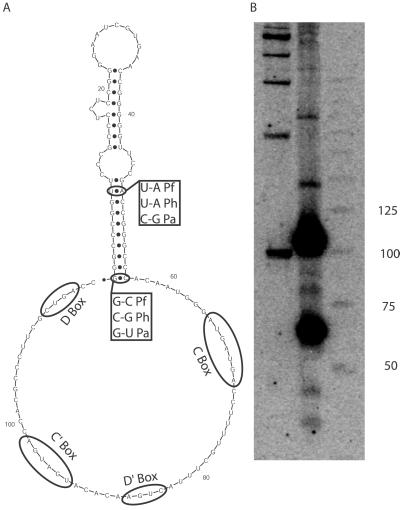
The transcript of candidate Pf1 unexpectedly overlapped a C/D box small nucleolar RNA (snoRNA) homologue, sR9 (23) (alternatively called sR19; ref. 24). The candidate region is upstream of the C/D box consensus sequences of sR9; it forms a putative stem-loop structure that is conserved among the three Pyrococcus species and shows covariation (Fig. 4A). We probed a Northern blot and performed 5′ RACE with an oligonucleotide complementary to the C/D box region of sR9 and found that sR9 is present in two abundant forms in the cell—a shorter form similar to other C/D box RNAs in Pyrococcus and a longer form that includes the stem-loop structure (Fig. 4B). The function of the stem-loop (if any) is unclear. sscA is also adjacent to a C/D box RNA, sR44, but the bands visible on Northern blots (Fig. 2 and data not shown) suggest that the abundant forms are physically separate in vivo. However, evidence from the 5′- and 3′-RACE experiments suggests that sscA is cotranscribed with either sR44 or ORF PF1375, which is the translation elongation factor eF-1 α-subunit (Fig. 3). We have no evidence to either support or contradict cotranscription of all three genes in a single operon.
Figure 4.
snoRNA sR9 with the additional stem-loop structure. (A) Predicted secondary structure folding for sR9, with covarying bases in the stem structure noted. The annotations for the C, C′, D, and D′ boxes come from ref. 23. (B) Northern blot probed with an oligonucleotide complementary to the 2′-O-methyl guide region.
There are over 50 known Archaeal C/D snoRNA homologues in P. furiosus (23, 24). Of these, only sR9, sR44, and four others were detected in our screens; all six of these are either adjacent to or in the intron of another structured ncRNA. All but one have a G+C content of 50–55%; sR40 has a G+C content of 64%. By themselves, the C/D snoRNA homologues seem to have little conserved intramolecular secondary structure. These observations suggest that both of our screens identify only a subset of highly structured ncRNAs and that they fail to reliably detect unstructured ncRNAs.
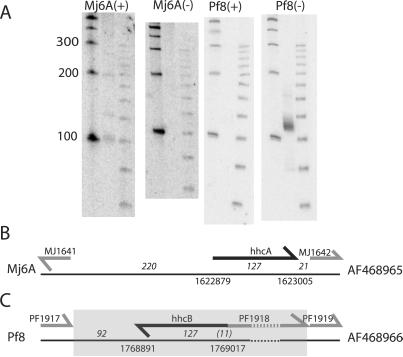
blastn searches of GenBank and the available Archaeal genomes failed to identify any significant similarity (P < 0.005) between these new genes and any known gene. hgcG is significantly similar to a region of the Archaeoglobus fulgidus genome. In a recent experimental screen for ncRNAs in A. fulgidus, this locus was identified as the second-most abundant transcript (Afu-4), further suggesting that hgcG is a real ncRNA conserved among at least two genera of Archaea (T.-H. Tang, J.-P. Bachellerie, H. Huber, M. Drungowski, T. Elge, J. Brosius, and A. Hüttenhofer, personal communication). hgcC shows significant similarity to another region of the M. jannaschii genome (candidate Mj6A) as well as to a region of the P. furiosus genome (candidate Pf8) that is identified as GC-rich in the Viterbi screen but was not pulled out in either comparative screen, because similar sequences do not exist in either P. abyssi or P. horikoshii (Table 1). To see whether Mj6A and Pf8 are also biologically relevant, we tested for expression by using oligonucleotide probes to Northern blots. The similar region in M. jannaschii shows only weak expression, whereas it appears there are high levels of expression of a 120-nt RNA from the homologous region in P. furiosus (Fig. 5A). We mapped the 5′ and 3′ ends of both genes and named them hhcA and hhcB (“homologue of hgcC”; Fig. 5 B and C). When the genomic locus for hhcB was compared with that of syntenic regions in P. abyssi and P. horikoshii, it became apparent that hhcB and the adjacent ORF PF1918 were an insertion at this locus in P. furiosus (or a deletion from the other two genomes) (Fig. 5C). The orthologues of PF1917 and PF1919 are separated by only eight nucleotides in P. abyssi and P. horikoshii. PF1918 is identified as a “probable transposase” (E = 0.00037) when it is searched against the Pfam database (25). The unexpected phylogenetic distribution of these three homologues along with the association of one with a transposase argues that this RNA species is associated with transposition events, whether by function or by chance.
Figure 5.
hgcC and its homologues. (A) Northern blot showing expression of homologues of hgcC in M. jannaschii and P. furiosus. (B) Genomic context of hhcA, as in Fig. 3. (C) Genomic context of hhcB, as in Fig. 3, except the full length of ORF PF1918 is included. The dashed line indicates a gap not drawn to scale. The shaded region is not present in the syntenic regions of P. abyssi and P. horikoshii.
Discussion
Here we have presented several screens for ncRNAs in the AT-rich hyperthermophiles M. jannaschii and P. furiosus. Each screen identified approximately five previously unidentified ncRNAs in each organism. Two independent screens in P. furiosus produced nearly identical sets of expressed ncRNA genes. Therefore, we believe these two screens have come close to saturation for a class of highly structured, conserved ncRNAs. These screens do not identify ncRNA genes without significant secondary structure, as canonical C/D box methylation guide RNA sequences were not identified unless they were adjacent to other, highly structured features (23, 24). We cannot exclude the possibility that more nonconserved ncRNA genes or ncRNA genes without significant secondary structure remain to be found.
A QRNA screen of E. coli resulted in an estimate of about 200 structural ncRNA genes in this organism (11), a number that is roughly consistent with the results of three other screens (8–10). Thirty-four different loci identified in these screens have been experimentally shown to express small stable RNAs thus far. Additionally, several other ncRNAs (other than the well known rRNAs and tRNAs) were already known in E. coli (8, 10). In contrast, we find far fewer new structural ncRNAs in screens of P. furiosus and M. jannaschii. The reasons for this discrepancy remain unclear. One possibility is that the constraints of high temperature environments select against the use of ncRNAs in hyperthermophiles. Another is that E. coli (which has both a genome size and predicted protein-coding gene count about twice that of P. furiosus or M. jannaschii) has more complex regulation and has more regulatory RNAs. It is also possible that these expressed ncRNA transcripts have no significant function, and that these numbers vary greatly from organism to organism because of nonadaptive mechanisms (although conserved RNA structure tends to argue against this). As more screens for ncRNAs are done in more prokaryotes, it should become easier to resolve which hypothesis is correct.
Another open question is that of function. In most cases we have no evidence suggesting a potential function. sR9 is clearly a 2′-O-methyl guide snoRNA, although the function of the stem-loop at the 5′ terminus is unclear. hgcC and its homologues are associated with a transposon, but their relationship to transposition is unknown. Genetic or biochemical studies will be needed to elucidate function. Because M. jannaschii and P. furiosus are not easily manipulable genetic systems presently (26), finding homologues of these genes in other organisms will be essential to apply reverse genetic approaches (e.g., knockouts).
A related question concerns evolutionary conservation and phylogenetic diversity. Many ncRNA genes, including all those previously known in M. jannaschii and P. furiosus, are known to exist across at least two of the three domains of life (i.e., ribosomal RNA, tRNA, RNase P, 7S RNA, and C/D box snoRNA homologues). Other ncRNAs are as yet only known in a phylogenetically restricted group. The novel ncRNAs detected here seem to have narrow phylogenetic distributions. With two exceptions discussed above, we did not detect any primary sequence similarity between these novel ncRNAs and other Archaeal genomic sequences, including between the ncRNAs identified in M. jannaschii and P. furiosus. However, because structural ncRNAs often evolve to conserve structure rather than sequence, it is possible that homologues cannot be detected through simple primary sequence searches. Secondary structure-based search methods may be able to identify homologues (27). To apply these methods we need a trusted secondary structure; toward this end we aim to collect homologous sequences from closely related species and solve the secondary structure of these RNAs by phylogenetic comparative analysis (28).
Acknowledgments
We thank Jim Brown for M. jannaschii cell paste and helpful discussions; Michael Terns (Univ. of Georgia) for P. furiosus RNA; Frank Robb (Center for Marie Biotechnology, Univ. of Maryland) for P. furiosus culture; Jan Amend and D'Arcy Meyer for assistance in anaerobic culturing; Genoscope and the Utah Genome Center, Department of Genetics, University of Utah for making their unpublished Pyrococcus sequences available on the web; Alexander Hüttenhofer for communicating results before publication; and members of the Eddy lab for helpful comments on this manuscript. This work was supported in part by National Institutes of Health Grant HG01363. R.J.K. is a Predoctoral Fellow of the Howard Hughes Medical Institute.
Abbreviations
- ncRNA
noncoding RNA
- snoRNA
small nucleolar RNA
- RACE
rapid amplification of cDNA ends
Note Added in Proof.
A similar screen has recently been reported in M. jannaschii by Schattner (29).
Footnotes
References
- 1.Eddy S R. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 2.Borodovsky M, McIninch J. Comput Chem. 1993;17:123–133. [Google Scholar]
- 3.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 4.Salzberg S L, Delcher A L, Kasif S, White O. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grogan D W. Mol Microbiol. 1998;28:1043–1049. doi: 10.1046/j.1365-2958.1998.00853.x. [DOI] [PubMed] [Google Scholar]
- 6.Galtier N, Lobry J R. J Mol Evol. 1997;44:632–636. doi: 10.1007/pl00006186. [DOI] [PubMed] [Google Scholar]
- 7.Daniel R M, Cowan D A. Cell Mol Life Sci. 2000;57:250–264. doi: 10.1007/PL00000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner E G, Margalit H, Altuvia S. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 9.Carter R J, Dubchak I, Holbrook S R. Nucleic Acids Res. 2001;29:3928–3938. doi: 10.1093/nar/29.19.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wassarman K M, Repoila F, Rosenow C, Storz G, Gottesman S. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivas E, Klein R J, Jones T A, Eddy S R. Curr Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 12.Rivas E, Eddy S R. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowe T M, Eddy S R. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durbin R, Eddy S, Krogh A, Mitchison G. Biological Sequence Analysis. Cambridge, U.K.: Cambridge Univ. Press; 1998. [Google Scholar]
- 15.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris J K, Haas E S, Williams D, Frank D N, Brown J W. RNA. 2001;7:220–232. doi: 10.1017/s1355838201001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams M W, Holden J F, Menon A L, Schut G J, Grunden A M, Hou C, Hutchins A M, Jenney F E, Jr, Kim C, Ma K, et al. J Bacteriol. 2001;183:716–724. doi: 10.1128/JB.183.2.716-724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voorhorst W G, Eggen R I, Luesink E J, de Vos W M. J Bacteriol. 1995;177:7105–7111. doi: 10.1128/jb.177.24.7105-7111.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuker M, Mathews D H, Turner D H. In: RNA Biochemistry and Biotechnology. Barciszewski J, Clark B F C, editors. Dordrecht, The Netherlands: Kluwer; 1999. pp. 11–43. [Google Scholar]
- 20.Mathews D H, Sabina J, Zuker M, Turner D H. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 21.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 22.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 23.Omer A D, Lowe T M, Russell A G, Ebhardt H, Eddy S R, Dennis P P. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 24.Gaspin C, Cavaille J, Erauso G, Bachellerie J P. J Mol Biol. 2000;297:895–906. doi: 10.1006/jmbi.2000.3593. [DOI] [PubMed] [Google Scholar]
- 25.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonhammer E L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sowers K R, Schreier H J. Trends Microbiol. 1999;7:212–219. doi: 10.1016/s0966-842x(99)01492-4. [DOI] [PubMed] [Google Scholar]
- 27.Eddy S R, Durbin R. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James B D, Olsen G J, Pace N R. Methods Enzymol. 1989;18:227–239. doi: 10.1016/0076-6879(89)80104-1. [DOI] [PubMed] [Google Scholar]
- 29.Schattner P. Nucleic Acids Res. 2002;30:2076–2082. doi: 10.1093/nar/30.9.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]