Abstract
Gene expression ratios derived from spotted-glass microarray experiments have become invaluable to researchers by providing sensitive and comprehensive indicators of the molecular underpinnings of cell behaviors and states. However, several drawbacks to this form of data have been noted, including the inability to relate ratios to absolute expression levels or to compare experimental conditions not measured with the same control. In this study we demonstrate a method for overcoming these obstacles. First, instead of cohybridizing labeled experimental and control samples, we hybridize each sample against labeled oligos complementary to every microarray feature. Ratios between sample intensities and intensities of the oligo reference measure sample RNA levels on a scale that relates to their absolute abundance, instead of to the variable and unknown abundances of a cDNA reference. We demonstrate that results from this type of hybridization are accurate and retain absolute abundance information far better than conventional microarray ratios. Next, to ensure the accurate measurement of sample and oligo reference intensities, which may differ by several orders of magnitude, we use a linear regression algorithm, implemented in a freely available perl script, to combine the linear ranges of multiple scans taken at different scanner sensitivity settings onto an extended linear scale. We discuss future applications of our method to measure RNA expression on the absolute scale of number of transcripts per cell from any organism for which oligo-based spotted-glass microarrays are available.
One of the most successful genomics technologies developed to date is the DNA microarray (1–4), which permits simultaneous detection of expression levels for every gene in an organism. Currently, spotted-glass microarray experiments are performed by using probes synthesized from two RNA samples (a reference and an experimental sample) that are labeled with different color fluors and cohybridized to an array. Differences in gene expression levels are reported as the ratio between the experimental and reference intensities (conventional microarray ratios). However, this technology, which has produced large databases of whole-genome expression data on subjects ranging from bacterial metabolism to human cancer, is not yet fully mature.
The use of ratios in two-color hybridization experiments controls for several sources of experimental variation inherent in the spotted-glass microarray technology, including variation across ORFs in labeled nucleotide frequency, amount and quality of DNA spotted onto the array, uneven hybridization, and spot size and morphology (2, 5). However, conventional microarray ratios have several properties that limit subsequent computational analysis, and thus the amount of information that can be extracted from these large data sets. First, reporting expression data as ratios between cDNA reference and experimental samples results in loss of the absolute transcript abundance information carried by the individual spot fluorescence intensities, thus obscuring important differences in levels of gene expression between ORFs. The use of cDNA reference samples, which are largely uncharacterized and not easily reproduced, also hampers comparisons between data sets that use different cDNA references. One example is the inability to cluster similar conditions between time-course experiments that use initial time points as reference samples (supplemental materials to ref. 6).
In this study, we develop a combination of experimental and computational methods that improves the accuracy and comparability of DNA microarray data by (i) measuring spot intensity relative to a calibrated reference sample and (ii) extrapolating data from multiple scans to increase the range of intensities reported. In this system, labeled cDNA from a sample of interest is hybridized in conjunction with a set of differentially labeled oligonucleotides of known abundance (calibrated oligo reference) containing sequences complementary to every spot on the array. After hybridization, spot intensities are measured by scanning at several detection sensitivity settings, and the results are combined onto a common linear scale by using a linear regression algorithm implemented in a perl script called masliner (MicroArray Spot LINEar Regression). Finally, RNA abundance is expressed as a ratio to the calibrated oligo reference, a sample that is easily reproduced and provides significant signal for every feature on the array.
Materials and Methods
Strains and Media.
The Saccharomyces cerevisiae strain used in this study is FY4, a MATa prototroph isogenic to a GAL2+ derivative of S288C (7). Media were prepared as described (8). Glucose cultures (Glu) were grown in yeast extract/peptone/dextrose (YPD; 2% glucose), raffinose cultures (Raff) were grown in SDRaf (2% raffinose), and ethanol cultures (EtOH) were grown in SDEtOH (2% ethanol). Galactose cultures (Gal) were grown in YPRaf (2% raffinose) and induced for 20 min by the addition of galactose to 2%. All cultures were grown at 30°C and harvested at densities of 1–2 × 107 cells per ml.
Microarray Labeling and Hybridization Reactions.
Total yeast RNA was prepared as described (9). Labeled cDNA probes were synthesized from 20 μg total yeast RNA by using the Atlas Glass Fluorescent Labeling kit (CLONTECH). The Cy3–RevORF oligo (GATCCCCGGGAATTGCCATG), used as the oligo reference sample for microarrays printed with the yeast ORF PCR product set, was synthesized with a 5′ Cy3 modification. Oligo reference hybridizations contained 200 pmols of this oligo. Other oligos used for hybridization were synthesized with 5′ C6-amino modifications and labeled as follows. A total of 500–1,000 pmols of an oligo were resuspended in 10 μl of 2X Atlas fluorescent labeling buffer (CLONTECH), mixed with 10 μl of 5 mM Cy3 or Cy5 monofunctional reactive dye (Amersham Pharmacia Biotech), and incubated in the dark at 25°C for 30 min. Labeled oligos were purified by ethanol precipitation and microcon YM-10 or YM-30 filtration (Amicon).
Microarray production and hybridization are described on our web site. Arrays were scanned on a ScanArray 5000 (GSI Lumonics, Wilmington, MA) at 90% laser power and photomultiplier tube gain (PMT) settings ranging from 45–85%. Constant PMT voltage and varied laser power gave similar results. Scans taken in increments of 5–10% PMT provided sufficient intensity ranges for masliner analysis. We did not observe significant photobleaching with repeated scans. The range of PMT settings for each slide was visually determined by finding the lowest setting required to bring the brightest spots into the linear range and the highest setting with reasonable background fluorescence.
Microarray Data Analysis.
Signal intensities were measured with the GENEPIX 3.0 image analysis software (Axon Instruments, Foster City, CA), and results files were used as input for masliner. For each fluor on each slide, scans were iteratively processed with masliner (from lowest to highest intensity scan) by using “straight” mode with a range of 2,000 (−ll) to 60,000 (−lh) and the default parameters. The linear range for the scanner used in these experiments was determined from scatter plots of spot background-subtracted intensity (BSI) values from scans at multiple sensitivities.
Within a set of experiments, the BSIs of all cDNA and oligo hybridizations were normalized to each other by the ratios of the total fluorescence values. Because there is no expectation that total fluorescence of a cDNA sample and a calibrated reference oligo sample should be equivalent, cDNA and oligo hybridizations were normalized separately. BSIs less than the value of 1 SD of the local background were assigned the SD value.
Spiked Oligo Experiment.
Oligos complementary to Operon oligo array features with low expression levels in glucose cDNA hybridizations were labeled and spiked into 80-μl hybridization reactions containing Cy3- and Cy5-labeled glucose cDNA. Cy3-labeled oligos were added as an equimolar mixture of 0.5 pmols of each oligo. Cy5-labeled oligos with were added as a 5-fold dilution series from 100 pmols (FBP1) to 0.256 fmols (HXT5). Unadjusted series data are taken from two pairs of high sensitivity scans: Cy5 at 75% PMT with Cy3 at 85% PMT, and Cy5 at 65% PMT with Cy3 at 75% PMT. If Mi represents the measured abundance of spiked oligo i in a measurement series (unadjusted or masliner-adjusted), and Ai the actual spiked abundance, we report the accuracy of the measurement series as the average over i of abs(log10(Mi) − log10(Ai)) (accuracy score). The antilog of the accuracy score is the geometric mean of the fold changes of the greater to the smaller measurement across the series of oligos (geometric average fold change). The consistency score, which measures the degree to which measured abundances of successive dilution series members are close to the target 5-fold ratio, is calculated as the average over i of abs(5 − (Mi/Mi + 1)). The accuracy and consistency scores of a perfect measurement series are 0. For data from single scans, saturated spots are those whose BSI is greater than 60,000; for masliner-adjusted data, the saturated spots are those flagged as saturated by masliner. Background spots are those whose BSI is less than the value of 2 SDs of all spot background intensities on the array. The background intensity of the masliner-adjusted data were derived from the highest intensity scans in the series.
Software and Supplementary Web Site.
Masliner accepts GENEPIX 3.0 files from consecutive scans of the same slide. A series of scans are combined by processing each GENEPIX 3.0 file for successively higher scan intensities through masliner along with the output file generated from the prior scans. Masliner processing is briefly described below, and the masliner software, additional information on processing, supplementary data, and additional methods are available on our web site (http://arep.med.harvard.edu/masliner/supplement.htm).
Results
Common Linear Scaling of Multiple Scans Improves Measurement Accuracy.
The relationship between fluorophore quantity in a microarray spot and spot intensity reported by a scanner is linear only within a certain range of intensities, being dominated by noise below and subject to saturation above that range. Thus, a single microarray scan usually cannot provide accurate information on the full range of expression levels of genes in a typical sample. Incorporating data from scans acquired at multiple scanner sensitivity settings has the potential to increase the range of signal intensities detectable, being limited ultimately by the noise resulting from background fluorescence on the slide and the number of binding sites for the labeled probe on the array. Although early microarray studies increased the dynamic range of detectable gene expression by using multiple scans at different scanner sensitivity settings (2), this recommendation is absent from current protocols (10, 11).
To facilitate an accurate comparison between a limited range of oligo intensities with the full range of gene expression values, we developed an algorithm that corrects for saturation by combining data from the linear ranges of scans collected at multiple laser power or PMT settings into a common extended linear range. An example is given in Fig. 1. Given data from a low- and high-sensitivity scan, the linear range measurements for each scan are combined by using a perl script called masliner. By using information on a scanner's linear range, masliner computes a linear regression (12) of the BSIs for the high versus low intensity scans for spots in the linear range of both and uses it to extrapolate adjusted BSIs for each spot with BSIs above the linear range in the higher sensitivity scan. Results are printed to an output file containing adjusted values within and unadjusted values below the bounds of the common linear scale. The output file also estimates the error of prediction associated with the linear regression(s) and indicates any spots too saturated to be calibrated accurately. Masliner may be run iteratively to derive a common linear scale for an entire series of scans taken at different sensitivities to substantially extend the signal intensity range acquired from an array. For example, a series of masliner adjustments using data from four scans increased the intensity of the brightest spot on the array in Fig. 1 from 65,535 (saturation) to 3.2 million units, a 49-fold range increase with a 0.4% estimated error associated with the linear regression (http://arep.med.harvard.edu/masliner/supplement.htm).
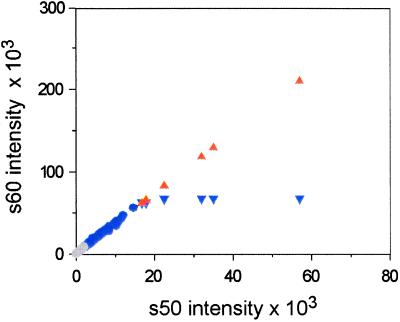
Figure 1.
Common linear scaling of microarray spot intensities. A single array was scanned twice on a ScanArray 5000 at constant laser power (90%) and two PMT settings (50% and 60%). For each spot, the BSI from the 50% scan (s50) is plotted against BSI from the 60% scan (s60) (circles and inverted blue triangles). Six spots are saturated in s60 that were not saturated in s50 (inverted blue triangles). Using masliner, a linear regression was computed for the 176 s50 vs. s60 values within the linear range of the scanner (blue circles, 2,000 ≤ BSI ≤ 60,000). The regression was used to extrapolate values for the six s60 saturated spots (orange triangles), effectively generating an extended common linear scale for spots over both scans. Some spots (n = 6,185) were below the linear range in both scans (gray circles). A total of 3,288 of these spots were brought into the common linear scale by additional scans at higher gains and executions of masliner. These data were taken from the cDNA intensities of the Glu/oligo experiment presented in Table 2.
To test whether our method of integrating data from multiple scans was able to accurately maintain relative abundance values over multiple scans, we conducted a spiked-oligo experiment (Fig. 2). In this experiment, oligos complementary to the features on microarrays spotted with the Yeast Genome Oligo Set (Qiagen Operon) were end-labeled with Cy3 or Cy5 and added to a hybridization reaction as an equimolar mixture of Cy3-labeled oligos and a five-fold dilution series of Cy5-labeled oligos. To perform calculations in the context of actual microarray expression data, oligos corresponding to low intensity spots in a glucose cDNA sample (http://arep.med.harvard.edu/masliner/supplement.htm) were added to a hybridization reaction containing Cy3- and Cy5-labeled cDNA probes synthesized from total yeast RNA of glucose-grown cells (Glu). The resulting arrays were scanned several times at a constant laser power with varied PMT settings, signal and background intensities from each scan were measured, saturated BSIs were adjusted by using masliner, and BSIs were normalized to total fluorescence. Results from this experiment contain two types of data for which the actual ratio is known, ratios of the spiked oligos, present in a 5-fold dilution series, and ratios of the self-versus-self hybridization of the Glu cDNA, present at a ratio of 1.
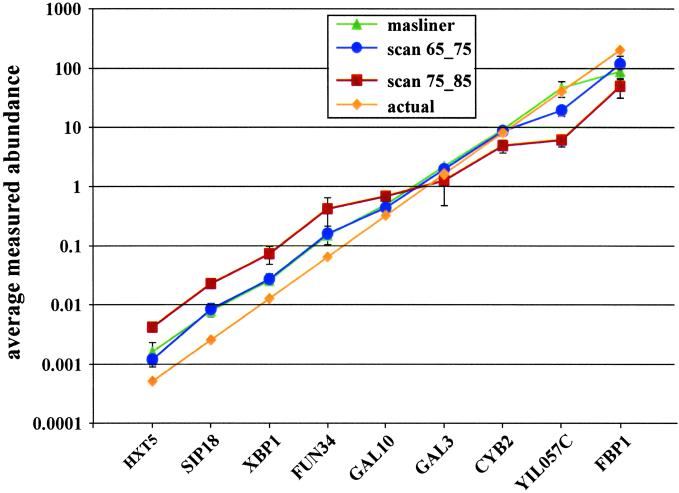
Figure 2.
Adjustment by masliner accurately maintains a 5-fold dilution series of Cy5 to Cy3 ratios over four orders of magnitude. A log plot of the average ratios of two slides in the spiked oligo experiment is shown ± SDs. Data are presented for masliner adjustment (triangles) and from single scans taken at Cy5 65% PMT and Cy3 75% PMT (circles) and Cy5 75% and Cy3 85% (squares). Actual spiked abundance values are also shown (diamonds). Of the four single intensity scan pairs examined, scans 65_75 and 75_85 had the largest dynamic ranges and the most accurate values.
The ratios of the spiked oligos in this experiment (Fig. 2) demonstrate that extrapolation of intensities from multiple scans using masliner was able to accurately maintain abundance measurements over a large range of ratios. Data processed through masliner produced a dynamic range of ratios greater than 4 orders of magnitude with an accuracy score (Materials and Methods) of 0.272. The limited range at the high end of the spectrum was the result of a low FBP1 ratio, most likely caused by a lack of available binding sites on the array for the high concentration of this oligo (http://arep.med.harvard.edu/masliner/supplement.htm). Thus, BSIs adjusted above saturation by the masliner script maintain accurate relative abundance values over several orders of magnitude.
Fig. 2 also shows the more variable results that can be obtained from conventional ratios based on individual scans in the series. One set of scans, scan 65_75, has effectively the same accuracy score as the masliner series (0.268), representing a geometric average fold change less than 1% different from that of masliner. The other set, scan 75_85, has an accuracy score considerably worse than either the masliner or the PMT 65% series (0.613, geometric average fold change ≈ 4.1). The accuracy score describes the overall accuracy of measured abundances for the dilution series but does not describe their consistency, i.e., how closely the measured abundances of successive series members are to the target 5-fold ratio. This property is measured by the “consistency score” (Materials and Methods). Masliner exhibits better consistency than either scan 65_75 or 75_85 (Table 1). Thus, over the range of spiked oligo ratios examined, the accuracy of the masliner-adjusted values was equal to or better than the data derived from any set of single scans.
Table 1.
Quality measure comparison between masliner and single-scan data from the oligo spiking experiment (averages of two replicates)
| Accuracy score | Consistency score | Background spots | Saturated spots | |
|---|---|---|---|---|
| masliner | 0.272 | 1.1 | 652.5 (10%) | 0 (0%) |
| scan 65_75 | 0.268 | 1.5 | 703.5 (11%) | 96.0 (2%) |
| scan 75_85 | 0.613 | 2.2 | 652.5 (10%) | 245.5 (4%) |
To further characterize the quality of masliner-adjusted values with those acquired from a single scan, we analyzed the values obtained for the remaining 6,298 spots on the spiked oligo arrays that correspond to the Glu cDNA self-versus-self hybridization. One means of evaluating the quality of scanning parameters in this type of experiment is to compare the number of spots with values that are either below the background noise of the hybridization (values below 2 SDs of all spot background intensities) or above the saturation threshold for detection by the scanner (60,000 for the instrument used in this study). A comparison of results from the three different data sets (Table 1) clearly demonstrates the advantage of masliner over any single scan. For example, masliner adjustment brings an additional 1% of the spots on the array above the background cutoff than scan 65_75 while at the same time retaining another 2% lost because of saturation. Although masliner and scan 75_85 have the same number of background spots because masliner accepts the values of spots below the linear range from the highest sensitivity scans in a series (the scan 75_85 pair), scan 75_85 loses 4% of spots to saturation. Additional comparisons are available on our web site. Thus, extrapolation of data from multiple scans with the masliner algorithm increases the number of spots with usable data by 3% relative to the best scoring single scan in the oligo spiking experiment.
A Common Oligo Reference Sample Improves Data Comparability and Transcript Abundance Estimation.
Many of the artifacts associated with conventional microarray ratios could be eliminated by measuring RNA levels relative to a calibrated reference sample, resulting in retention of transcript abundance information and direct comparability of all samples measured with the reference. To serve as a reference for RNA abundance calibration, a sample must produce detectable signal at each array feature, contain all species in the mixture at known concentrations, and be easily and accurately reproduced. Several nucleic acid mixtures satisfy these criteria to varying extents, including genomic DNA for organisms with low-complexity genomes (13), equimolar mixtures of PCR products or oligos complementary to all array features, and oligos complementary to common sequences present in all array features. Whole-genome microarrays spotted with ORFs amplified by using the GenePairs yeast primers (Research Genetics, Huntsville, AL) contain a universal sequence tag common to every spot on the array, originally engineered to facilitate PCR amplification. Use of a labeled oligo complementary to this common sequence as a reference sample would control for differences in target DNA quantity, spot morphology, and uneven hybridization, but not labeling or sequence-specific hybridization differences between transcripts. Nonetheless, we chose this sequence as a simple, inexpensive way to test our hypotheses concerning microarray data measured relative to a calibrated reference.
To evaluate the accuracy of the common oligo reference measurements relative to conventional microarray ratios, we compared data from hybridizations by using cDNA and oligo reference samples for yeast grown in four well-characterized media conditions, rich glucose (Glu), rich galactose (Gal), minimal raffinose (Raff), and minimal ethanol (EtOH). Gene expression levels in the galactose, raffinose, and ethanol samples were measured conventionally relative to the glucose sample directly or by reconstructing the same ratios by using the oligo reference values. A sample comparison is shown for two examples in four classes of genes: highly induced in Gal, moderately induced in Gal, moderately induced in Glu, and equally expressed in both Glu and Gal (Fig. 3). The reconstructed ratios were extremely similar to the values measured directly by conventional hybridization. Comparison between the conventional microarray ratios and the reconstructed ratios for all spots on the array by two-tailed t tests and by a set of eight Wilcoxon rank sum tests, produced results consistent with the hypothesis that the two sets of ratios were statistically equivalent (http://arep.med.harvard.edu/masliner/supplement.htm). For example, 5.5% of all spots had significantly different values between the conventional microarray and reconstructed ratios for a t test at P < 0.05, as would be expected for identical distributions. These results not only demonstrate that the results from the oligo reference experiments are accurate, but also that users who wish to visualize data as expression ratios may generate these ratios from any two sets of oligo reference data.
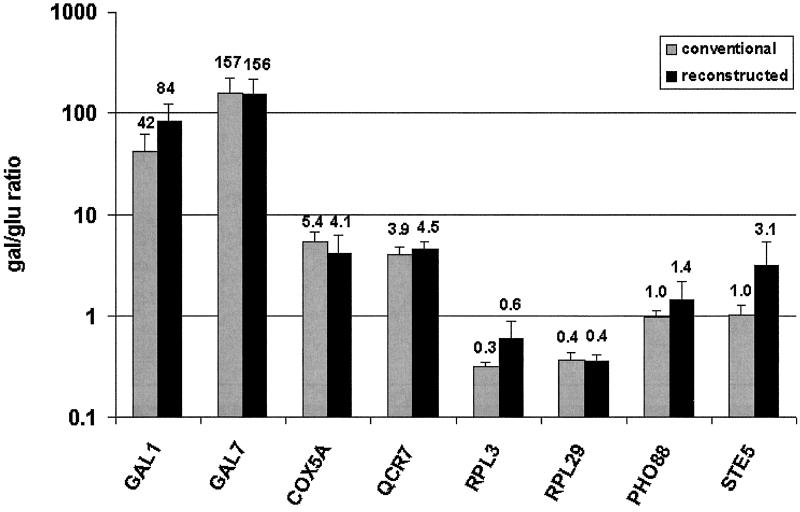
Figure 3.
Accurate reconstruction of cDNA ratios from oligo reference values. Directly measured Gal/Glu ratios from conventional hybridizations (gray) and ratios reconstructed from oligo reference hybridizations (black) are shown for two examples in four gene classes: highly induced in Gal (GAL1, GAL7), moderately induced in Gal (COX5A, QCR7), moderately induced in Glu (RPL3, RPL29), and equally expressed in both (PHO88, STE5). Conventional Gal/Glu ratios are expressed as the average of the ratios from four hybridizations ± SE. Reconstructed ratios were calculated as the average of 24 nonredundant combinations of the 4 Gal/oligo and Glu/oligo values ± SE. Average values are also printed.
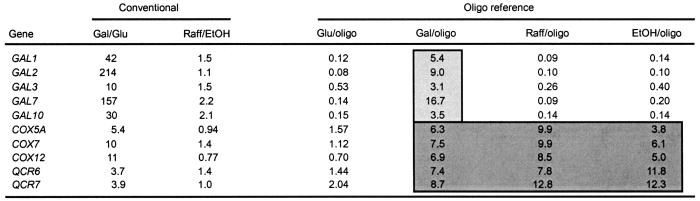
To test whether this common oligo reference could facilitate better comparisons of gene expression across multiple conditions than conventional cDNA references, we compared results from conventional cDNA cohybridizations of galactose to glucose and raffinose to ethanol samples with common oligo reference hybridizations with each of the four cDNA samples. Table 2 shows results for two distinct classes of genes, genes involved in galactose metabolism that are highly expressed in galactose, but none of the other carbon sources, and genes involved in mitochondrial function that are moderately induced in the three semi- or nonfermentable carbon sources relative to glucose (10, 14). In this example, use of conventional cDNA reference ratios obscures expression pattern differences between the two classes. With conventional ratios, one sees only that both classes appear to be induced (ratios > 3) in the Gal/Glu hybridization and uninduced (ratios ≈ 1) in the Raff/EtOH hybridization. In contrast, the oligo reference data shows a clear distinction between the GAL genes, which are induced only in galactose, and the mitochondrial genes, which are also induced in raffinose and ethanol. This type of artifact also hampers subsequent computational analysis, e.g., clustering is able to separate the two gene classes given the oligo reference, but not the conventional ratio data in Table 2 (http://arep.med.harvard.edu/masliner/supplement.htm). Although correct grouping of these two gene expression classes can be achieved with conventional ratios by using certain combinations of experimental and reference cDNAs, such data requires either prior knowledge of the genes and conditions to determine the correct hybridization pairs or multiple hybridization pair combinations. Thus, although conventional cDNA reference hybridizations work well for direct, pair-wise comparisons, measuring RNA expression levels relative to a common oligo reference sample facilitates comparison of gene expression patterns across multiple conditions, allowing accurate grouping of genes or conditions without prior knowledge about the sample.
Table 2.
Conventional versus oligo reference hybridization results for subsets of genes involved in galactose metabolism and mitochondrial function
Oligo reference values with a statistically significant induction over the glucose values are shaded (one-sided Wilcoxon rank sum test, P ≤ 0.006), for the galactose metabolism (light gray) and mitochondrial function (dark gray) gene data.
Our initial examination of the oligo reference data for several candidate genes suggested a good correlation between the calibrated oligo abundance measurements and absolute levels of expression. For example, the calibrated abundance measurements for GAL1, GAL7, and GAL10, which are strongly transcriptionally repressed in the absence of galactose, all have extremely low calibrated abundance values (≈0.1) in the three non-galactose conditions (Table 2). To estimate how related the calibrated abundance values were to absolute mRNA transcript abundances, we performed two types of analyses. First, we assessed whether differences in calibrated abundance values between two groups of candidate genes, the GAL and mitochondrial genes presented in Table 2, were statistically significant. By one-sided Wilcoxon test, the expression levels of the GAL genes are significantly less than expression of the mitochondrial genes in the Glu (P = 0.004), Raff (P = 0.006), and EtOH (P = 0.006) conditions. Next, we compared our results to published sage-derived transcript counts per cell for yeast grown in comparable conditions (15, 16). The results (http://arep.med.harvard.edu/masliner/supplement.htm) demonstrate a modest, but statistically significant Pearson correlation coefficient between these two series of values (0.62, P < 0.004), although differences in strain and growth conditions between the two experiments may be masking a better correlation. Together, these data suggest that the common oligo reference data retain absolute abundance information far better than expression levels based on conventional microarray ratios and allow at least a rough approximation of absolute transcript abundance.
Discussion
One of the next challenges in functional genomics is the development of experimental techniques capable of measuring key components of cellular function, such as RNA expression, protein abundance, and metabolite concentration, in terms that reflect copy number per cell by using methods that are both cost effective and high throughput. This type of data permits comparison between different experiments, laboratories, experimental systems, and data types, a crucial aspect of the database-dependent analysis of biological systems that the field of functional genomics requires. In addition, this type of information will facilitate the exploration of areas not currently possible given the limitations of data derived with current techniques, including the calculation of in vivo rate and binding constants, characterization of threshold- and gradient-based regulatory switches, analysis of codon adaptation indices, analysis of translational efficiency, measurement of promoter and transcription factor strength, and cross-species comparisons of RNA, protein, or metabolite abundance.
In this study, we have made three important contributions to the measurement of absolute RNA expression levels by using spotted-glass microarrays. First, we have demonstrated that mRNA expression values measured relative to a common oligo reference sample produce transcript abundance values that can be compared across multiple conditions and experiments. This type of data also facilitates more accurate analysis using computational methods, such as gene or condition clustering. Next, we facilitate accurate comparisons between a limited range of oligo intensities and the full range of gene expression values by combining intensity measurements onto a large, common linear scale using a linear regression algorithm. This method may be used with any microarray system that has the ability to acquire scans at multiple illumination intensities and/or detector sensitivities, which in theory includes Affymetrix chips. Finally, we have laid the foundation for a system that will accurately report the absolute abundance of RNA transcripts in a way that will be convertible to units of transcripts per cell. Although our common oligo reference does not control for some important aspects of the spotted-glass microarray system, including sequence-specific hybridization and labeling differences between transcripts, it is a simple and affordable method of retaining a level of information lost in conventional ratios. Moreover, the results of our oligo-spiking experiment (Fig. 2) demonstrate that referencing cDNA intensities to an equimolar mixture of oligos, which do control for these aspects, permits the accurate measurement of abundances. Thus, the use of a calibrated reference containing an equimolar mixture of oligos (equimolar oligo reference) complementary to every feature on an oligo array, such as the whole-genome oligo arrays used in this study, would facilitate RNA abundance measurements on an absolute scale that could be easily converted to units of transcripts per cell. A direct comparison between this type of calibrated microarray abundance data and data from a transcript counting method, such as sage or mpss (17), should provide a more accurate assessment of the ability of our system to measure absolute transcript abundance with the cost-effective, high-throughput technology of spotted-glass microarrays.
Although our data suggest that a single scan with optimal scanner settings can accurately measure ratios over much of the same range as the masliner adjusted values, other considerations suggest advantages of the masliner system. First, the scanner sensitivity settings required to produce accurate results from a single scan were not known at the time of scanning, but only identified in subsequent analysis of multiple scans. Therefore, a strategy of relying on single scans depends critically on identifying optimal sensitivities over a wide range of experiments, equipment, and array designs. In contrast, our results show that masliner was able to leverage the content of the optimal scans from a series of scans in an entirely automated and unsupervised manner. Second, identification of individual scans that were optimal in the spiked oligo experiment required knowledge of the spiked oligo abundances, whereas masliner was able to generate accurate results without use of this prior knowledge. A strategy identifying single optimal scans may therefore require development of appropriate spiked controls. Finally, masliner-like regression analysis of multiple scans allows detection of variability in individual scans and may allow for correction of error introduced by the scanning process (Jean O'Malley, personal communication; and http://arep.med.harvard.edu/masliner/supplement.htm).
An important consideration for any new technology is cost. Our combination of experimental and computational techniques allow us to easily compare intensities generated from 0.5 pmol of an end-labeled oligo with the entire range of cDNA intensities generated by a typical microarray experiment. As a result, the retail cost of an equimolar mixture of 6,300 oligos corresponding to the reverse complement of the Operon oligo set for yeast should be less than 9 dollars per slide. Although the large-scale synthesis and normalization of such a large set of oligos is beyond the capacity of an academic laboratory, we hope that commercial sources will make such products available to the academic and pharmaceutical communities. Another cost factor is the ≈2-fold increase in variance incurred in the calibrated oligo reference system (http://arep.med.harvard.edu/masliner/supplement.htm), which can be addressed by replicates, and which we consider a tradeoff for the increased information content and comparability of data obtained by the system.
Recently, much attention has been given to improving the computational analysis of RNA expression data, and several algorithms have been developed in an attempt to improve data normalization, error estimation, data comparability, and result confidence estimates. Previous work from our laboratory examined these issues for conventional microarray data and concluded that many of the bioinformatic problems related to ratio data were the result of properties inherent to the cDNA-reference experimental system itself (6). The experimental modifications to the current microarray protocol and bioinformatic tools described in this study resolve many of these issues and offer other notable advantages. For instance, by producing significant reference signal for each array feature, use of a calibrated oligo reference sample provides both a quality control measure for every microarray feature and “landing lights” for improved alignment by image analysis software. Data obtained by using our system also has several bioinformatic advantages, including retention of RNA abundance information, more accurate measurement of highly expressed genes that often produce saturated intensity measurements, estimation of error based on the feature intensity, and increased ability to compare data between different experiments and laboratories. Although this study demonstrated the utility of our system in a well-characterized organism with a relatively simple genome, the application to higher eukaryotes with more complex genomes, such as humans and mice, will be equally applicable if not more important.
Acknowledgments
We are extremely grateful Dan Stetson for assistance with microarray printing. We are also grateful to Rob Mitra, Brad Cairns, Patrik d'Haeseleer, Saeed Tavazoie, Jean O'Malley, and Petri Toronen for helpful discussions, and to Rob Mitra and two anonymous reviewers for critical comments on the manuscript. A.M.D. was supported by an appointment to the Alexander Hollaender Distinguished Postdoctoral Fellowship Program (United States Department of Energy). This work was generously supported by United States Department of Energy Grant DE-FG02-87ER60565, the Lipper Foundation, and National Institutes of Health Grants U01HL66678-02 and 1U01HL66582-01.
Abbreviations
- PMT
photomultiplier tube gain
- BSI
background-subtracted intensity
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gress T M, Hoheisel J D, Lennon G G, Zehetner G, Lehrach H. Mamm Genome. 1992;3:609–619. doi: 10.1007/BF00352477. [DOI] [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 3.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 4.Hughes T R, Mao M, Jones A R, Burchard J, Marton M J, Shannon K W, Lefkowitz S M, Ziman M, Schelter J M, Meyer M R, et al. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 5.Shalon D, Smith S J, Brown P O. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 6.Aach J, Rindone W, Church G M. Genome Res. 2000;10:431–45. doi: 10.1101/gr.10.4.431. [DOI] [PubMed] [Google Scholar]
- 7.Winston F, Dollard C, Ricupero-Hovasse S L. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 8.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, N.Y.: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 9.Swanson M S, Malone E A, Winston F. Mol Cell Biol. 1991;11:4286. doi: 10.1128/mcb.11.8.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 11.Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes J E, Snesrud E, Lee N, Quackenbush J. BioTechniques. 2000;29:548–562. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- 12.Sokal R R, Rohlf F J. Biometry: The Principles and Practices of Statistics in Biological Research. New York: Freeman; 1995. [Google Scholar]
- 13.Wei Y, Lee J M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston M, Carlson M. The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 193–281. [Google Scholar]
- 15.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 16.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 17.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd D H, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]