Abstract
Bacteriophage T4 tail fibers have a quaternary structure of bent rigid rods, 3 × 160 nm in size. The four proteins which make up these organelles are able to self-assemble in an essentially irreversible manner. To use the self-assembly domains of these proteins as elements in construction of mesoscale structures, we must be able to rearrange these domains without affecting the self-assembly properties and add internal binding sites for other functional elements. Here we present results on several alterations of the P37 component of the T4 tail fiber that change its length and add novel protein sequences into the protein. One of these sequences is an antibody binding site that is used to inactivate phage carrying the modified gene.
To realize the potential for nanotechnology, methods for practical directed assembly of mesoscale structures must be developed. We use a biological paradigm to develop the science and engineering needed to implement a practical bottom-up manufacturing system. Living cells normally assemble mesoscale structures (e.g., muscle fibers, mitotic spindles, flagella, virus particles) following well-studied mechanisms, including vectorial assembly and specific interaction moieties. Our approach is to create a set of nanoscale subunits of precise size, shape, and functionality that can be assembled in a massively parallel manner. Our subunits are based on the tail fiber proteins of bacteriophage T4. These proteins make up a self-assembling, precisely defined, highly stable structure (1, 2) and, as we show below, are readily amenable to re-engineering without losing these properties.
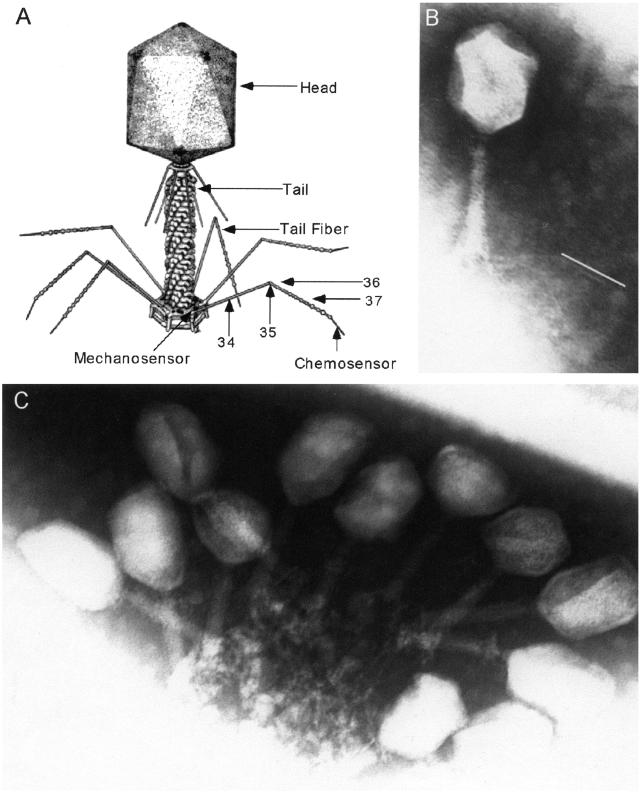
Bacteriophage (phage) T4 is one of the archetypical members of the family Myoviridae or T-even phage. These viruses are characterized by a large, elongated icosohedral head (which contains the phage DNA), a contractile tail (to stabilize the phage perpendicular to the cell and penetrate the cell wall), and tail fibers (which contain the phage receptors and trigger infection) (3, 4) (Fig. 1A). The tail fiber proteins have an unusual quaternary structure of long, thin (3 nm × 160 nm), rigid rods (5). Their function is to transduce chemical recognition of the Escherichia coli host into a mechanical force on the phage base plate, essentially acting as a set of cooperative levers. This mechanical stress triggers a series of protein conformational changes that lead to entry of the phage DNA into the cell (6, 7).
Figure 1.
Phage images. (A) Structure of bacteriophage T4. (B) Electron micrograph of T4 37SΔ1 phage. The white line next to the distal portion of the tail fiber indicates the length of a wild-type distal tail fiber at the same magnification. (C) Electron micrograph of T4 37SΔ1ras2 treated with mAb and secondary antiserum.
The three main tail fiber proteins, P34, P36 and P37¶, are thought to be principally composed of dimeric‖ antiparallel β-sheets (8). Gp35, which forms the angle in the tail fiber, probably has a more complex structure. The joints between the homodimeric segments are also likely to have a more complex structure but there is no evidence that the central rod regions have any tertiary structure (i.e., interactions between distant amino acid residues) at all (5). The extended antiparallel β-sheet secondary structure should directly support the rigid rod quaternary structure. If correct, we thought that deletions or additions to the central rod regions which maintain the β-sheet structure should permit alteration in tail fiber length without greatly affecting overall structural integrity. Furthermore the binding domains at the ends of the proteins should form separate functional domains from the central, rigid rod domain. Finally the β-sheet structure should contain turns and loops that can be expanded with functional peptides without disrupting the quaternary structure. In this paper we describe modifications of P37 that support some of these hypotheses.
Materials and Methods
E. coli and Phage Strains and Reversion Assay.
T4 37amA481 (11) was the mutant used to derive all phage strains discussed in this paper. E. coli B40 (suI) (lab strain, courtesy of P. Strigini, Harvard Medical School, Boston) was used to grow and titer phage containing an amber mutation, and E. coli BB (su0) (12) was used for all non-amber phages. T4 37amA481 pseudorevertants were identified by their ability to form plaques on BB, and stocks were prepared by standard techniques (13). Plasmids were produced, and recombined with phage using E. coli MC1061 (F− araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (Strr) hsdR2 (rK− mK+) mcrA mcrB1) (14) as the host strain.
PCR Primers and Product Cloning.
Primers cysF (CTATTAACGGACTTTTGAGA) and cysR (TTCAATACGTCCAATAGTTT) amplify the central rod region of phage T4 gene 37 including the location of the SΔ1 deletion and we used them to screen pseudorevertant phage as well as for sequencing. These primers amplify a 1.4-kb fragment from wild-type T4 DNA but only a 0.36-kb product from T4 37SΔ1 DNA. Primers recF (GACGAGCTCCTTCGGGTTCCCTTTTTCTTTA) and 37B-2R (TTGGGTAACTCGACATGA) amplify a 3.2-kb segment of the tail fiber gene cluster including the 3′ end of gene 35, gene 36, and the first two-thirds of gene 37. When these primers are used to amplify T4 37SΔ1, a 2.1-kb fragment is produced in which the deletion junction is approximately in the middle. We cloned this 2.1-kb PCR product into pGEM-T (Promega) for sequencing, further modification (see below), and to transfer modified genes into T4 phage by recombination between the plasmid and infecting phage. The construct containing this 2.1-kb insert was designated p37SΔ1.
Recombination of Phage and Plasmid.
We transferred modified genes into phage by infecting plasmid bearing cells with T4 37amA481 (whose amber mutation is located in the segment of DNA that is missing in T4 37SΔ1 and its derivatives) and growing the phage to produce a stock. Because MC1061 is not an amber-suppressing strain, only cells where recombination between the plasmid and phage genome occurred would produce viable pseudorevertant phage. We selected recombinant phage from the lysates by plating on BB (su0) and screened plaques by PCR to identify which plaques contained the 37SΔ1 deletion.
Measuring Adsorption Rates.
Adsorption rates were measured by using a single time point method (15). Briefly, phage were incubated with log phase cells for a fixed time, usually 5 or 10 min at 37°C (within the phage eclipse period). At that time we diluted the phage/cell mixture into buffer saturated with chloroform to lyse the cells. The number of infectious phage remaining is determined and the adsorption constant is calculated as Kads = (2.3/Ct)log(Po/Pt), where C is the cell concentration (ml−1), t is the incubation time (in minutes), Po is the infectious phage concentration (ml−1) at time 0, and Pt is the infectious phage concentration (ml−1) at time t.
Construction of SΔ1G5, SΔ1UCS, SΔ1ras1, and SΔ1ras2.
The pentaglycine coding segment in SΔ1G5 was added to the cloned DNA in p37SΔ1/T by using overlapping PCR primers (16). Primers 37SΔ1-1F (GGCGATGGTGGCGGTGGCGGCAATGTACAATTTTACGCTG) and 37SΔ1-1R (TACATTGCCGCCACCGCCACCATCGCCATTTAATCTCAA) contain complementary sequences corresponding to the Gly-5-containing SΔ1 junction. They were used with the flanking recF and 37B-2R primers to produce two modified half segments that were then recombined using the complementary ends to fuse the two segments and the flanking primers to amplify the whole segment. The entire segment was then cloned into pGEM-T.
SΔ1UCS (universal cloning site) was creating by amplifying half segments of the SΔ1 clone with primers 37SΔ1–2F (GGCGATGAGACGGTACCGTCTCAATGTACAATTTTACGCTG) and 37SΔ1–2R (TACATTGAGACGGTACCGTCTCATCGCCATTTAATCTCAA). Each primer contains a BsmBI and KpnI site. The two half segments were joined using the KpnI site to create a single segment with two BsmBI sites around the central KpnI site inserted into the SΔ1 junction. BsmBI cuts at positions 7/11 outside of the recognition site and the two BsmBI sites in p37SΔ1UCS/T are arranged so that the two cuts drop out the center segment (containing both BsmBI sites) leaving the original construct sequence with two different cohesive ends. This arrangement allows for the insertion (with an unambiguous orientation) of any double-stranded oligonucleotide with the correct cohesive ends. Thus, any oligopeptide can be cloned into junction of the SΔ1 deletion. We inserted the ras1 control (nonepitopic for Y13–259; see below) sequence by combining the oligonucleotides SΔ1R-1F (GCGATGGTGGCGGTGGCGCCCGCGGCGTGGGAAAGAGTGCCCTGACCATCCAGCTGATCGGTGGCGGTGGCA) and SΔ1R-1R (ACATTGCCACCGCCACCGATCAGCTGGATGGTCAGGGCACTCTTTCCCACGCCGCGGGCGCCACCGCCACCA). Similarly, the ras2 mAb epitope coding sequence was inserted by using the oligonucleotides SΔ1R-2F (GCGATGGTGGCGGTGGCGAAGAATACTCCGCAATGCGCGACCAGTACATGCGCACCGGTGAAGGTGGCGGTGGCA) and SΔ1R-2R (ACATTGCCACCGCCACCTTCACCGGTGCGCATGTACTGGTCGCGCATTGCGGAGTATTCTTCGCCACCGCCACCA). To anneal each oligonucleotide pair, we mixed the appropriate oligonucleotides in equimolar amounts, boiled the mixtures briefly, and cooled the mixtures slowly to form the appropriate double-stranded oligonucleotides with the correct single-stranded extensions. These oligonucleotides were ligated directly into BsmBI-digested p37SΔ1UCS/T. The insertions were confirmed by sequencing with the cysF primer.
mAb Inactivation Experiments.
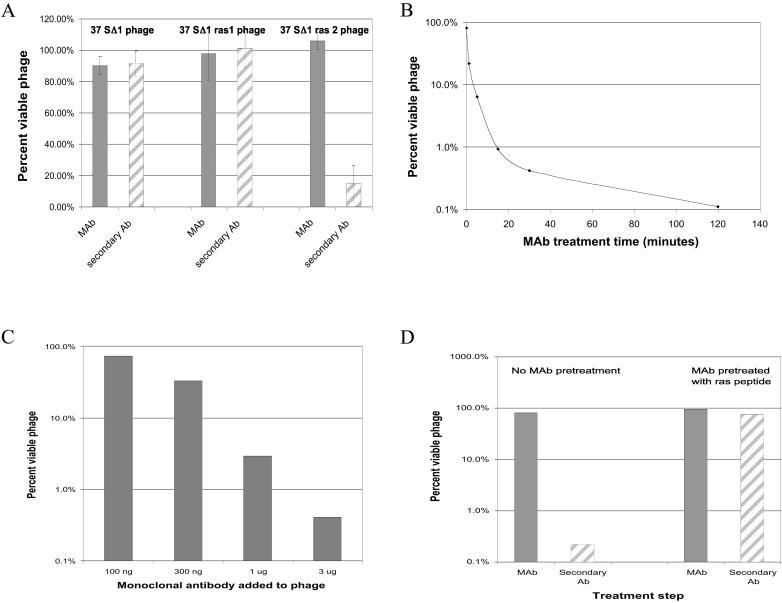
We purchased mAb Ab-1 (Y13–259; ref. 17) and inactivating peptide from Calbiochem and rabbit anti-rat whole IgG serum from Sigma. The mAb and peptide were resuspended in Dulbecco's PBS and the anti-serum was used as supplied. For inactivation experiments, we diluted phage to 1010 cells/ml in 10 mM phosphate pH 7.4/10 mM MgSO4. We added mAb (from a 0.1 mg/ml stock) to 500 μl of diluted phage and incubated the mixture for 30 min (unless otherwise indicated) at room temperature on a rotisserie mixer. Then we added 4 μg of secondary antiserum (from a 2 mg/ml stock) and incubated for 30 min at room temperature. For the initial experiments shown in Fig. 2A we used 1 μg of mAb, whereas 3 μg or the indicated amount was used for the remaining experiments. For the free epitope inhibition experiment shown in Fig. 2D, we mixed the peptide (EEYSAMRDQVMRTGE) and mAb at a 10:1 molar ratio and incubated for 30 min at room temperature. The mAb/peptide mixture was then added to phage as described above.
Figure 2.
Inactivation of phage by a monoclonal antibody. (A) Treatment of phage with mAb and secondary anti-serum. Each phage type was treated with 1 μg of mAb as described in Materials and Methods. (B) Time course of mAb treatment. SΔ1ras2 phage were treated with 3 μg of mAb for the indicated time before a 30 min incubation with secondary anti-serum. (C) Dose-response study of SΔ1ras2 phage with varying amounts of mAb. (D) Effect of treating mAb with free epitope before inactivation of SΔ1ras2.
Electron Microscopy of Phage.
Phage and phage/antibody complexes were stained with 1% phosphotungstate (pH 7) on carbon grids. Grids were examined at 100 kV by using a Philips CM10 transmission electron microscope. The final micrograph images were at a magnification of ×73,000.
Results and Discussion
Identification and Characterization of a Large Deletion in P37.
The tail fiber acts as a trigger to signal initiation of the tail sheath contraction process that precedes phage DNA injection. The reversible, noncovalent binding of a number of tail fiber distal ends to their specific receptor sites on the cell surface leads to a cooperative mechanical stress in the base plate. This stress triggers base plate expansion and initiates the tail sheath contraction, which extends the tail core through the cell wall (18, 19). The tail fibers' critical function for phage viability provides a sensitive assay for rigidity in tail fiber structure because any major loss of rigidity in the structure should impair the tail fibers' triggering function.
We used PCR analysis to screen spontaneous pseudorevertants of a gene 37 amber mutation (amA481), and identified a phage that appeared to have approximately 1 kb of DNA deleted from the middle of gene 37. This gene codes for the protein forming the distal end of the tail fibers, and its C terminus forms the phage receptor. Sequence analysis confirmed that a single contiguous segment of DNA coding for 346 of 1,026 amino acid residues (34%) was deleted in this phage, which was designated SΔ1 (spontaneous deletion 1). Table 1 shows the protein sequences of the deletion junctions and the corresponding wild-type protein. The deleted region begins at amino acid 73, which is 23 residues downstream from the conserved N-terminal domain of P37. This conserved region is thought to form the stiff butt end joint with the P36 C-terminal conserved domain (20). Thus, this deletion falls completely within the P37 rod-like region. Phage carrying the SΔ1 mutation produce plaques of normal size and appearance indicating that they are able to infect and grow normally. We also measured the adsorption rate of the SΔ1 phage (a measure of the rate of irreversible binding to the cell surface) and found that it was the same as wild-type phage (9.2 vs. 9.5 × 10−10 ml/min; SΔ1/wild-type = 0.97).
Table 1.
Partial protein sequences of naturally occurring and engineered gene 37 proteins
| Phage | Partial protein sequence of gene 37 at SΔ1 junction |
|---|---|
| Wild-type T4 | GLLRLNGDYVQ//GSNNVQFYADG |
| 37 SΔ1 | GLLRLNGD|NVQFYADG |
| 37 SΔ1G5 | GLLRLNGDGGGGGNVQFYADG |
| 37 SΔ1ras1 (control) | GLLRLNGDGGGGARGVGKSALTIQLIGGGGNVQFYADG |
| 37 SΔ1ras2 (mAb epitope) | GLLRLNGDGGGGEEYSAMRDQYMRTGEGGGGNVQFYADG |
Sequences flanking the SΔ1 junction are in italics, double slash represents 340 deleted amino acid residues, vertical line marks the position of the junction, inserted sequences are in boldface.
It was possible that the deletion mutation is compensated for by a second (duplication/insertion) mutation so that the overall tail fiber length was unchanged. To test this possibility, we cloned a 2-kb segment of DNA from the SΔ1 phage that surrounds the deletion site and placed it in a nonexpressing plasmid. Restriction and sequence analysis confirmed that this clone contained the expected DNA segments surrounding the 1,038 bp SΔ1 deletion and no additional DNA sequences. Homologous recombination was used to transfer the SΔ1 deletion into T4 phage containing the A481 amber mutation (which is located in the segment corresponding to the SΔ1 deletion segment). The SΔ1 deletion transferred at high efficiency, indicating that there is no other suppressor mutation needed to produce a viable phage.
We examined phage carrying the SΔ1 mutation by electron microscopy. The shortened distal portion of the tail fiber is clearly visible in the electron micrograph of SΔ1 phage (Fig. 1B). To compare wild-type tail fiber to SΔ1 tail fiber we calculated the ratio of the lengths of the distal half fiber/proximal half fiber (D/P) by using measurements from enlarged electron micrographs. We found that for wild-type fibers D/P = 0.99 ± 0.06 (n = 11) and for SΔ1 fibers D/P = 0.54 ± 0.14 (n = 6). This finding confirms that the viable SΔ1 phage have shortened but otherwise functional tail fibers.
Inserting a Peptide Into a β-loop In P37.
In the β-sheets forming the central rod regions of the tail fibers, the loop regions contribute little to maintaining the H-bond network, nor to the van der Waals interaction in the hydrophobic layer within the rod (21, 22). We postulate that the loops can be more variable and flexible than other regions of the tail fiber proteins. This postulate suggests that the junction of the SΔ1 deletion is in a loop (rather than in a β-strand) of the rod portion of gene 37. Surface loops in proteins can often be expanded to include additional peptide sequences with minimal effects on protein structure, function or stability (23). Thus, if the SΔ1 junction is in a loop, we should be able to insert additional sequences into the junction, expanding the loop, without severely disrupting the structural integrity of the tail fiber.
To test this we added DNA sequences encoding a pentaglycine peptide into the SΔ1 junction (Table 1) in the cloned gene segment. This modified sequence also transferred readily into phage by homologous recombination. [The SΔ1G5 phage produced poorer stocks, although the adsorption constant was almost the same as for the wild-type and SΔ1 phage (12 × 10−10 ml/min; SΔ1G5/wild type = 1.3). Poorer stocks might indicate a mild interference with phage development.] This finding confirms that the SΔ1 junction is able to accept peptide insertions without any significant loss of structural integrity and fits our hypothesis that the junction identifies a loop in the β-structure.
Inserting and Characterizing an Antibody Epitope into a Tail Fiber Protein.
To use tail fiber derived proteins as mesoscale assembly units, we will also need to attach specific functions to the assembled arrays of structural units. They may be attached before or after maturation of the final structure or at an intermediate step. The attachment may be covalent (e.g., disulfide bridges) or noncovalent (e.g., his tags). Incorporation of a peptide epitope may also be used to attach a functionality linked to the appropriate antibody. Fusions between antibodies and functional peptides have been extensively developed (24, 25). In the case of our nanoarchitectures, the compound would be fused to a mAb that is specific for an epitope in the structural unit. Thus, we set out to show that an antibody epitope could be incorporated into a tail fiber protein.
Next, we inserted two different 15 aa sequences from the human H-ras gene into the putative loop at the SΔ1 fusion junction (Table 1). Both peptides were flanked by four glycines on each side. One construct, SΔ1ras1, containing a nonepitope segment of H-ras, was created as a control, whereas the other, SΔ1ras2, contains the epitope specifically recognized by the rat monoclonal IgG antibody Y13-259 (17). Each of these modified genes readily transferred into phage by homologous recombination.
To test whether the epitope was accessible for interactions with the exogenous antibody, we treated SΔ1, SΔ1ras1, and SΔ1ras2 phage with the anti-ras mAb. If the mAb can bind to the H-ras epitope, it might inactivate the phage by linking together tail fibers on a single phage, thereby preventing proper binding to the cell surface. Alternatively, several phage might be linked together to form large noninfectious complexes. However, as Fig. 2A shows, mAb treatment alone (gray bars) did not result in phage inactivation. When the phage/mAb mixtures were further treated with an anti-rat IgG serum (striped bars) (which binds to the Fc region of the mAb), 85% of the SΔ1ras2 phage were inactivated. Because the SΔ1ras1 control phage were unaffected and because the anti-rat IgG antiserum alone has no effect on the SΔ1ras2 phage (data not shown), this finding demonstrates that the ras2 epitope is exposed on the surface of the tail fiber and accessible to the mAb. The requirement for the secondary antibody for phage inactivation may reflect the axial symmetry of P37. Because each mature fiber contains more than one epitope in close proximity on each tail fiber, it is likely that both binding sites in the mAb become bound to a single fiber. This would not be expected to inactivate the phage. Hence, the need for the secondary antibodies to crosslink tail fibers by binding to two mAbs bound to two different fibers and inactivate the phage. Regardless of the specific mechanism of inactivation, these experiments show that a functional peptide can be added to the rod region of a tail fiber protein without disrupting the tail fiber structure or function.
We further investigated the interaction of the SΔ1ras2 phage with the mAb. Fig. 2B shows that inactivation depends on the time allowed for mAb binding before addition of the secondary antiserum, reaching a maximum of 99.9% by 120 min. Fig. 2C shows that inactivation also has a simple dose-response relationship with the amount of Y13–259 mAb used. Fig. 2D shows that the SΔ1ras2 phage could be protected from inactivation by pretreating the mAb with a free 15-aa peptide of the same sequence as the 15-residue epitope inserted into the tail fiber protein. Although 99.8% of the phage were inactivated in the control treatment (with buffer only), there was no significant inactivation when the mAb was pretreated with the peptide. This finding demonstrates that the inactivation requires a specific interaction of the antibody with its specific epitope sequence.
We examined how the mAb interacts with the tail fiber by imaging mAb-treated phage. Fig. 1C shows a typical cluster of inactivated phage. The phage form a “bouquet” with the tail fibers linked together. It is unlikely that phage in such a bouquet could orient properly on the cell surface to allow the tail fibers to function cooperatively and trigger infection.
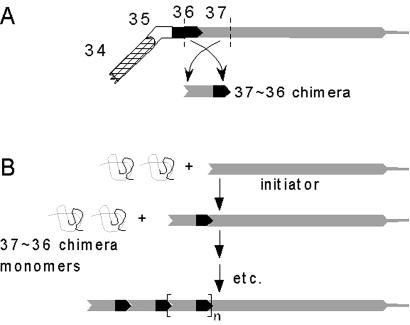
Taken together, these results demonstrate that rearrangements, fusions, and insertions can be made to a tail fiber protein without disrupting the functional integrity of the mature protein structure. They also support our hypothesis that fusion sites can be used for insertion of foreign peptides in such a way that they are available for binding. Further, these results support our hypothesis that the binding domain at the N-terminal end of P37 (and, presumably, the binding domains of other tail fiber proteins) is functionally separable from the central rod region. This finding suggests that chimeric proteins composed of the P37-binding domain of P36 joined by a central rod domain to the P36-binding end of P37 will form homo-polymeric fibers as shown in Fig. 3. The fusion site of these chimeric proteins should accept a functional peptide just as the SΔ1 junction does. This will provide the potential for attaching immunoconjugated functional moieties at precise locations along the fiber. The length of the chimeric proteins can be adjusted by using more or less of the rod region from either of the parent proteins, allowing the spacing of the functional moieties to be controlled. Furthermore, other β-loops within the central rod domain can be used as insertion sites for the addition of antigenic peptides that can subsequently be recognized by antibodies to add either functional or structural capabilities including crosslinking of the polymeric fibers into open two- and three-dimensional arrays.
Figure 3.
Self-assembly of fibers from tail fiber derived chimeric proteins. (A) Protein domains used to create a chimeric protein subunit (matured protein complexes shown). (B) Self-assembly of chimeric proteins onto a P37 initiator. The initiator is required to allow the gp36 monomer domains to combine to form the mature P36 domain. As in the wild-type fiber, this assembly propagates along the axis of the central rod domain. In the case of the chimeric proteins, this results in the formation of another P37 domain, supporting the next round of polymerization.
This approach will let us engineer protein fibers to place functional moieties in predesigned positions relative to one another to construct nanocomponents and nanodevices that exhibit functions not attainable with single nanoparticles or nanostructured materials. Taken together, the capability demonstrated here provides great potential for fabrication of a broad range of nanostructures that will exhibit anticipated, and in many cases, still unanticipated functions.
Acknowledgments
We thank Fred Eiserling for the drawing of bacteriophage T4, Cathy Linsenmayer and Melissa McCoy for technical assistance, and Debu RayChaudhuri for criticism and encouragement. E.G. thanks Pat Durham for his excellent technical assistance at the start of this project. We especially thank Phil Harriman for his support of this work in its nascent phase. This work was supported in part by National Science Foundation Grant DBI 9834603 and Office of Naval Research Grant N00014-98-1-0784 (to E.G.).
Note Added in Proof.
In collaboration with Dr. W. Stafford (Boston Biomedical Research Institute), we have recently used analytical ultracentrifugation to confirm the trimeric nature of the gene 34, 36, and 37 tail fiber segments as first proposed by the lab of A. Steven (10) (unpublished data).
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Nanoscience: Underlying Physical Concepts and Phenomena,” held May 18–20, 2001, at the National Academy of Sciences in Washington, DC.
This paper was submitted directly (Track II) to the PNAS office.
GpX (gene product) refers to the monomeric product of gene X, whereas PX refers to the matured, multimeric complex of gpX's that has assembled into the structure that is found in the phage T4 virion.
There is conflicting evidence as to whether the gene 34, 36, and 37 segments of the tail fibers are homodimers (8, 9) or homotrimers (10). For the purposes of this work either case is possible because the monomers are arranged in an overall parallel fashion (i.e., N termini together at one end of the mature protein and C termini at the other end). This means that insertions, deletions, or modifications in the protein monomers will be located at identical positions in the mature dimer/trimer. For simplicity, we will refer to the proteins as forming dimers.
References
- 1.Wood W B, Eiserling F A, Crowther R A. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol. Press; 1994. pp. 282–290. [Google Scholar]
- 2.Henning U, Hashemolhosseini S. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol. Press; 1994. pp. 291–298. [Google Scholar]
- 3.Wood W B. Harvey Lect. 1979;73:203–223. [PubMed] [Google Scholar]
- 4.Eiserling F A, Black L W. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol. Press; 1994. pp. 209–212. [Google Scholar]
- 5.Beckendorf S K. J Mol Biol. 1973;73:37–53. doi: 10.1016/0022-2836(73)90157-5. [DOI] [PubMed] [Google Scholar]
- 6.Arscott P G, Goldberg E B. Virology. 1976;69:15–22. doi: 10.1016/0042-6822(76)90190-2. [DOI] [PubMed] [Google Scholar]
- 7.Crawford J T, Goldberg E B. J Mol Biol. 1980;139:679–690. doi: 10.1016/0022-2836(80)90054-6. [DOI] [PubMed] [Google Scholar]
- 8.Earnshaw W C, Goldberg E B, Crowther R A. J Mol Biol. 1979;132:101–131. doi: 10.1016/0022-2836(79)90498-4. [DOI] [PubMed] [Google Scholar]
- 9.Ward S, Dickson R C. J Mol Biol. 1971;62:479–492. doi: 10.1016/0022-2836(71)90149-5. [DOI] [PubMed] [Google Scholar]
- 10.Cerritelli M E, Wall J S, Simon M N, Conway J F, Steven A C. J Mol Biol. 1996;260:767–780. doi: 10.1006/jmbi.1996.0436. [DOI] [PubMed] [Google Scholar]
- 11.Fisher K M, Bernstein H. Mol Gen Genet. 1970;106:139–150. doi: 10.1007/BF00323832. [DOI] [PubMed] [Google Scholar]
- 12.McFall E, Stent G W. J Gen Microbiol. 1958;18:346–363. doi: 10.1099/00221287-18-2-346. [DOI] [PubMed] [Google Scholar]
- 13.Carlson K, Miller E S. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol. Press; 1994. pp. 421–441. [Google Scholar]
- 14.Casadaban M J, Cohen S N. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 15.Adams M H. Bacteriophages. New York: Interscience; 1959. [Google Scholar]
- 16.Sambrook J, Russel D W. Molecular Cloning. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 17.Sigal I S, Gibbs J B, D'Alonzo J S, Scolnick E M. Proc Natl Acad Sci USA. 1986;83:4725–4729. doi: 10.1073/pnas.83.13.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford J T, Goldberg E G. J Mol Biol. 1977;111:305–313. doi: 10.1016/s0022-2836(77)80053-3. [DOI] [PubMed] [Google Scholar]
- 19.Crowther R A. J Mol Biol. 1980;137:159–174. doi: 10.1016/0022-2836(80)90323-x. [DOI] [PubMed] [Google Scholar]
- 20.Riede I, Drexler K, Eschbach M-L. Nucleic Acids Res. 1985;13:605–616. doi: 10.1093/nar/13.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Branden C, Tooze J. Introduction to Protein Structure. 2nd Ed. New York: Garland; 1999. [Google Scholar]
- 22.Xu G, Wang W, Groves J T, Hecht M H. Proc Natl Acad Sci USA. 2001;98:3652–3657. doi: 10.1073/pnas.071400098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regan L. Curr Opin Struct Biol. 1999;9:494–499. doi: 10.1016/S0959-440X(99)80070-0. [DOI] [PubMed] [Google Scholar]
- 24.Vitetta E S, Fulton R J, May R D, Till M, Uhr J W. Science. 1987;238:1098–1104. doi: 10.1126/science.3317828. [DOI] [PubMed] [Google Scholar]
- 25.Byers V S, Baldwin R W. Immunology. 1988;65:329–335. [PMC free article] [PubMed] [Google Scholar]