Abstract
Nuclear mtDNA sequences (numts) are a widespread family of paralogs evolving as pseudogenes in chromosomal DNA [Zhang, D. E. & Hewitt, G. M. (1996) TREE 11, 247–251 and Bensasson, D., Zhang, D., Hartl, D. L. & Hewitt, G. M. (2001) TREE 16, 314–321]. When trying to identify the species origin of an unknown DNA sample by way of an mtDNA locus, PCR may amplify both mtDNA and numts. Indeed, occasionally numts dominate confounding attempts at species identification [Bensasson, D., Zhang, D. X. & Hewitt, G. M. (2000) Mol. Biol. Evol. 17, 406–415; Wallace, D. C., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 14900–14905]. Rhesus and cynomolgus macaque mtDNA haplotypes were identified in a study of oral polio vaccine samples dating from the late 1950s [Blancou, P., et al. (2001) Nature (London) 410, 1045–1046]. They were accompanied by a number of putative numts. To confirm that these putative numts were of macaque origin, a library of numts corresponding to a small segment of 12S rDNA locus has been made by using DNA from a Chinese rhesus macaque. A broad distribution was found with up to 30% sequence variation. Phylogenetic analysis showed that the evolutionary trajectories of numts and bona fide mtDNA haplotypes do not overlap with the signal exception of the host species; mtDNA fragments are continually crossing over into the germ line. In the case of divergent mtDNA sequences from old oral polio vaccine samples [Blancou, P., et al. (2001) Nature (London) 410, 1045–1046], all were closely related to numts in the Chinese macaque library.
With the advent of genomics it is increasingly recognized that horizontal gene transfer has been going on throughout evolution. Fragments of mitochondrial DNA (mtDNA) have migrated regularly to chromosomal DNA (1–4), many of which are located near the centromeres (5). These nuclear mtDNA sequences (numts) can be as small as 100 bases and as large as 270 kb, or 75% of the Arabidopsis thaliana plant mtDNA genome (5). In silico scanning of the human genome sequence revealed ≈300–400 numts corresponding to between 0.6 and 88% of human mtDNA (2, 6).
Although numts behave as pseudogenes and fix mutations at a rate higher than nuclear coding sequences, they evolve more slowly than mtDNA sequences. Indeed the fixation rate of vertebrate mtDNA is ≈20-fold greater than that of numts (7). From the perspective of PCR, mtDNA and numts represent the equivalent of a multigene family with components in both the high and low molecular weight fractions. Not surprisingly, the proportion of numts in a DNA sample varies with the tissue (3) and the extraction protocol (8, 9). For example, peripheral blood contains mainly resting lymphocytes in which the copy number of mtDNA is low compared with activated cells. For some samples, mostly numts were recovered (10).
Given their high copy number mtDNA frequently is chosen for the identification of species DNA. If the samples are old, small fragments generally are amplified. Furthermore, given its faster fixation rate it provides far greater resolution, certainly of more recent events such as comparing conspecific populations (11). However, the presence of numts may confound analyses of normal mtDNA haplotypes (12, 13). At best they represent a form of noise that must be controlled for. If not, mistaking numts for mtDNA may lead to erroneous conclusions based on their different evolutionary trajectories (14–16). Finally, PCR-mediated recombination is a technical problem associated with amplifying multigenic targets (17). All these factors come together if mtDNA is to be used to identify the species origin of DNA from an unidentified sample.
It has been suggested that the AIDS epidemic might have had its origins in the use of chimpanzee primary kidney tissue cultures necessary for the preparation of an experimental attenuated oral poliovirus (OPV) in the late 1950s in Central Africa (18–21). However, PCR-based studies of 40-year-old frozen samples of vaccine failed to detect any SIVcpz nucleic acids or chimpanzee DNA (22, 23). By contrast rhesus and cynomolgus macaque (Macaca mulatta and Macaca fascicularis) mtDNA was found in abundance, effectively scotching the hypothesis (22). Among the collection of mtDNA sequences, some were highly divergent and unrelated to any known mtDNA haplotype and were presumed to be numts. However, given that a macaque genome has not been sequenced yet there was no way of proving this presumption. To address this question a library of more than 100 numts was derived from a single rhesus macaque. The data demonstrate that numts derived from the oral polio vaccine samples were derived uniquely from macaque genomes.
Materials and Methods
Peripheral blood mononuclear cells from a rhesus macaque (M. mulatta) of Chinese origin were lysed in 0.5 ml of lysis buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA/100 mM NaCl/0.5% SDS) and treated with 100 μg/ml proteinase K for 2 h at 56°C. After phenol extraction and ethanol precipitation, total DNA was resuspended in 10 mM Tris⋅HCl, pH 8.0/1 mM EDTA.
Mitochondrial 12S rDNA primers 12Sa and 12So have been described (24). Hot-start PCR was applied in all reactions. The buffer conditions were: 2.5 mM MgCl2/50 mM KCl/10 mM Tris⋅HCl, pH 8.3/200 μM of each dNTP/100 μM of each primer/2.5 units of Taq DNA polymerase (Perkin–Elmer) in a final volume of 100 μl. Denaturation, annealing, extension, and conditions were: 95°C for 5 min, then 35× (95°C for 30 sec, 55°C for 30 sec, 72°C for 5 min) followed by a final step at 72°C for 10 min. Albeit only 101 bp long, elongation times were used to reduce the incidence of PCR recombination (17). PCR products were purified from 1.5% agarose gels (Qiaex II kit, Qiagen, Chatsworth, CA) and ligated into the TOPO TA cloning vector (Invitrogen). After transformation of Escherichia coli XL1-Blue, colonies were picked and prepared for DNA sequencing. Sequencing was performed by using thermosequenase (USB, Amersham Pharmacia).
Sequences were aligned by using CLUSTAL W (25). Nucleotide sequence distances were determined with DNADIST of the PHYLIP 3.5 package (26). The tree was derived by neighbor-joining analysis applied to pairwise sequence distances calculated by using a variety of methods including the Kimura two-parameter method to generate unrooted trees. The final output was generated with TREEVIEW (27). The accession numbers are AF502296–AF502367. Alternatively, sequences can be found at ftp://pasteur.fr/pub/retromol/numts02.
Results and Discussion
A Library of numts from a Single Rhesus Macaque.
Amplification of the small fragment of the 12S mitochondrial rDNA gene was performed on total DNA isolated from peripheral blood mononuclear cells of a Chinese rhesus macaque (M. mulatta). Because peripheral blood mononuclear cells are mainly resting cells, the ratio of numts to normal mtDNA sequences is relatively high. Hence the protocol is biased in favor of detecting numts. A total of 139 clones was sequenced and aligned to the most abundant sequence (37 clones), which by comparison to extant sequences corresponded to a known rhesus macaque mitochondrial haplotype. The remaining 102 sequences (73%) differed by 1–29 mutations compared with the animal's mtDNA sequence. Because the PCR error was low in this study (<1 mutation in 37 clones sequenced as exemplified by the normal macaque haplotype sequences) it is presumed that these sequences corresponded to bona fide numts. Formally even a normal mtDNA sequence could represent a recent mitochondrial immigrant to the nucleus (i.e., numt), for the integration sites were not sequenced. Hence the 73% value probably constitutes a lower limit.
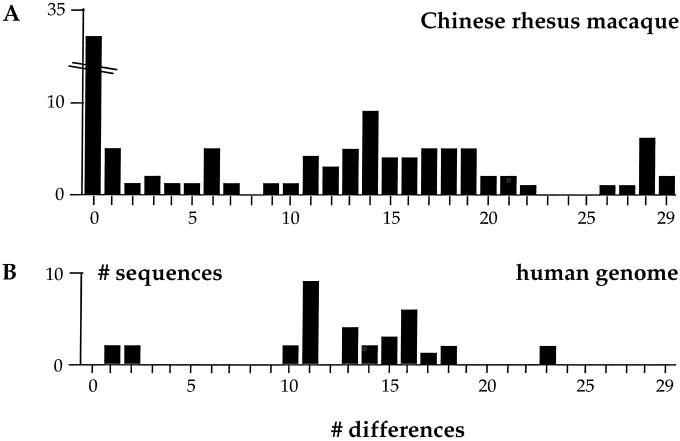
Among the collection of 102 numts, 78 were unique (76%), and the remainder was found in 2–4 copies. The frequency distribution of numts as a function of sequence divergence is given in Fig. 1A. There was a broad distribution centered around 14 substitutions per numt. However, without knowing the dynamics of gain and loss of numts it is not possible to comment on the fact that the distribution was not smooth. To appreciate better the distribution, the corresponding fragment of the human 12S sequence was used to screen the human genome (http://www.ncbi.nlm.nih.gov/genome/seq/) for numts. A total of 36 numts were identified, resulting in 22 distinct sequences (Fig. 1B). This number agrees well with a generalized computer search for numts in the human genome (6). Albeit fewer in number than for the macaque genome, the distribution was comparable; the collection was centered around 13–14 differences per clone with a maximum of 23. It is possible that the collection of human numts is incomplete. The human genome sequence is ≈93% complete, covers mainly gene-rich DNA, and generally lacks the highly repeated centromeric regions. For the Arabidopsis plant genome, many numts were located in the gene-poor centromeric regions. If a similar distribution pertained to the human genome, it might explain the smaller number of numts found.
Figure 1.
Frequency distribution of divergent numts from a Chinese rhesus macaque (A) and the human genome (B). The numts correspond to a small segment of the mitochondrial 12S rDNA gene used to analyze old OPV samples (see below). For the macaque library, the major form (37/139 clones) corresponds to a known macaque mtDNA haplotype. For those gleaned from the human genome, a human haplotype (GenBank accession no. X89832) was taken as reference.
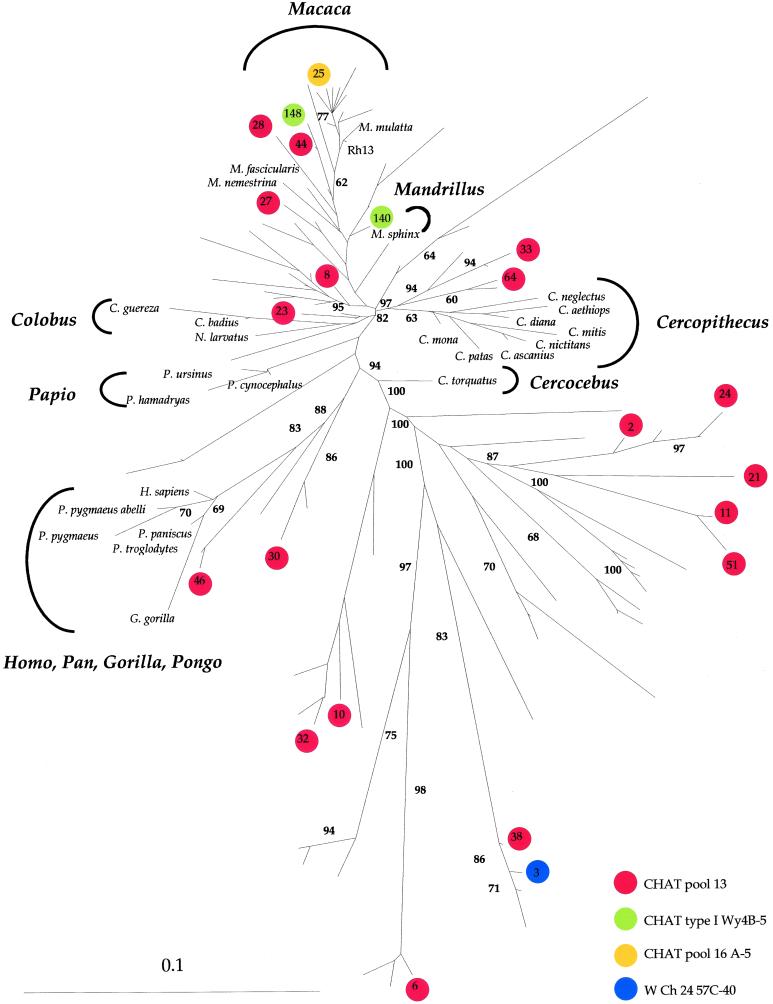
A neighbor-joining tree of the library of numts is shown in Fig. 2 along with reference mtDNA haplotypes and putative numts from the OPV samples (see below). Three points can be made about the library of macaque numts. First, some numts are interspersed within extant macaque mtDNA haplotypes, indicating that numt formation is an ongoing process as has been noted previously (3). Second, no macaque numts clustered with mitochondrial haplotypes of the Cercocebus, Cercopithecus, Mandrillus, Colobus, Papio, or Homo/Pan lineages, which is consistent with the enormity of sequence space and the fact that mtDNA is fixing mutations faster than numts (7). A pseudogene not under selection cannot follow the trajectory of a more rapidly evolving gene under selection. Third, most of the highly divergent numts fell into two or more distinct lineages with very long branch lengths unaccompanied by any known haplotype. The clustering of some sequences at the tips may be understood via DNA duplication followed by mutation, a feature that has been described already.
Figure 2.
Phylogenetic tree of a wide collection of 12S rDNA sequences. They include the numts from a single Chinese rhesus macaque (M. mulatta), 26 mtDNA sequences corresponding to primate lineages for reference, and 22 putative numts from OPV samples (22). The latter are indicated by colored circles, with each color corresponding to a different vaccine lot. The numbers refer to distinct numt clones. M. mulatta refers to the normal mtDNA haplotype of the Chinese rhesus macaque. Rh13 corresponds to the normal rhesus macaque allele from CHAT pool 13. Sequences were aligned by CLUSTAL W. The tree was derived by neighbor-joining analysis applied to pairwise sequence distances calculated with DNADIST of the PHYLIP 3.5 package using the Kimura two-parameter method to generate unrooted trees. Horizontal branch lengths are drawn to scale with the bar indicating 0.1 nucleotide replacements per site. The final output was generated with TREEVIEW. The number at each node represents the percentage of bootstrap replicates (out of 100). Only bootstrap values of ≥60 are given.
Identification of numts from OPV Samples.
During the poliomyelitis vaccination campaigns in Central Africa during the late 1950s, numerous OPV lots were used. CHAT pool 13 in particular was used to vaccinate 75,000 people in Léopoldville, now Kinshasa, between August 1958 and April 1960. From more than 250 12S rDNA sequences amplified from old OPV samples made in the late 1950s, 22 were putative numts (22); 18 were derived from a vaccine lot called CHAT pool 13, 1 from CHAT type Wy4B-5, 1 from CHAT pool 16 A-5, and 1 from WCh24 57C-40. These sequences are included in Fig. 2 and are color-coded to facilitate identification. In all cases, they cluster closely with numts from the single Chinese rhesus macaque. Six were intermixed with known macaque haplotypes, meaning that they are recent nuclear immigrants. The remaining 13 were situated in divergent clusters of macaque numts. In no case do any map within the Cercocebus, Cercopithecus, Mandrillus, Colobus, Papio, or Homo/Pan lineages.
In a previous publication, it was suggested that some mitochondrial 12S rDNA sequences from the OPV samples were closely related (Δ = 2-bp/101-bp fragment) to normal haplotypes of the mona monkey (Cercopithecus mona; ref. 12). Exactly the same 101-bp fragment was amplified in the present study and our earlier study of the OPV vaccine lots (22). No such sequences were found in the OPV samples, whereas the closest Chinese macaque numt differed by 4 bases from the C. mona mtDNA sequence. Some of the OPV-derived numts were not identical to Chinese macaque numts. Although not shown here, the genus macaque shows a considerable degree of polymorphism in this locus (≈3–4%; Fig. 1; ref. 22). Both Chinese and Indian rhesus macaque kidneys (M. mulatta) were used to make vaccine lots, as were cynomolgus macaques (M. fascicularis) albeit to a lesser extent. Hence, lack of identity may well reflect numt polymorphisms among extant macaque subspecies. Finally, inspection of the ensemble of numts suggested that the OPV-derived clone 55 might well be a PCR-mediated recombinant of normal macaque haplotype and a numt (17). Out of precaution it was not included in Fig. 2. Taken together, the above findings substantiate the earlier conclusion that these were in fact macaque numts (22).
As the phylogenetic analysis of the Chinese macaque numts has shown, functional mtDNA lineages are not tracked by numt pseudogenes. The only time that numts and normal mtDNA sequences cluster is when they are both from the same lineage, for numt formation is an ongoing process. The same observation can be made from an analysis of human numts extracted from the human genome (Fig. 1; ref. 2). Hence in identifying species origin from mtDNA sequences, any sample from an existing species must be accompanied by recent nuclear immigrants that differ by very few mutations from a known haplotype. When a sequence is highly divergent from any extant mtDNA, perhaps it should be considered an numt, if only out of prudence. In the special case where only numts are found, identification still is possible, particularly if a numt library is available. Perhaps the only conundrum will be the description of extinct species.
It is not difficult to generate a library of numts by PCR. With high-throughput sequencing a numt reference set can be generated within days. Indeed, the PCR-based approach generated more numts than was gleaned from a 93% copy of the human genome. Such an approach certainly constitutes a far faster and cheaper means to resolving ambiguities arising from the existence of numts.
In conclusion, the divergent mtDNA sequences derived from the OPV samples proved to be numts of macaque origin, which reinforces the original conclusion that only macaque kidney tissues were used to prepare lots of OPV (22, 23, 28, 29).
Acknowledgments
We thank Céline Elbé-Renoux for macaque DNA and Michel Henry for sequencing. This work was supported by grants from the Institut Pasteur and the Agence Nationale pour la Recherche sur le SIDA.
Abbreviations
- numts
nuclear mtDNA sequences
- OPV
oral poliovirus
Footnotes
References
- 1.Zhang D E, Hewitt G M. TREE. 1996;11:247–251. doi: 10.1016/0169-5347(96)10031-8. [DOI] [PubMed] [Google Scholar]
- 2.Bensasson D, Zhang D, Hartl D L, Hewitt G M. TREE. 2001;16:314–321. doi: 10.1016/s0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- 3.Wallace D C, Stugard C, Murdock D, Schurr T, Brown M D. Proc Natl Acad Sci USA. 1997;94:14900–14905. doi: 10.1073/pnas.94.26.14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard J L, Schmidt G W. J Mol Evol. 1995;41:397–406. doi: 10.1007/BF00160310. [DOI] [PubMed] [Google Scholar]
- 5.Lin X, Kaul S, Rounsley S, Shea T P, Benito M I, Town C D, Fujii C Y, Mason T, Bowman C L, Barnstead M, et al. Nature (London) 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 6.Mourier T, Hansen A J, Willerslev E, Arctander P. Mol Biol Evol. 2001;18:1833–1837. doi: 10.1093/oxfordjournals.molbev.a003971. [DOI] [PubMed] [Google Scholar]
- 7.Zischler H, Geisert H, Castresana J. Mol Biol Evol. 1998;15:463–469. doi: 10.1093/oxfordjournals.molbev.a025943. [DOI] [PubMed] [Google Scholar]
- 8.Sorenson M D, Quinn T W. Auk. 1998;115:214–221. [Google Scholar]
- 9.Greenwood A D, Pääbo S. Mol Ecol. 1999;8:133–137. doi: 10.1046/j.1365-294x.1999.00507.x. [DOI] [PubMed] [Google Scholar]
- 10.Bensasson D, Zhang D X, Hewitt G M. Mol Biol Evol. 2000;17:406–415. doi: 10.1093/oxfordjournals.molbev.a026320. [DOI] [PubMed] [Google Scholar]
- 11.Avise J C. Science. 2001;294:86–87. doi: 10.1126/science.294.5540.86. [DOI] [PubMed] [Google Scholar]
- 12.Poinar H, Kuck M, Pääbo S. Science. 2001;292:743–744. doi: 10.1126/science.1058463. [DOI] [PubMed] [Google Scholar]
- 13.van der Kuyl A C, Kuiken C L, Dekker J T, Goudsmit J. J Mol Evol. 1995;40:173–180. doi: 10.1007/BF00167111. [DOI] [PubMed] [Google Scholar]
- 14.Woodward S R, Weyand N J, Bunnell M. Science. 1994;266:1229–1232. doi: 10.1126/science.7973705. [DOI] [PubMed] [Google Scholar]
- 15.Zischler H, Hoss M, Handt O, von Haeseler A, van der Kuyl A C, Goudsmit J. Science. 1995;268:1192–1193. doi: 10.1126/science.7605504. [DOI] [PubMed] [Google Scholar]
- 16.van der Kuyl A C, Kuiken C L, Dekker J T, Perizonius W R, Goudsmit J. J Mol Evol. 1995;40:652–657. doi: 10.1007/BF00160513. [DOI] [PubMed] [Google Scholar]
- 17.Meyerhans A, Vartanian J P, Wain-Hobson S. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtis T. Rolling Stone. 1992. , March 19 (626) 54–60. [Google Scholar]
- 19.Stricker R B, Elswood B F. Lancet. 1992;339:867. [PubMed] [Google Scholar]
- 20.Cribb J. The White Death. Sydney: Harper Collins; 1996. [Google Scholar]
- 21.Hooper E. The River. London: Allen Lane; 1999. [Google Scholar]
- 22.Blancou P, Vartanian J P, Christopherson C, Chenciner N, Basilico C, Kwok S, Wain-Hobson S. Nature (London) 2001;410:1045–1046. doi: 10.1038/35074171. [DOI] [PubMed] [Google Scholar]
- 23.Berry N, Davis C, Jenkins A, Wood D, Minor P, Schild G, Bottiger M, Holmes H, Almond N. Nature (London) 2001;410:1046–1047. doi: 10.1038/35074176. [DOI] [PubMed] [Google Scholar]
- 24.Poinar H N, Hofreiter M, Spaulding W G, Martin P S, Stankiewicz B A, Bland H, Evershed R P, Possnert G, Pääbo S. Science. 1998;281:402–406. doi: 10.1126/science.281.5375.402. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J, Higgins D, Gibson T. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 27.Page R D M. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 28.Plotkin S A, Koprowski H. Science. 1999;286:2450. doi: 10.1126/science.286.5449.2449d. [DOI] [PubMed] [Google Scholar]
- 29.Weiss R A. Nature (London) 2001;410:1035–1036. doi: 10.1038/35074222. [DOI] [PubMed] [Google Scholar]