Abstract
During neuronal maturation, dendrites develop from immature neurites into mature arbors. In response to changes in the environment, dendrites from certain mature neurons can undergo large-scale morphologic remodeling. Here, we show a group of Drosophila peripheral sensory neurons, the class IV dendritic arborization (C4da) neurons, that completely degrade and regrow their elaborate dendrites. Larval dendrites of C4da neurons are first severed from the soma and subsequently degraded during metamorphosis. This process is controlled by both intracellular and extracellular mechanisms: The ecdysone pathway and ubiquitin-proteasome system (UPS) are cell-intrinsic signals that initiate dendrite breakage, and extracellular matrix metalloproteases are required to degrade the severed dendrites. Surprisingly, C4da neurons retain their axonal projections during concurrent dendrite degradation, despite activated ecdysone and UPS pathways. These results demonstrate that, in response to environmental changes, certain neurons have cell-intrinsic abilities to completely lose their dendrites but keep their axons and subsequently regrow their dendritic arbors.
Keywords: metamorphosis, pruning, ecdysone receptor
Dendrites are the primary site where neurons receive synaptic and/or sensory inputs. Recently, significant progress has been made in understanding how dendrites develop from unspecified neurites and mature into projections that receive inputs from the surrounding environment (1, 2). In the mature nervous system, neurons continue to respond to both changes in the environment and/or altered activities in the neural circuit by displaying morphological and functional plasticity according to experience (3, 4). Despite the progress being made in understanding the plasticity of dendritic spines (5, 6), less is known about large-scale remodeling of mature dendrites.
One opportunity to observe large-scale remodeling of dendrites is during metamorphosis, when insects such as Manduca and Drosophila morph from larva to pupa and then to adults. Many of the larval organs, including the nervous system, are degraded and replaced by newly formed adult structures. As to the surviving larval neurons, extensive remodeling is necessary to renew their functional connections (7). In the CNS, these neurons include the Manduca femoral depressor motoneurons (8), the Drosophila mushroom body γ-neurons (9-11), and a set of fly olfactory projection neurons (12). In the Drosophila peripheral nervous system, dendritic arborization (da) neurons are thought to function as sensory neurons for the developing embryo and larvae (13, 14). Some larval da neurons survive into adulthood (15, 16), and one such neuron, ddaE, changes its da during metamorphosis (17).
Whereas dendritic remodeling has been commonly observed with concomitant axonal remodeling (8, 10-12), it is not known whether neurons have the cell-intrinsic abilities to selectively remodel their dendrites while retaining their axons. To address this question, we set out to identify a group of Drosophila neurons that persists through metamorphosis and demonstrates clear reorganization of dendrites. Screening through a number of upstream activating sequence (UAS)/Gal4 lines expressing EGFP, we were able to identify a group of pickpocket-expressing neurons (18), the class IV da (C4da) neurons, that remodel their dendrites during metamorphosis. We show that these C4da neurons degrade their extensive mature larval dendrites during metamorphosis but keep their axons intact. And this dendrite-specific process is regulated by both cell-intrinsic and extrinsic mechanisms involving the matrix metalloproteases, the ubiquitin-proteasome system (UPS), and ecdysone receptor (EcR) signaling.
Methods
Fly Stocks. The fly stocks used in this study include ppk-EGFP5 (18); ppk-Gal4 (a gift from R. Yang and W. Grueber, University of California, San Francisco); ppk-Gal4, UAS>CD2>CD8-GFP, hsFLP (a gift from W. Grueber); Mmp1Q273, Mmp2W307, and UAS-Timp (19); FRTG13 MARCM (mosaic analysis with a repressible cell marker) alleles of Uba1 and Mov34 (11); FRTG13 MARCM allele of Mmp1Q112Mmp2W307 (a gift from A. Page-McCaw, Rensselaer Polytechnic Institute, Troy, NY); FRT19A MARCM allele of Usp3 (10); UAS-UBP2 (20); UAS-p35 (21); UAS-mCD8::GFP (Bloomington Stock Center, Indiana University, stock no. 5137); UAS-EcR-B1-ΔC655-F645A and UAS-EcR-B1-ΔC655-W650A (UAS-EcR-DN, Bloomington Stock Center nos. 6869 and 6872); and UAS-EcR-A, UAS-EcR-B1, and UAS-EcR-B2 (Bloomington Stock Center nos. 6470, 6469, and 6468).
MARCM and UAS>CD2>CD8-GFP Analysis. MARCM analyses were performed as described in ref. 22. To generate mosaic clones, w; p[FRTG13] or Mmp1Q112Mmp2W307, FRTG13/CyO or Uba1, FRTG13/CyO or Mov34, FRTG13/CyO or Rpn6, FRTG13/CyO flies were mated with w; elav-Gal4, UAS-mCD8::GFP, hsFLP; FRTG13, tub-Gal80/CyO flies, and Usp3, FRT19A/Fm7C flies were mated with w; 109 (2)80-Gal4, UAS-mCD8::GFP, hsFLP/Cyo; FRT19A, tub-Gal80 flies. Live imaging was obtained on a Bio-Rad MRC 600 confocal microscope. Generation of single C4da neuron clones in ppk-Gal4, UAS>CD2>CD8-GFP, hsFLP flies followed the protocol described in ref. 23 with the following modification: Heat shock was performed at 38°C for 30 min after 6 h of egg collection at 25°C.
Dissection and Immunocytochemistry. To image the ventral nerve cord (VNC) after head eversion, pupal cases were first removed. Using double-sided tape, pupae were immobilized ventral side down, and epidermis was carefully filleted. Liquefied tissue immediately under the epidermis was removed. The entire prep was immediately mounted for confocal imaging on the Bio-Rad MRC 600 microscope. For immunocytochemistry, larvae and pupae were stained with the following antibodies: rabbit anti-EGFP, 1:2,000 (kindly provided by Y. Hong, University of California, San Francisco); rat anti-mCD8a, 1:100 (Caltag, South San Francisco, CA); mouse anti-EcR-C Ag10.2, 1:10 [Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA]; mouse anti-EcR-A 15G1a, 1:10 (DSHB); mouse anti-EcR-B1 AD4.4, 1:10 (DSHB); mouse anti-Armadillo N2 7A1, 1:100 (DSHB); mouse anti-ubiquitin ab7254, 1:1,000 (Abcam, Cambridge, MA); donkey anti-rabbit Cy2-conjugated secondary antibody, 1:500 (The Jackson Laboratory); and donkey anti-mouse rhodamine red X-conjugated secondary antibody, 1:200 (The Jackson Laboratory). For EcR staining, fixed samples were blocked with 5% goat serum in PBS plus 0.3% Triton X-100 (PBST) for 1 h at room temperature (RT), followed by RT overnight incubation with 1:10 dilution of primary antibody in PBST. Samples were washed extensively in PBST (1 h five times at RT), followed by secondary antibody incubation overnight at 4°C. The samples were washed 1 h three times at RT, dehydrated, and mounted in DPX mounting medium. Fluorescence images were obtained on a Leica TCS SP2.
Results and Discussion
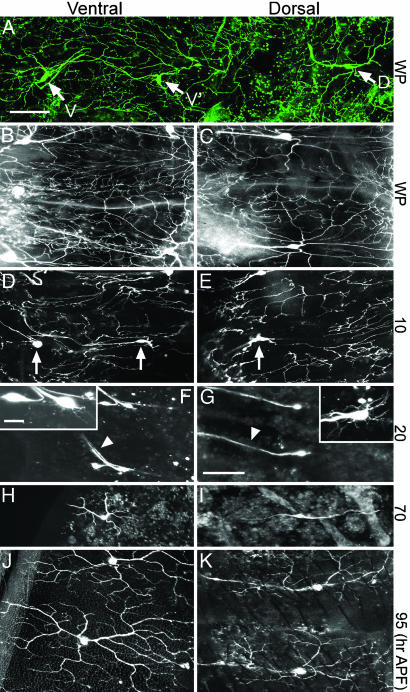
Dendritic Remodeling of C4da Neurons During Pupariation. To visualize abdominal C4da neurons during Drosophila metamorphosis, we used the pickpocket (ppk)-EGFP reporter line described in ref. 18. Filleted white pupae (WP), at the onset of metamorphosis, were stained with an anti-EGFP antibody to reveal three C4da neurons, vdaB (V), v′ada (V′), and ddaC (D), in each hemisegment (Fig. 1A). Because the soma and dendritic projections of these neurons remained very close to the body surface during pupariation, we used live imaging to follow these neurons throughout metamorphosis.
Fig. 1.
Dendritic remodeling of C4da neurons during metamorphosis. (A) Anti-EGFP antibody staining of ppk-EGFP WP fillet. One abdominal hemisegment is shown. Arrows point to V, V′, and D C4da neurons. (B-K) Live confocal images of abdominal segments of pickpocket-EGFP pupae during metamorphosis. For each time point, paired images show ventral (Left) and dorsal (Right) views. (B and C) WP stage. Note the intact dendritic fields of all three neurons. (D and E) Ten hours APF. Arrows point to cell bodies with detached dendrites. (F and G) Twenty hours APF. Arrowheads point to axon projections. Inset in F shows enlarged view of V and V′ neurons. V′ (Right) has thin dendritic projections. Inset in G shows enlarge view of D neurons with thin dendrites. (H and I) Seventy hours APF. Both V′ neurons in H and D neurons in I extend adult dendritic arbors. (J and K) Ninety-five hours APF. Adult dendritic fields of V′ and D neurons just before eclosion. Dorsal is right and anterior is up in all images. (Scale bars: A-K, 100 μm; Insets, 20 μm.)
Initially at the WP stage, the C4da neurons exhibited intact, complex class IV dendritic branches that covered much of the pupal surface (Fig. 1 B and C). Shortly after the white pupal stage, 2 h after puparium formation (APF), fine terminal dendritic branches began to disappear (data not shown). By 10 h APF, most major dendritic branches were severed from the soma (arrows, Fig. 1 D and E). This severing of dendrites was also observed in a recent study of da neuron remodeling (24). During the next 8 h, which coincided with head eversion during metamorphosis, these severed and blebbing dendrites were degraded (data not shown). By 20 h APF, the process of larval dendrite removal was complete, leaving C4da neurons with their axonal projections but devoid of larval dendrites (arrowheads, Fig. 1 F and G). Axons from all three C4da neurons project into the VNC and will be addressed later in this report. By this time, V′ and D neurons began to extend fine dendritic projections (Fig. 1 F and G Insets). The V neurons, which did not show new dendritic projections, disappeared between 30 and 35 h APF (data not shown), leaving V′ as the surviving neuron in the ventral hemisegment.
Compared with the rapid sequence of larval dendritic pruning, the process of pupal dendrite regrowth is slow. By 70 h APF, both V′ and D neurons began to take on the shape of their respective adult neurons (Fig. 1 H and I). By 95 h APF, shortly before adult eclosion, the dendritic patterns of abdominal V′ neurons closely resembled larval C4da neurons before pupariation (Fig. 1J). In contrast, the D neurons took on a more elongated dendritic field (Fig. 1K), perhaps reflecting a functional divergence between these two neurons in the adult fly. These results show that C4da neurons can completely degrade their elaborate larval dendrites during early metamorphosis, survive these changes, and subsequently regrow their dendritic arbors.
Dendrite Degradation Defects in Mmp Mutants During Metamorphosis. During Drosophila metamorphosis, most larval organs are replaced by adult structures. To understand the cellular environment during C4da dendrite degradation, we examined the expression of Armadillo, an adhesive junction protein that outlines the epithelial monolayer (25) during early metamorphosis. High-level Armadillo staining at the WP stage was completely abolished by 13 h APF but subsequently returned at 20 h APF when the pupal epithelium was reformed (data not shown). Thus, the pruning of C4da neuron dendrites occurred concurrently with epithelial remodeling during metamorphosis. To determine whether the degradation of larval dendrites is a result of local tissue remodeling or neuron-intrinsic signaling, we first focused on enzymes that are important for tissue remodeling.
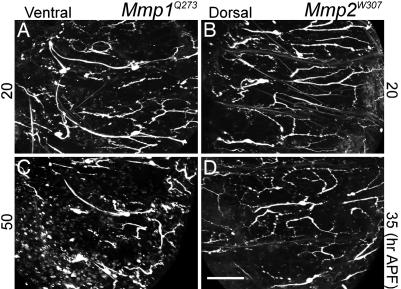
Drosophila matrix metalloproteases (metalloproteinases) Mmp1 and Mmp2 regulate tissue remodeling during metamorphosis (19). The weaker alleles of both genes, Mmp1Q273 and Mmp2W307, survive past head eversion to midpupariation (19), making it possible to visualize dendritic pruning of ppk-EGFP-expressing C4da neurons. Remarkably, there were abundant C4da neuron larval dendrites in both Mmp1 and Mmp2 mutants after head eversion (Fig. 2). Whereas in WT pupae at 20 h APF all larval dendrites from C4da neurons were cleared from the extracellular space (Fig. 1 F and G), in both Mmp1 and Mmp2 mutants, larval dendrites that were severed from the soma remained (Fig. 2 A and B). These larval dendrites persisted to much later stages at 50 and 35 h APF, just before the lethal phases of Mmp1Q273 and Mmp2W307 mutants, respectively (Fig. 2 C and D). The ineffective removal of larval dendrites in Mmp mutants was not caused by a generalized delay in metamorphosis because Mmp mutant pupae had completed head eversion and epidermal remodeling (data not shown), thus indicating a specific defect in dendrite degradation. Because Mmp1;Mmp2 double mutants do not survive to pupariation (19), it was not possible to look at dendritic pruning in the double mutant background.
Fig. 2.
Dendrite degradation defects in Mmp1 and Mmp2 mutants. Live confocal images of the ppk-EGFP transgene in Mmp1Q273 (A and C) and Mmp2W307 (B and D) backgrounds during pupariation are shown. Ventral and dorsal fields of mutants showed identical phenotype, so a representative is shown. (A and B) Twenty hours APF in Mmp1Q273 and Mmp2W307 backgrounds, respectively. (C) Fifty hours APF in Mmp1 mutant. (D) Thirty-five hours APF in Mmp2 mutant. Dorsal is right and anterior is up in all images. (Scale bar: 100 μm.)
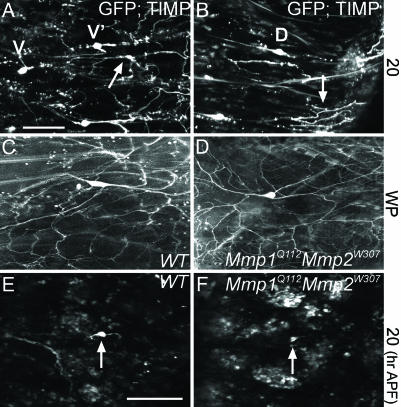
Extracellular Mmp Activity Is Required for Removal of Severed Dendrites. To determine whether Mmps functioned on the cell surface of dendrites to regulate degradation, we first expressed an Mmp inhibitor in C4da neurons to see whether the survival of larval dendrites can be prolonged. Using the ppk promoter to express transcriptional activator Gal4 (ppk-Gal4), we used the UAS-Gal4 system (26) to express the Drosophila tissue inhibitor of metalloproteases (TIMP) in C4da neurons. Fly TIMP is closely related to mammalian TIMP-3 (27), which associates with extracellular membrane surfaces to modulate Mmp activities (28). In control animals expressing GFP at 20 h APF, we saw identical pruning of larval dendrites as in ppk-EGFP flies (Fig. 1 F and G and data not shown). In contrast, when TIMP was overexpressed in C4da neurons, larval dendrites remained in the extracellular space at 20 h APF (arrows, Fig. 3 A and B).
Fig. 3.
Non-cell-autonomous requirement of Mmp activity in dendrite degradation. (A and B) Live confocal images of ppk-Gal4 driving expression of UAS-mCD8::GFP and UAS-Timp in C4da neurons at 20 h APF. C4da neurons are marked as V and V′ in ventral view (A) and as D in dorsal view (B). Arrows point to undegraded larval dendrites. (C-F) Live time-lapse images of WT and Mmp1Q112Mmp2W307 C4da MARCM clones. (C and D) WP stage. (E and F) Twenty hours APF. Arrows point to soma of C4da clones. Anterior is up and dorsal is right in all images. (Scale bars: 100 μm.)
The fact that TIMP inhibition can successfully delay the degradation of larval dendrites confirmed Mmp's involvement in this process. But these enzymes could be synthesized by either the C4da neurons or by the surrounding cells. To identify the source of this Mmp activity, we performed MARCM studies (29) to generate C4da clones that were deficient in both Mmps. Mmp1Q112Mmp2W307 double mutant C4da clones not only showed dendritic branching patterns similar to WT clones during early pupariation (Fig. 3 C and D), but live time-lapse imaging revealed complete larval dendrite removal after head eversion at 20 h APF just like WT controls (Fig. 3 E and F). These results showed that cell-intrinsic Mmps were not required for dendritic pruning and that extracellular Mmp activity was sufficient for degrading severed larval dendrites during metamorphosis. A possible source of this extracellular activity could be phagocytic blood cells, because they have been recently shown to engulf dendritic debris during metamorphosis (24).
EcR Activity Is Critical for the Initiation of Dendritic Remodeling. Whereas removal of severed dendrites required extrinsic metalloproteases, C4da neurons in Mmp mutants still retained their ability to sever larval dendrites from the soma during metamorphosis. To look for cell-intrinsic pathways in cleaving larval dendrites, we examined the role of ecdysone, a steroid hormone that regulates much of Drosophila metamorphosis (30). Binding of ecdysone to its nuclear receptor heterodimers, consisting of Ultraspiracle (Usp) and one of three EcR isoforms (EcR-A, EcR-B1, and EcR-B2), mediates a transcriptional hierarchy that regulates tissue responses during metamorphosis (31). To determine whether EcR signaling plays a role in initiating dendritic pruning, we first examined EcR expression patterns in the ppk-EGFP transgenic line that specifically labels C4da neurons.
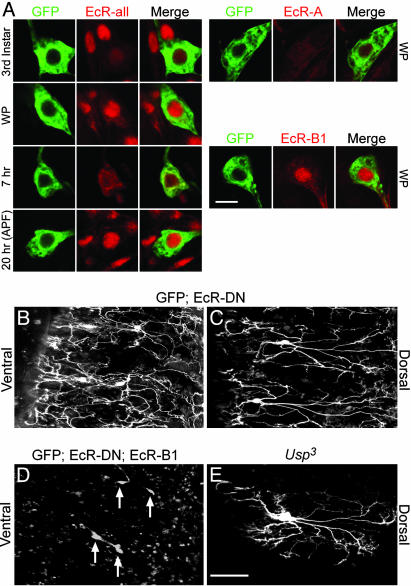
Fig. 4A shows staining with the EcR-C antibody (EcR-all), which recognizes the common regions of EcR family members, and staining with EcR-A and EcR-B1 specific antibodies during different stages of late larval through early pupal development (EcR-B2-specific antibody is not available). All three C4da neurons exhibited similar staining patterns, so a representative is shown for each time point. In third-instar larvae, when the ecdysone level is low before the onset of pupariation, EcR expression in C4da neurons was relatively low when compared with surrounding cells that already exhibit a high level of nuclear EcR (Fig. 4A, 3rd Instar). At the WP stage, with a transient rise in ecdysone level, EcR in C4da neurons became concentrated in the nucleus (Fig. 4A, WP). Over the next 7 h, EcR was gradually redistributed throughout the soma of C4da neurons (Fig. 4A, 7 hr), which corresponded to a rapid drop-off in ecdysone levels in the pupae after initiation of metamorphosis (30). Strong nuclear EcR localization in C4da neurons returned at 20 h APF [Fig. 4A, 20 hr (APF)], correlating with the onset of midpupal ecdysone release (30). Antibodies specific to either EcR-A or B1 showed that whereas EcR-A expression was diffuse and weak throughout metamorphosis, EcR-B1 expression in C4da neurons corresponded to the dynamic nuclear localization patterns seen with the EcR-C antibody (Fig. 4A and data not shown).
Fig. 4.
Ecdysone signaling is required for initiation of dendrite degradation during metamorphosis. (A) Antibody staining of ppk-EGFP fillets with anti-EcR (red) antibodies during late larval and early pupal development. EGFP (green) fluorescence comes from ppk-EGFP transgene expression. Columns show ppk-EGFP C4da neurons (GFP), anti-EcR staining alone (EcR-all, -A, and -B1), or merge of GFP and anti-EcR images. Rows show staining patterns for the following stages: third instar, WP, 7 and 20 h APF (EcR-all), and WP (EcR-A and -B1). A representative C4da neuron is shown for each time point. (B and C) Live confocal images of C4da neurons expressing both mCD8::GFP and EcR-DN proteins during pupariation. Shown are ventral and dorsal views, respectively, at 20 h APF. (D) Ventral live confocal view of C4da neurons expressing mCD8::GFP, EcR-DN, and WT EcR-B1 proteins under the control of ppk-Gal4 at 20 h APF. Arrows point to V and V′ C4da neurons. (E) Doral live confocal view of Usp MARCM C4da neuron at 20 h APF. Anterior is up and dorsal is right in B-E. (Scale bars: A, 5 μm; B-E, 100 μm.)
To examine the functional significance of EcR expression, we attempted to disrupt ecdysone signaling specifically in C4da neurons. EcR mutants either do not survive to the pupal stage or die shortly after the onset of metamorphosis (32); therefore, it was not possible to look at dendritic remodeling in those mutants. The cytological location of EcR genes also precluded MARCM studies (29); therefore, we used a set of previously described dominant-negative (DN) EcR proteins to inhibit EcR activity (33). Unlike control pupae (Fig. 1 F and G and data not shown), when ecdysone signaling was inhibited by EcR-DN proteins, C4da neurons lost their ability to initiate larval dendrite pruning at 20 h APF (Fig. 4 B and C). Two independent EcR-DN alleles showed identical phenotypes (Fig. 4 B and C and data not shown). To determine whether the defects were specific to the ecdysone signaling pathway, we first tried to rescue the EcR-DN phenotype. Coexpression of both EcR-DN and WT EcR-B1 proteins in C4da neurons resulted in complete rescue of dendritic pruning defects in all three neurons (arrows in Fig. 4D and data not shown). This rescue was complete with two copies of ppk-Gal4 in C4da neurons, showing that the rescue was not caused by reduced expression of DN protein in the coexpression experiments.
Because dimerization of EcR-B1 with its obligatory hormone receptor partner Usp (34) is essential for transcriptional regulation, we examined the involvement of Usp in dendrite remodeling. Usp mutants do not survive to metamorphosis (35); however, we were able to generate Usp MARCM clones for analysis. At 20 h APF, Usp mutant C4da clones failed to prune their larval dendrites (Fig. 4E), and this genetic mutation showed an identical phenotype to the EcR-DN experiments (Fig. 4C). Given the severity and full penetrance of this phenotype, together with the timing of EcR-B1 nuclear localization (Fig. 4A), we conclude that the ecdysone signaling pathway plays an important cell-intrinsic role in initiating dendritic pruning in C4da neurons during metamorphosis.
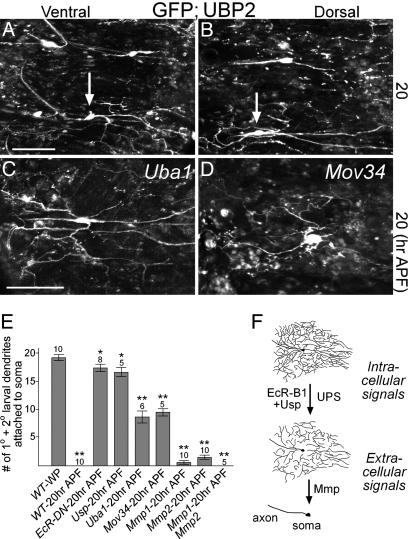
The Ubiquitin Proteasome System Is Required for Dendrite Breakage from Neurons. What might be the cellular machineries that carry out dendrite pruning in C4da neurons? One attractive model is a caspase-mediated local digestion and degradation of dendrites. However, overexpression of p35, an effective inhibitor of fly caspases (21), in C4da neurons did not prevent or delay larval dendrite degradation during metamorphosis (data not shown). Another protein degradation pathway, the UPS, has been shown to regulate both axon and dendrite pruning of mushroom body neurons during fly metamorphosis (11). To test the involvement of UPS in C4da neuron remodeling, we used ppk-Gal4 to overexpress UBP2, a yeast ubiquitin protease, in the C4da neurons. By reversing the process of substrate ubiquination, UBP2 is an effective UPS inhibitor in the fly (20). Compared with control neurons (Fig. 1 F and G and data not shown), some of the C4da neurons expressing UBP2 aberrantly retained their larval dendritic arbors (arrows, Fig. 5 A and B). Note that this pruning defect is very different from that seen in Mmp mutants (Fig. 2). Whereas Mmp mutants accumulated severed larval dendrites in the extracellular space, UBP2 inhibition prevented efficient severing of dendrites from the soma.
Fig. 5.
Requirement of the UPS pathway and quantitation of mutant phenotypes in C4da neurons during dendrite degradation. (A and B) Live confocal images of ppk-Gal4 driving expression of both UAS-mCD8::GFP and UAS-UBP2 in C4da neurons at 20 h APF. Arrows point to the V′ neuron in A and the D neuron in B, showing attached larval dendritic branches. (C and D) Live confocal images of Uba1 and Mov34 C4da MARCM clones at 20 h APF. Anterior is up and dorsal is right in all images. (E) Number of 1° and 2° larval dendrites attached to soma in WT and mutant C4da neurons during metamorphosis. Number of samples (n) in the group is above each bar. *, P < 0.05; **, P < 0.001, Wilcoxon two-sample test. Error bars represent SEM. (F) Regulation of large-scale dendritic pruning in C4da neurons during metamorphosis. (Scale bars: 100 μm.)
To further examine the involvement of the UPS machinery in dendritic pruning, we used the MARCM system to generate C4da clones that were either deficient in ubiquitin activation enzyme 1 (Uba1) or had mutation in the 19S particle of the proteasome (Mov34) (11). Time-lapse imaging of Uba1 and Mov34 mutant C4da clones at WP stage and 20 h APF showed that, unlike WT clones (Fig. 3E), both mutant clones failed to efficiently sever their larval dendrites during metamorphosis (Fig. 5 C and D and data not shown). These results confirmed the requirement for an activated UPS in the severing of larval dendrites from C4da neurons during metamorphosis.
To compare the defects in larval dendritic pruning caused by different mutations, we counted the number of large (primary and secondary) dendritic branches that remain attached to C4da neuron soma. In WT pupae at the start of metamorphosis, C4da neurons extended close to 20 large dendritic branches, none of which was retained after head eversion at 20 h APF (Fig. 5E). Mutations that disrupted ecdysone signaling, such as EcR-DN expression or Usp-deficient clones, resulted in the retention of 85-90% of large dendritic branches after head eversion (Fig. 5E). Mutations in the UPS pathway, such as Uba1 and Mov34, resulted in the retention of 45-49% of large dendritic branches at 20 h APF (Fig. 5E). Mmp1 or Mmp2 mutants only retained 3-8% of large dendritic branches after head eversion, and Mmp1;Mmp2 mutant clones did not retain larval dendrites at 20 h APF (Fig. 5E). These data suggest that dendrite remodeling in C4da neurons starts with signals from ecdysone and UPS that result in the cleavage of larval dendrites from the soma, which then allows for the degradation of severed dendrites by Mmp activity in the extracellular matrix (Fig. 5H).
It is possible that UPS is an upstream regulator of EcR and can lead to EcR misexpression in UPS mutants; however, we observed normal EcR expression patterns in both Uba1 and Mov34 C4da MARCM clones (data not shown). It is also conceivable that EcR signaling is upstream of the UPS cascade, but this idea is difficult to demonstrate experimentally. We reasoned that in this case, inhibition of EcR signaling should result in lower levels of protein ubiquination. However, staining in EcR-DN-expressing C4da neurons showed no significant differences in the level of ubiquitin/polyubiquitin between undegraded larval dendrites and WT dendrites before degradation (data not shown). This finding does not rule out an EcR function upstream of UPS during dendritic remodeling, because EcR signaling may regulate critical factors in the UPS cascade after protein ubiquination at the level of ubiquitin ligases (36). The identities of such ligases are currently unknown.
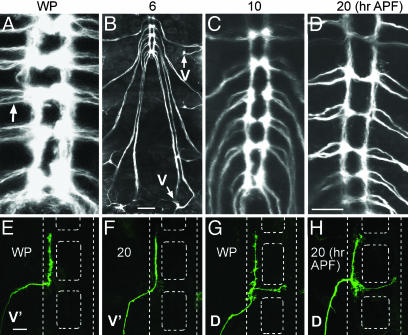
C4da Neurons Retain Larval Axons During Dendrite Pruning. To test whether dendritic pruning in C4da neurons involves concurrent axonal remodeling as described in refs. 8 and 10-12, we looked at axon tracks of C4da neurons in the Drosophila VNC during early metamorphosis. Direct live imaging of the ppk-EGFP transgenic line at the WP stage showed axon tracks from three C4da neurons (arrow, Fig. 6A). Axon tracing of EGFP-expressing C4da neurons at 6 h APF showed continuous axon tracks between all three C4da neurons and the VNC (arrows in Fig. 6B and data not shown). At 10 h APF, the VNC appeared more compact, presumably as a result of the various remodeling events that occurred in the nervous system during metamorphosis (Fig. 6C). At 20 h APF, axon tracks of EGFP-expressing C4da neurons could still be clearly identified at the VNC (Fig. 6D) and were continuous with the soma (data not shown), despite the complete removal of dendritic arbors of these same neurons (Fig. 1 F and G).
Fig. 6.
VNC axon projections of C4da neurons during metamorphosis. (A) Direct live confocal image of VNC in ppk-EGFP pupae at WP stage. Arrow points to axons from three C4da neurons entering the VNC per hemisegment. (B) Six hours APF. Photo-collage of live z-stack confocal images tracing ventral axon projections from ppk-EGFP-positive C4da neurons. Arrows point to the soma of V C4da neurons. For clarity, surface dendrites were not projected. (C and D) Z-stack confocal images of axonal projections from ppk-EGFP-positive C4da neurons into the VNC at 10 and 20 h APF, respectively. (E-H) Single V′ and D C4da neuron clones expressing mCD8::GFP stained with anti-mCD8a antibody. (E and F) V′ C4da neuron axon terminals at WP stage and 20 h APF, respectively. (G and H) D C4da neuron axon terminals at WP stage and 20 h APF, respectively. White dashed lines represent locations of the VNC. Anterior is up in all images. (Scale bars: A, C, and D, 50 μm; B, 100 μm; E-H, 15 μm.)
Drosophila peripheral sensory neurons generally have simple axon projections into the VNC that terminate locally (37). To visualize C4da neuron axon terminals during metamorphosis, we used the UAS>CD2>CD8-GFP system (23), together with ppk-Gal4, to generate single clones of surviving V′ and D neurons. Consistent with previous observations (37), the V′ C4da neuron projected its axon ipsilaterally upon entering the VNC to the segment immediately anterior during the WP stage (Fig. 6E). At 20 h APF, after complete pruning of larval dendrites, the V′ C4da neuron kept this axonal projection intact in the VNC (Fig. 6F). The D C4da neuron axon, in addition to having an ipsilateral branch that projected to the anterior segment, sent a commissural branch that crossed the midline at the segment where the axon entered the VNC (Fig. 6G). Likewise, at 20 h APF, the D C4da neuron kept both axonal terminal branches intact (Fig. 6H). These data showed that C4da neurons did not significantly modify their larval axons during concurrent dendrite degradation, despite the activated ecdysone and UPS pathways, which are known to facilitate axon remodeling and degradation (10, 11).
What might account for the dendrite-specific remodeling in C4da neurons, as opposed to the previously reported concurrent remodeling of both axons and dendrites (8, 10-12)? It is possible that local environments may play a role. A recent study in Manduca found central versus peripheral hormonal differences affecting axon versus dendrite remodeling (38). However, it remains to be tested whether the ecdysone levels are different in the fly epidermis and the VNC during metamorphosis. Anatomically, C4da neurons have distinct axon-dendrite polarity in that the cell bodies send out multiple primary dendritic arbors to the surrounding environment while each extends a single axon toward the VNC. This morphology is in contrast to most insect neurons, such as femoral depressor motoneurons and mushroom body γ-neurons, which extend a single branch from the soma that later gives rise to both dendrites and axons. As such, C4da neurons may have developed separate mechanisms at the soma to remodel just the dendrites. Just what these mechanisms might include is currently unknown.
Implications for Dendritic-Specific Remodeling. We have provided evidence that certain mature neurons have the ability to selectively degrade their dendritic projections in vivo and regrow new ones. Although fly metamorphosis is a specialized developmental process, dendrite-specific remodeling may provide a paradigm for neurons to retain part of their connections in the neuronal circuitry while responding to environmental changes such as tissue degeneration near their dendrites. Certain conditions in mammalian systems, such as trauma and injury, can induce localized degeneration and remodeling and may mimic the active tissue remodeling during metamorphosis. In the human CNS, for example, significant reorganization of granule cell projections in the dentate gyrus after human temporal lobe epilepsy has been observed (39). Thus, it would be of great interest to examine whether dendritic-specific remodeling of C4da neurons in Drosophila represents an evolutionarily conserved mechanism for neurons to respond to drastic changes in their environment, and to determine whether mammalian neurons have similar capacities to remodel their dendrites.
Acknowledgments
We thank A. Page-McCaw, L. Cherbas (Indiana University, Bloomington), and L. Luo (Stanford University, Stanford, CA) for generously providing fly stocks; R. Yang for donating ppk-Gal4; W. Grueber for donating ppk-Gal4, UAS>CD2>CD8-GFP, hsFLP; the Bloomington Stock Center and Developmental Studies Hybridoma Bank for providing fly stocks and antibodies; R. Watts for helpful suggestions on UPS studies; and S. Younger and members of L.Y.J. and Y.N.J.'s laboratory for expertise and advice throughout this work. This work was supported by National Institutes of Health Grant R01 NS40929 (to Y.N.J.). C.T.K. is a Howard Hughes Medical Institute (HHMI) postdoctoral fellow. Y.N.J. and L.Y.J. are HHMI Investigators.
Author contributions: C.T.K. designed research; C.T.K. performed research; C.T.K., L.Y.J., and Y.N.J. analyzed data; C.T.K., L.Y.J., and Y.N.J. wrote the paper; and Y.N.J. was the principal investigator.
Abbreviations: da, dendritic arborization; C4da, class IV da; UPS, ubiquitin-proteasome system; UAS, upstream activating sequence; MARCM, mosaic analysis with a repressible cell marker; VNC, ventral nerve cord; EcR, ecdysone receptor; WP, white pupae; APF, after puparium formation; TIMP, tissue inhibitor of metalloproteases; DN, dominant negative; Uba1, ubiquitin activation enzyme 1.
References
- 1.Scott, E. K. & Luo, L. (2001) Nat. Neurosci. 4, 359-365. [DOI] [PubMed] [Google Scholar]
- 2.Jan, Y. N. & Jan, L. Y. (2003) Neuron 40, 229-242. [DOI] [PubMed] [Google Scholar]
- 3.Cline, H. T. (2001) Curr. Opin. Neurobiol. 11, 118-126. [DOI] [PubMed] [Google Scholar]
- 4.Chklovskii, D. B., Mel, B. W. & Svoboda, K. (2004) Nature 431, 782-788. [DOI] [PubMed] [Google Scholar]
- 5.Li, Z. & Sheng, M. (2003) Nat. Rev. Mol. Cell. Biol. 4, 833-841. [DOI] [PubMed] [Google Scholar]
- 6.Collingridge, G. L., Isaac, J. T. & Wang, Y. T. (2004) Nat. Rev. Neurosci. 5, 952-962. [DOI] [PubMed] [Google Scholar]
- 7.Truman, J. W. (1990) J. Neurobiol. 21, 1072-1084. [DOI] [PubMed] [Google Scholar]
- 8.Truman, J. W. & Reiss, S. E. (1995) J. Neurosci. 15, 4815-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng, X., Wang, J., Haerry, T. E., Wu, A. Y., Martin, J., O'Connor, M. B., Lee, C. H. & Lee, T. (2003) Cell 112, 303-315. [DOI] [PubMed] [Google Scholar]
- 10.Lee, T., Marticke, S., Sung, C., Robinow, S. & Luo, L. (2000) Neuron 28, 807-818. [DOI] [PubMed] [Google Scholar]
- 11.Watts, R. J., Hoopfer, E. D. & Luo, L. (2003) Neuron 38, 871-885. [DOI] [PubMed] [Google Scholar]
- 12.Marin, E. C., Watts, R. J., Tanaka, N. K., Ito, K. & Luo, L. (2005) Development (Cambridge, U.K.) 132, 725-737. [DOI] [PubMed] [Google Scholar]
- 13.Gao, F. B., Brenman, J. E., Jan, L. Y. & Jan, Y. N. (1999) Genes Dev. 13, 2549-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grueber, W. B., Jan, L. Y. & Jan, Y. N. (2002) Development (Cambridge, U.K.) 129, 2867-2878. [DOI] [PubMed] [Google Scholar]
- 15.Shepherd, D. & Smith, S. A. (1996) Development (Cambridge, U.K.) 122, 2375-2384. [DOI] [PubMed] [Google Scholar]
- 16.Williams, D. W. & Shepherd, D. (1999) J. Neurobiology 39, 275-286. [PubMed] [Google Scholar]
- 17.Williams, D. W. & Truman, J. W. (2004) J. Neurosci. 24, 1541-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grueber, W. B., Ye, B., Moore, A. W., Jan, L. Y. & Jan, Y. N. (2003) Curr. Biol. 13, 618-626. [DOI] [PubMed] [Google Scholar]
- 19.Page-McCaw, A., Serano, J., Sante, J. M. & Rubin, G. M. (2003) Dev. Cell 4, 95-106. [DOI] [PubMed] [Google Scholar]
- 20.DiAntonio, A., Haghighi, A. P., Portman, S. L., Lee, J. D., Amaranto, A. M. & Goodman, C. S. (2001) Nature 412, 449-452. [DOI] [PubMed] [Google Scholar]
- 21.Hay, B. A., Wolff, T. & Rubin, G. M. (1994) Development (Cambridge, U.K.) 120, 2121-2129. [DOI] [PubMed] [Google Scholar]
- 22.Emoto, K., He, Y., Ye, B., Grueber, W. B., Adler, P. N., Jan, L. Y. & Jan, Y. N. (2004) Cell 119, 245-256. [DOI] [PubMed] [Google Scholar]
- 23.Wong, A. M., Wang, J. W. & Axel, R. (2002) Cell 109, 229-241. [DOI] [PubMed] [Google Scholar]
- 24.Williams, D. W. & Truman, J. W. (2005) Development (Cambridge, U.K.) 132, 3631-3642. [DOI] [PubMed] [Google Scholar]
- 25.Muller, H. A. & Wieschaus, E. (1996) J. Cell. Biol. 134, 149-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401-415. [DOI] [PubMed] [Google Scholar]
- 27.Wei, S., Xie, Z., Filenova, E. & Brew, K. (2003) Biochemistry 42, 12200-12207. [DOI] [PubMed] [Google Scholar]
- 28.Yu, W. H., Yu, S., Meng, Q., Brew, K. & Woessner, J. F., Jr. (2000) J. Biol. Chem. 275, 31226-31232. [DOI] [PubMed] [Google Scholar]
- 29.Lee, T. & Luo, L. (1999) Neuron 22, 451-461. [DOI] [PubMed] [Google Scholar]
- 30.Riddiford, L. M. (1993) in The Development of Drosophila melanogaster, eds. Bate, M. & Martinez Arias, A. (Cold Spring Harbor Lab. Press, Woodbury, NY), Vol. 2, pp. 899-939. [Google Scholar]
- 31.Thummel, C. S. (1996) Trends Genet. 12, 306-310. [DOI] [PubMed] [Google Scholar]
- 32.Bender, M., Imam, F. B., Talbot, W. S., Ganetzky, B. & Hogness, D. S. (1997) Cell 91, 777-788. [DOI] [PubMed] [Google Scholar]
- 33.Cherbas, L., Hu, X., Zhimulev, I., Belyaeva, E. & Cherbas, P. (2003) Development (Cambridge, U.K.) 130, 271-284. [DOI] [PubMed] [Google Scholar]
- 34.Yao, T. P., Segraves, W. A., Oro, A. E., McKeown, M. & Evans, R. M. (1992) Cell 71, 63-72. [DOI] [PubMed] [Google Scholar]
- 35.Henrich, V. C., Szekely, A. A., Kim, S. J., Brown, N. E., Antoniewski, C., Hayden, M. A., Lepesant, J. A. & Gilbert, L. I. (1994) Dev. Biol. 165, 38-52. [DOI] [PubMed] [Google Scholar]
- 36.Ciechanover, A. & Brundin, P. (2003) Neuron 40, 427-446. [DOI] [PubMed] [Google Scholar]
- 37.Merritt, D. J. & Whitington, P. M. (1995) J. Neurosci. 15, 1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knittel, L. M. & Kent, K. S. (2005) J. Neurobiol. 63, 106-125. [DOI] [PubMed] [Google Scholar]
- 39.Parent, J. M. & Lowenstein, D. H. (1997) Curr. Opin. Neurol. 10, 103-109. [DOI] [PubMed] [Google Scholar]