Abstract
In eukaryotes, a single translational release factor, eRF1, deciphers three stop codons, although its decoding mechanism remains puzzling. In the ciliate Tetrahymena thermophila, UAA and UAG codons are reassigned to Gln codons. A yeast eRF1-domain swap containing Tetrahymena domain 1 responded only to UGA in vitro and failed to complement a defect in yeast eRF1 in vivo at 37°C. This finding demonstrates that decoding specificity of eRF1 from variant code organisms resides at domain 1. However, the wild-type eRF1 hybrid fully restored the growth of eRF1-deficient yeast at 30°C. Tetrahymena eRF1 contains a variant sequence, KATNIKD, at the tip of domain 1. The TASNIKD variant of hybrid eRF1 rendered the eRF1-nullified yeast viable, although in an in vitro assay, the same hybrid eRF1 responded only to UGA. Nevertheless, the yeast eRF1 bearing the KATNIKD motif instead of the TASNIKS heptapeptide present in higher eukaryotes remains omnipotent in vivo. Collectively, these data suggest that variant genetic code organisms like Tetrahymena have an intrinsic potential to decode three stop codons in vivo, and that interaction within domain 1 between the KAT tripeptide and other sequences modulates the decoding specificity of Tetrahymena eRF1.
The termination of protein synthesis takes place on the ribosomes in response to a stop codon, rather than a sense codon, at the “decoding” site. Polypeptide release factors (RFs) are essential to this process. Prokaryotes generally have two codon-specific factors that have overlapping specificities: RF1 recognizes UAA and UAG, and RF2 recognizes UAA and UGA (1). In contrast, eukaryotic eRF1s from organisms with a canonical genetic code recognize all three stop codons (2). By virtue of their functions, RFs have long been thought to mimic tRNA (3, 4). Recently, a functional mimic of the anticodon of tRNA—referred to as the tripeptide anticodon—in RF1 and RF2, which is responsible for stop codon recognition, has been identified (5). The recognition of stop codons by eRF1, however, remains unknown.
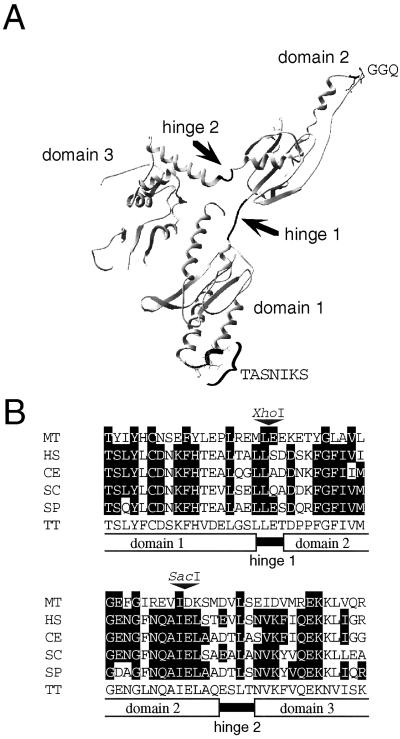
The crystal structure of human eRF1 to 2.8 Å has been published (6). It was pointed out that the overall shape and dimensions of eRF1 resemble those of a tRNA molecule, with domains 1 and 2 of eRF1 corresponding to the tRNA's anticodon stem and aminoacyl acceptor stem, respectively (see Fig. 1A). This domain assignment relies on the assumptions that the universal GGQ motif (7) located at the tip of domain 2 is a structural counterpart of the tRNA aminoacyl group on the CCA-3′ acceptor stem, and that domain 1, in which a codon-specific defect can be created (8), may be equivalent to the anticodon arm of tRNA (6). Of these three domains, domain 3 is known to interact with eRF3 (9–11).
Figure 1.
Strategy of domain swapping between Sp-eRF1 and Tt-eRF1. (A) The three-dimensional structure of human eRF1 (PDB ID code 1DT9). Each domain was swapped at the hinge regions indicated by arrows. The TASNIKS heptapeptide and the GGQ tripeptide are shown at the tips of domains 1 and 2, respectively. (B) Conserved amino acid sequences at the junctions of domains 1–2 and 2–3 of eRF1s. Amino acids identical to those of Tetrahymena eRF1 are shown by outlined characters; restriction enzyme sites used for domain swapping are shown by triangles. Species abbreviations: MT, Methanobacterium thermoautotrophicum; HS, Homo sapiens; CE, S. cerevisiae; SP, S. pombe; and TT, T. thermophila.
In contrast to the bacterial RFs, the omnipotence of eRF1s in deciphering stop codons impedes the identification of a “functional anticodon” moiety, if any, of eRF1s in eukaryotes. Capture of an eRF1 variant of stop-codon selectivity or of preference in other organisms would facilitate the study of eRF1s. Ciliates might provide us with such a tool based on the fact that some of them are known to possess UAA and UAG (or UGA) reassigned as a sense codon instead of a stop codon during evolution; for example, in Euplotes octacarinatus, UGA is decoded as Cys (12), and UAA and UAG are decoded as Gln in Tetrahymena thermophila (13–16). Kervestin et al. (17) showed recently, in an in vitro assay based on mammalian ribosomes, that eRF1 from the ciliate Euplotes aediculatus responds to UAA and UAG as stop codons and lacks the capacity to decipher UGA, which encodes Cys in this organism. This finding is the first in vitro indication of an eRF1 variant of stop codon selectivity.
Ciliate eRF1 genes have been cloned from T. thermophila (18), E. octacarinatus (19), E. aediculatus (17, 20), and Oxytricha trifallax (UAA and UAG for Gln; ref. 20). Extensive comparisons between universal-code eRF1s and variant-code eRF1s from ciliates highlight the sequence variations at several regions (17, 21), one of which is the tip region of domain 1; the TASNIKS heptapeptide (and its surrounding peptide) sequence is highly conserved in universal-code eRF1s but differs significantly from ciliate eRF1s (20). The ciliate-specific diversity at TAS and NIKS sites also has been pointed out independently (1, 22, 23). Recently, the role of NIKS motif in RF activity and ribosome binding has been shown for human eRF1 in in vitro experiments (24). Therefore, it is tempting to speculate that the TASNIKS heptapeptide region is functionally essential and can modulate stop codon discrimination in eukaryotes.
In this study, we examined whether domain 1 of eRF1 and the TASNIKS heptapeptide are involved in the recognition of stop codons by using Tetrahymena eRF1 (referred to as Tt-eRF1). The authentic Tt-eRF1 is unable to catalyze polypeptide termination in vitro with mammalian ribosomes because of its inefficient binding to the heterologous ribosomes (18). Therefore, based on the structural information, domains 1–3 were swapped between Tt-eRF1 and the fission yeast Schizosaccharomyces pombe eRF1 (referred to as Sp-eRF1; ref. 9), and the activity of the wild-type and tripeptide-variant hybrids was examined. The data clearly indicated that domain 1 is responsible for the deciphering of stop codons on the ribosome, and that the decoding capacity can be modulated by interaction between the TAS tripeptide and other sequences within the domain 1 of eRF1.
Materials and Methods
Strains, Plasmids, Chemicals, and Genetic Manipulations.
The Saccharomyces cerevisiae strain used was MT557/1d MATa sal4–2 (= sup45 ts) ura3–1 ade2–1 leu2–3,112 (25). A LEU2-insertion knockout allele (Δsup45∷LEU2) of S. cerevisiae SUP45 was constructed by substituting the LEU2 marker for the HpaI-EcoRV segment of SUP45 cloned in plasmid pUKC802 (26). Sp-eRF1 and Tt-eRF1 genes were recloned from pET-Sp-eRF1 (9) and pTT-eRF1–4/12 (18), respectively, into the BamHI or BamHI-EcoRI site of the URA3-marked centromere plasmid pYX112 (Novagen), in which the SUP45 sequences were placed under the TPI promoter. Yeast cultures were grown in yeast extract/peptone/dextrose liquid medium, or in synthetic minimal (SD) or synthetic complete media, as described (27). Bacteria were grown in LB broth (28) supplemented with the relevant antibiotics for selection (50 μg/ml ampicillin or 50 μg/ml kanamycin). l-[35S]methionine was purchased from NEN, and AUG and tetraplets containing stop codons and UGGA were synthesized by A. Veniaminova and M. Ryabkova (Institute of Biorganic Chemistry, Novosibirsk, Russia).
eRF1 Domain Swapping.
Intervals of domains 1–3 of Sp-eRF1 and Tt-eRF1 were marked with restriction enzyme sites, i.e., XhoI and SacI sites at 1–2 and 2–3 junctions, respectively, by using designed PCR primers (see Fig. 1). Each domain fragment was amplified by PCR by using the following primers: Tt-eRF1 domain 1, 5′-GGGGAATTCTCTAGAACCATGGAAGAGAAAGATCAACGT-3′ and 5′-CCCCTCGAGAAGTGAACCCAATTCATCAAC-3′; Tt-eRF1 domain 2, 5′-CCCCTCGAGACCGACCCTCCTTTTGGTTTC-3′ and 5′-GGGGAGCTCAATAGCTTGGTTAAGACCATTTTC-3′; Tt-eRF1 domain 3, 5′-GGGGAGCTCGCTCAAGAATCTTTAACTAACGTC-3′ and 5′-GGGGAGCTCTTATATGAAGCCTTCTTCTTCTTCGTAG-3′; Sp-eRF1 domain 1, 5′-GGGAATTCTCTAGAACCATGGATGAGACTGCTGAGAAAGCTATCG-3′ and 5′-CCCCTCGAGCAATTCTGCTAAAGCTTCAGT-3′; Sp-eRF1 domain 2, 5′-CCCCTCGAGAGTGATCAACGCTTCGGATTT-3′ and 5′-GGGGAGCTCTATAGCCTGGTTAAAACCAGC-3′; Sp-eRF1 domain 3, 5′-GGGGAGCTCGCTGCTGATACTTTGTCAAAT-3′ and 5′-GGGGAGCTCAAATTAGTCGGAGTCGGA-3′. The amplified DNAs of domains 1 through 3 were digested by NcoI-XhoI, XhoI-SacI, and SacI, respectively, and ligated into NcoI-SacI sites of pYX112 to give rise to hybrid eRF1 genes. One of the hybrids composed of Tetrahymena domain 1 and Schizosaccharomyces domains 2–3 was referred to as ΨeRF1 and examined in this study.
Site-Directed Mutagenesis.
Domain 1 variants of ΨeRF1 were constructed by site-directed mutagenesis via PCR by using the common primer 5′-CCCCTCGAGAAGTGAACCCAATTCATCAAC-3′ and the following primers containing relevant substitutions, designed according to the standard method (28): ΨeRF1 (TASNIKS) variant, 5′-GGGGAATTCAGTACGGCCTCTAATATTAAAUCCAGAGTCAACCGTCAATCTG-3′; ΨeRF1 (TASNIKD) variant, 5′-GGGGAATTCAGTACGGCCTCTAATATTAAAGACAGAGTCAACCGTCAATCT- G-3′; ΨeRF1 (TATKIKD) variant, 5′-GGGGAATTCAG- TACGGCCACTAATATTAAAGACAGAGTCAACCG- TCAATCTG-3′; and ΨeRF1 (KASNIKD) variant, 5′-GGG- GAATTCAGTAAGGCCTCTAATATTAAAGACAGA- GTCAACCGTCAATCTG-3′. The amplified DNAs were digested with EcoRI and XhoI and ligated into the same restriction sites of pYX112-ΨeRF1.
Expression and Purification of eRF1s.
The cDNA encoding the full-length human eRF1 was inserted into the NdeI-XhoI sites of the expression vector pET23b(+) (Novagen). The human eRF1 containing a His-tag at the C terminus was expressed in Escherichia coli and purified by using a metal affinity column, as described (7, 11). An Sp-eRF1 overexpression plasmid was constructed by subcloning the NcoI-NheI segment of Sp-eRF1 into the NcoI-BamHI sites of pET15b (Novagen) by linker ligation to give rise to pET15b-Sp-eRF1. The EcoRI-Bpu1102I segments carrying the wild-type and mutant ΨeRF1 sequences were substituted for the equivalent segment in the pET30b-based Tt-eRF1 expression plasmid, pTT-eRF1–38/1 (29), to give rise to pET30b-ΨeRF1 and its mutant derivatives. E. coli strain BL21 (DE3) was transformed with pET15b-Sp-eRF1 and pET30bΨeRF1, and the transformants were grown at 37°C in 0.2 liter of LB medium containing ampicillin (100 μg/ml) until A600 0.7 was reached. After addition of isopropyl β-d-thiogalactoside (IPTG) to a final concentration of 0.4 mM, the cells were grown for 3–4 h at 25°C and harvested by centrifugation. In the case of wild-type and mutant ΨeRF1s, the majority of the expressed proteins were in insoluble form (inclusion bodies), and the soluble fraction contained less than 10–15% of the total expressed ΨeRF1s. Cells were lysed by ultrasonication in 15 ml of buffer A (50 mM Hepes, pH 7.0/0.05 M KCl/0.1 mM EDTA) containing 10% (vol/vol) glycerol, 0.1% Nonidet P-40, 2 mM β-mercaptoethanol, 1 mM PMSF, and Protease Inhibitor Mixture (Roche Molecular Biochemicals). The lysate was centrifuged at 10,000 × g for 20 min at 4°C, and the supernatant was applied to a 5-ml HiTrap SP Sepharose HP column (Amersham Pharmacia) equilibrated with buffer A. The column was extensively washed with buffer A, and bound eRF1 was eluted with a linear KCl gradient (40 ml) from 50 to 800 mM in buffer A by using the FPLC System (Amersham Pharmacia). Fractions (1.0 ml) were collected, and 15-μl aliquots were analyzed by SDS/10% PAGE. Fractions containing eRF1 were combined, dialyzed against buffer B (0.05 M Tris⋅HCl, pH 8.0/50 mM KCl/1 mM EDTA) and applied to a 1-ml HiTrap Q Sepharose HP column (Amersham Pharmacia) equilibrated with buffer B. The column was washed with 20 ml of buffer B, and the proteins were eluted with a linear KCl gradient (20 ml) from 50 to 800 mM in buffer B. Fractions (0.5 ml) were collected, and 10-μl aliquots from the gradient fractions were analyzed by SDS/10% PAGE. Fractions containing eRF1 were combined and concentrated by using an Ultrafree-4 centrifugal filter unit Biomax-10 (Millipore).
Ribosomes and in Vitro RF Assay.
Rabbit reticulocyte ribosomal subunits were kindly provided by P. Simonenko (Institute of Protein Research, Pushchino, Russia) and purified according to the published method (30) as described (17). The eRF1 activity was measured at 25°C as described (17).
Results
Domain Swapping Between Tetrahymena and Fission Yeast eRF1 Proteins.
Domains 1 through 3 of eRF1 are structurally separated and connected by hinges 1 and 2 (Fig. 1A). These hinges and their adjacent sequences are relatively conserved in Tt-eRF1, Sp-eRF1, and other eRF1s (Fig. 1B). Hence, restriction enzyme sites were marked at these conservative sites to give rise to XhoI (at the junction of domains 1 and 2) and SacI (at the junction of domains 2 and 3). Thus, the hybrid eRF1s having domains swapped at these sites may not suffer from any sequence (hence, topological) disorder. Combinatory sets of six eRF1 hybrids were examined for their ability to complement a temperature-sensitive eRF1 mutant of the budding yeast S. cerevisiae (MT557/1d sup45 ts; ref. 25). Of these, the Sp-eRF1 derivative whose domain 2 was substituted with Tetrahymena domain 2 maintained the ability to restore the growth of the MT557/1d strain at 37°C. This result suggests that domain 2, a speculated mimic of the acceptor stem of tRNA, interacts with the conserved region of (heterologous) ribosomes. In contrast, the other hybrid eRF1s failed to complement the sup45 ts allele (data not shown). Taking these findings into consideration, a hybrid eRF1 construct composed of Tetrahymena domain 1, a speculated mimic of the anticodon arm of tRNA, and Schizosaccharomyces domains 2–3—hereafter referred to as ΨeRF1—were used as a parental construct to investigate the RF activity in a heterologous system.
Release Activity of Hybrid eRF1 in Vitro.
The release activity of the purified human eRF1, Sp-eRF1, and wild-type ΨeRF1 was measured with the three stop codons and the near-cognate tryptophan UGG codon in an in vitro RF assay. As was determined in a previous study (31), human eRF1 in the given assay system responded to the three stop codons (Table 1). Sp-eRF1 responded similarly. However, under the same conditions, ΨeRF1 responded only to UGA but not to UAG or to UAA, which encodes Gln in Tetrahymena. No activity was observed with the sense UGG codon by human eRF1, Sp-eRF1, or wild-type ΨeRF1 (Table 1), indicating the maintenance of the discriminating capacity of human, Schizosaccharomyces, and hybrid eRF1s toward the near-cognate codon. Swapping of Tetrahymena domains 2 and 3 for those of S. pombe allowed proper, but slightly (or insignificantly) lower, interaction with rabbit ribosomes, compared with interaction with human eRF1. These results demonstrate that, at least in the in vitro heterologous system, swapping Schizosaccharomyces domain 1 for Tetrahymena domain 1 could switch recognition specificity from omnipotence to UGA only. This finding strongly suggests the decoding capacity of domain 1.
Table 1.
Release activity of fission yeast eRF1 and wild-type and mutant ΨeRF1 proteins in an in vitro RF assay
| eRF1 | f[35S]Met released, cpm
|
|||
|---|---|---|---|---|
| UGAA | UAGA | UAAA | UGGA | |
| Human eRF1 | 4840 | 4850 | 4340 | 0 |
| S. pombe eRF1 | 4000 | 3800 | 3700 | 0 |
| Hybrid ΨeRF1 | ||||
| Wild type (KATNIKD) | 4200 | 0 | 0 | 0 |
| Mutant (KASNIKD) | 4500 | 0 | 0 | 0 |
| Mutant (TATNIKD) | 4300 | 0 | 0 | 0 |
| Mutant (TASNIKD) | 3800 | 0 | 0 | 0 |
| Mutant (TASNIKS) | 2000 | 0 | 0 | 0 |
The background level varied from 600 to 1200 cpm depending on the eRF1 preparation present in the incubation mixture. Zero means that the amount of cpm was ±15% of the background level. Average values from three independent experiments are presented.
In Vivo Complementation Activity of Hybrid eRF1 Variants.
The Sp-eRF1 and wild-type ΨeRF1 genes were cloned to plasmid pYX112 (under the TPI promoter) and transformed into the sup45 ts strain (MT557/1d). Ura+ transformants were selected at permissive temperature 30°C and examined for their growth at nonpermissive temperature 37°C. As shown in Fig. 2B Upper, Sp-eRF1-expressing transformants grew normally at 37°C, whereas ΨeRF1-expressing transformants failed to grow under the same condition. This finding was consistent with the in vitro finding of the UGA-specific decoding capacity of ΨeRF1, which could not compensate for the disabled decoding of UAG and UAA codons in the sup45 ts strain.
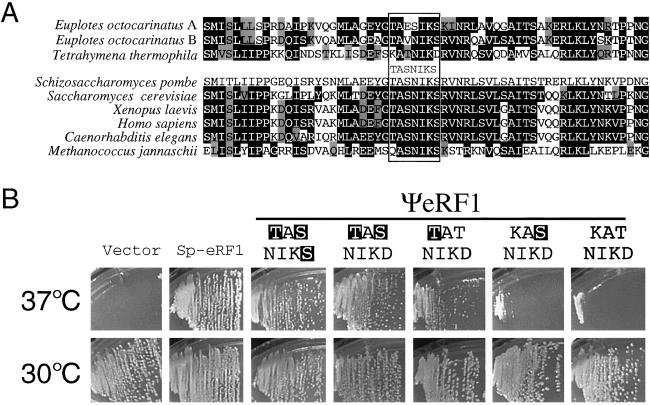
Figure 2.
In vivo complementation test of the temperature-sensitive eRF1 (sup45 ts) strain of S. cerevisiae by wild-type and variant hybrid eRF1s. (A) Amino acid sequence comparison of the TASNIKS heptapeptide (boxed) and surrounding regions of eRF1s from human, yeast, and ciliates. Residues identical and similar to those in S. pombe eRF1 are shown by outlined characters and gray boxes, respectively. (B) Growth of transformants at permissive (30°C) and nonpermissive (37°C) temperatures. MT557/1d (sup45 ts) cells were transformed with pYX112 derivatives encoding Sp-eRF1 and wild-type or variant ΨeRF1 proteins. Ura+ transformants were selected at 30°C, and their growth was monitored at 37°C. Amino acid changes introduced into the wild-type (KATNIKD) heptapeptide of ΨeRF1 are shown by outlined characters.
The Tetrahymena-specific KATNIKD heptapeptide sequence differs in three residues from that (i.e., TASNIKS) of the universal-code eRF1s (see Fig. 2A). Four ΨeRF1 variants were made, in which the heptapeptide was changed to KASNIKD, TATNIKD, TASNIKD, and TASNIKS, respectively, by the PCR manipulation using the designed primers. pYX112 derivatives encoding these variant ΨeRF1s were transformed into the sup45 ts strain, and the transformant growth was monitored at 37°C. Quite evidently, the TASNIKD variant rendered the sup45 ts strain perfectly viable, and the additional D→S change (i.e., TASNIKS) only slightly enhanced the growth (Fig. 2B Upper). It is noteworthy that the single substitution variants, KASNIKD and TATNIKD, restored, not perfectly but significantly, the viability of the sup45 ts strain at 37°C. These results suggest that the primary, but not entire, cause of the UAG/UAA-blindness of ΨeRF1, and thereby of Tetrahymena eRF1, could be a TAS-to-KAT change at the tripeptide element.
When the reciprocal, TAS→KAT, change was introduced into the S. pombe eRF1, the resulting KAT variant of Sp-eRF1 was still able to restore the viability of the sup45 ts strain at 37°C (data not shown). Therefore, the tripeptide variation can change the decoding capacity for UAA and UAG codons only in the Tetrahymena domain 1 but not in the universal-code domain 1. This result suggests that other Tetrahymena-specific variations also might be required for the UGA-only release activity.
Disruption of the eRF1 Gene of S. cerevisiae Expressing Variant ΨeRF1 Proteins.
To establish firmly the ability of the KAT→TAS tripeptide variant ΨeRF1s to function in S. cerevisiae, we aimed to knockout the chromosomal copy of the sup45 ts gene in the presence of the TAS variant proteins. MT557/1d transformants expressing Sp-eRF1, ΨeRF1 (KATNIKD), and its heptapeptide variants (TASNIKD and TASNIKS) were transformed by linear DNAs encoding a LEU2-insertion allele (Δsup45∷LEU2) of S. cerevisiae eRF1, and Leu+ cells were selected at 30°C and 37°C. Theoretically, if plasmid-borne variant ΨeRF1s acquired the capacity to decipher the UAG and UAA codons, Leu+ transformants should appear, and such Leu+ transformants should not appear in MT557/1d cells if ΨeRF1 (and its variants) are indeed blind to UAG and UAA codons, as was shown in the above in vitro assay.
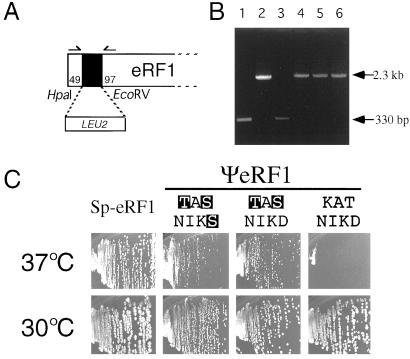
In accordance with the former prediction, many Leu+ colonies appeared in the presence of not only Sp-eRF1 (see Fig. 3C, left column of panels) but also of TASNIKS and TASNIKD variants of ΨeRF1s (see Fig. 3C, middle two columns of panels). To check for disruptions in the chromosomal copy of the eRF1 gene, DNAs were isolated from several Leu+ colonies, and it was examined whether the Δsup45∷LEU2 sequence replaced the sup45 ts allele. From these DNAs, eRF1 sequences were amplified by PCR by using primers coded for the 5′- and 3′-flanking sequences of S. cerevisiae sup45 and subjected to gel electrophoresis (Fig. 3A). Because these primer sequences do not crossreact with Sp-eRF1 and Tt-eRF1 sequences, it is expected that the chromosomal replacement will give rise to a 2.3-kb segment (Fig. 3B, lane 2), whereas the native chromosome will give rise to a 330-bp segment (Fig. 3B, lane 1). As shown in Fig. 3B (lanes 4–6), most of the independent Leu+ colonies thus far examined substituted the Δsup45∷LEU2 sequence for the sup45 ts sequence; this finding strongly points out that the chromosomal eRF1 gene is nullified, and that the plasmid-borne ΨeRF1-TAS variants are sufficient to support the viability. These findings are interpreted as indicating that the KAT→TAS mutant ΨeRF1 has a potential to decipher three stop codons in vivo.
Figure 3.
The capacity of wild-type and variant ΨeRF1 proteins to complement the nullified eRF1 gene of S. cerevisiae. (A) The gene disruption of S. cerevisiae eRF1. The LUE2 marker was inserted into the HpaI-EcoRV sites of the SUP45 sequence cloned in plasmid pET-Sc-eRF1. Numbers refer to the initiator codon of the eRF1 gene. The DNA containing the nullified Δsup45∷LEU2 allele was amplified by PCR using multicloning site primers that flank the sup45 insert and transformed into MT557/1d (sup45 ts) cells in the presence of pYX112 plasmids encoding Sp-eRF1 and wild-type (KATNIKD) or variant (TASKINS and TASNIKD) ΨeRF1s; Leu+ (Ura+) transformants were selected, and those whose chromosomal copy of the eRF1 (sup45 ts) sequence was replaced by the Δsup45∷LEU2 allele were isolated. (B) DNA analyses of the disruption of chromosomal copy of eRF1 in S. cerevisiae transformants. The DNAs containing the insert were amplified from Leu+ transformants obtained in A by PCR using primers 5′-TATTGAGATCTGGAAGGTCAAGAAGTTGG-3′ and 5′-GTTGATAGGTTTGTAAGGTTCGATGTC-3′ shown by arrows in A. These two primer sequences were chosen from S. cerevisiae eRF1 and do not crossreact with Tetrahymena eRF1 or S. pombe eRF1 sequences. Samples used for PCR amplification: lane 1, plasmid DNA encoding the wild-type eRF1 of S. cerevisiae (control); lane 2, plasmid DNA encoding the Δsup45∷LEU2 eRF1 of S. cerevisiae (control); lanes 3–6, Leu+ transformant DNAs selected as in A, in which the chromosomal copy of eRF1 was (lanes 4–6) or was not (lane 3) disrupted by the LEU2 insert. Lanes 4, 5, and 6 represent Leu+ transformants expressing wild-type (KATNIKD) and variant (TASNIKS, TASNIKD) ΨeRF1 proteins, respectively. (C) The growth of the eRF1-nullified S. cerevisiae cells in the presence of plasmids encoding Sp-eRF1 and wild-type or variant ΨeRF1 proteins at 30°C and 37°C.
Potential Capacity of ΨeRF1 for Deciphering Three Stop Codons.
In the course of this study, we encountered unexpected and highly interesting findings. Contrary to the predicted blindness to UAA and UAG of wild-type ΨeRF1, many Leu+ colonies appeared in yeast cells at 30°C, but not at 37°C, upon transformation with plasmid-encoding wild-type ΨeRF1. When the transformants that formed at 30°C were restreaked on the same selective plate at 37°C, no colonies formed (see Fig. 3C, right column of panels). The PCR analysis of these Leu+ colonies revealed that, like the other ΨeRF1 sup45 knockout variant transformants, all colonies thus far tested substituted the Δsup45∷LEU2 sequence for the sup45 ts sequence (see Fig. 3B, lane 6). These observations are interpreted as indicating that the wild-type ΨeRF1 possesses an intrinsic potential to decipher three stop codons at 30°C but not at 37°C, and interaction within the domain 1 between the KAT tripeptide and other sequences modulates the decoding specificity of Tetrahymena eRF1.
In Vitro Specificity of Wild-Type and Mutant ΨeRF1 Proteins.
The in vivo complementation results indicated that ΨeRF1 acquired omnipotent stop-codon decoding capacity by the tripeptide KAT→TAS change at 37°C or by reducing the complementation temperature to 30°C. This temperature is unphysiologically lower than the optimal temperature (around 37°C) for T. thermophila (32). When the wild-type ΨeRF1 protein was overexpressed in E. coli, purified and examined for the protein's capacity, it, however, did not respond to UAA and UAG codons at 25°C in the in vitro release assay (data not shown). Likewise, when the four mutant ΨeRF1 proteins were purified and examined under the same condition, these responded only to UGA in vitro, not to UAA and UAG (Table 1). Therefore, the E. coli-expressed ΨeRF1 proteins, with or without the tripeptide changes, remain specific to UGA at 25°C in vitro. The difference between the in vivo (i.e., omnipotent) and in vitro (i.e., unipotent) activity of eRF1s can be explained by assuming that the eRF1 proteins synthesized in E. coli or the in vitro release conditions do not reproduce the in vivo conditions.
Discussion
In Tetrahymena, how can Gln be incorporated efficiently into the reassigned codons UAG and UAA? This can be explained by assuming that Tt-eRF1 is a UGA-only RF, and does not respond to UAG and UAA. Alternatively, given that Tt-eRF1 can respond to three stop codons, Tetrahymena must employ a mechanism to enable cognate glutaminyl-tRNAs to win efficiently the competition with Tt-eRF1, either by weakening polypeptide termination or by increasing suppression with tRNAs (18). The authentic Tt-eRF1, unlike Euplotes eRF1 (17), is inactive to catalyze polypeptide termination in vitro on mammalian ribosomes (18). Nevertheless, when domain 1 of Sp-eRF1 was replaced by Tetrahymena domain 1, the resulting hybrid, ΨeRF1, became a UGA-specific RF and did not respond to UAG and UAA codons in the heterologous in vitro system. To our knowledge, this is the first report of active domain swapping between eukaryotic RFs from organisms of canonical and variant genetic codes. These findings indicate that of two potential scenarios, the more likely one is that variant-code eRF1s are blind to the reassigned codons, as shown with Euplotes eRF1 in vitro (17), and that domain 1 of Tetrahymena eRF1 determines the selective reading of stop codons.
To identify amino acid determinant(s) that confer the UGA-only decoding ability on Tt-eRF1, we used an in vivo complementation test using temperature-sensitive and knockout eRF1 mutants of S. cerevisiae. We considered that if the test ΨeRF1 variant restored the viability of the eRF1-null strain, the variant eRF1 had acquired the omnipotent capacity of deciphering three stop codons in yeast. The TASNIKS heptapeptide of domain 1 is one of the regions that are highly conserved in universal-code eRF1s but very divergent in ciliate (variant-code) eRF1s (1, 17, 20–22). We found here that the “UGA-only” ΨeRF1 (KATNIKD) acquired the capacity to complement the nullified eRF1 strain at 37°C by substitution of TAS for the KAT tripeptide. This indication is the first of apparent “gain-of-function” in deciphering a specific codon (i.e., reassigned stop codons here) by specific substitutions in eukaryotic RFs. This finding strongly suggests that the variant TAS tripeptide can modulate the decoding capacity. Hence, we refer to the TAS element as a tripeptide modulator for stop codon recognition.
The finding that ΨeRF1 fully complements the eRF1-null strain of S. cerevisiae at 30°C is intriguing. We assume that this capacity is not specific, or artificial, to ΨeRF1 but, rather, reflects the authentic property of Tetrahymena eRF1, and hence of its domain 1, because domain swapping was performed at domain junctions of highly conservative sequences. This result means that Tetrahymena eRF1, like universal-code eRF1s, can potentially decipher three stop codons, and that this omnipotent capacity is modulated by the KAT tripeptide at 37°C and not at 30°C.
Contrary to these in vivo activities of complementing the knockout eRF1 mutant by wild-type (at 30°C) or mutant (at 37°C) ΨeRF1s, the purified ΨeRF1 proteins, with or without the KAT-to-TAS change in the Tetrahymena domain 1, remain specific to UGA and do not respond to UAA and UAG codons in the in vitro release assay (see Table 1). Translation termination measured in vivo and in vitro considerably differs in many essential features. In vitro, the assay includes only the ribosome, eRF1, oligonucleotide as a template and the substrate (fMet-tRNA) and is optimized to reveal the functional capacity of eRF1. In contrast, in an in vivo system, uncountable numbers of other essential components are present. First of all, the class-II termination factor, eRF3, Upf proteins known to interact with termination factors in vivo and in vitro (33), natural mRNA (it is well known that termination in vivo strongly depends on the context of stop codons; ref. 34), numerous tRNAs that compete with eRF1 for the A site binding. Finally, eRF1 in vivo can be posttranslationally modified (phosphorylation, etc.). Therefore, it is not surprising that the decoding potential of eRF1 revealed in in vitro experiments could be modulated in vivo because of the combined action of all these intracellular factors. Moreover, 37°C seems to be closer to an optimal growth condition for T. thermophila (32) than 30°C when the in vitro and in vivo data appeared to be biased. We speculate that at 30°C, changes in affinity constants and/or rate constants of the molecules interacting with eRF1 in vivo cause a nonphysiological response of eRF1 toward stop codons. Also, the possibility cannot be excluded at present that the omnipotence is caused by the KAT-to-TAS change in ΨeRF1 via less accurate, as opposed to actively altered, recognition of codons. In E. coli, it is known that Glu-to-Lys substitutions near the tripeptide anticodon (5) in RF2 induce loss-of-specificity in the decoding capacity and render RF2 to terminate translation not only at cognate stop codons but also at noncognate stop codons, and even at sense codons (35, 36).
The apparent high temperature bias toward the exclusive reading of UGA—i.e., disabled recognition of UAG and UAA—is physiologically consistent with the nature of T. thermophila, the optimum growth temperature of which is around 37°C (32). This result in turn suggests that the TAS or variant KAT tripeptide may not represent a peptide anticodon, but it may influence the functioning of a hypothetical omnipotent peptide anticodon in Tetrahymena eRF1. It is of particular interest whether Euplotes eRF1 can complement the sup45 defect under several physiological conditions. If this were true, we might speculate that unlike codon-specific two-peptide anticodons in bacteria (5), the selective recognition of stop codons by variant-code eRF1s can be achieved by a modulator element that restricts reassigned-codon recognition by a putative omnipotent peptide anticodon of eukaryotic RFs.
Two models have been proposed that stop codons bind the three “cavities” on the domain 1 surface of eRF1 but in the opposite orientation (8, 37). If either of the cavity-binding models were true, this would represent the eukaryotic peptide anticodon, and variant codon specificity would be modulated by interactions between stop codon nucleotides, or the ribosome, and amino acid residues that are adjacent to the cavities (37). In summary, variant genetic code organisms like Tetrahymena have an intrinsic potential to decode three stop codons in vivo, and that interaction within domain 1 between the KAT tripeptide and other sequences, including rRNA, modulates the decoding specificity of Tetrahymena eRF1.
Acknowledgments
This work was supported in part by grants from the Ministry of Education, Sports, Culture, Science and Technology of Japan, the Human Frontier Science Program (awarded in 1997), the Basic Research for Innovation Biosciences Program of the Bio-oriented Technology Research Advancement Institution (BRAIN), the Mitsubishi Foundation, the Organization for Pharmaceutical Safety and Research (OPSR), the Russian Foundation for Basic Research, the Program for Support of Russian Scientific Schools, and European Union INTAS Grant 00-041.
Abbreviation
- RF
release factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nakamura Y, Ito K, Ehrenberg M. Cell. 2000;101:349–352. doi: 10.1016/s0092-8674(00)80845-4. [DOI] [PubMed] [Google Scholar]
- 2.Kisselev L L, Buckingham R H. Trends Biochem Sci. 2000;25:561–567. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura Y, Ito K, Isaksson L A. Cell. 1996;87:147–150. doi: 10.1016/s0092-8674(00)81331-8. [DOI] [PubMed] [Google Scholar]
- 4.Tate W P, Poole E S, Mannering S A. Prog Nucleic Acids Res. 1996;52:293–335. doi: 10.1016/s0079-6603(08)60970-8. [DOI] [PubMed] [Google Scholar]
- 5.Ito K, Uno M, Nakamura Y. Nature (London) 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 6.Song H, Mugnier P, Das A K, Webb H M, Evans D R, Tuite M F, Hemmings B A, Barford D. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 7.Frolova L Y, Tsivkovskii R Y, Sivolobova G F, Oparina N Y, Serpinsky O I, Blinov V M, Tatkov S I, Kisselev L L. RNA (New York, NY) 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertram G, Bell H A, Ritchie D W, Fullerton G, Stansfield I. RNA (New York, NY) 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito K, Ebihara K, Nakamura Y. RNA (New York, NY) 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merkulova T I, Frolova L Y, Lazar M, Camonis J, Kisselev L C. FEBS Lett. 1999;443:41–47. doi: 10.1016/s0014-5793(98)01669-x. [DOI] [PubMed] [Google Scholar]
- 11.Frolova L Y, Merkulova T I, Kisselev L L. RNA (New York, NY) 2000;6:381–390. doi: 10.1017/s135583820099143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer F, Schmidt H J, Plumper E, Hasilik A, Mersmann G, Meyer H E, Engstrom A, Heckmann K. Proc Natl Acad Sci USA. 1991;88:3758–3761. doi: 10.1073/pnas.88.9.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horowitz S, Gorovsky M A. Proc Natl Acad Sci USA. 1985;82:2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cupples C G, Pearlman R E. Proc Natl Acad Sci USA. 1986;83:5160–5164. doi: 10.1073/pnas.83.14.5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuchino Y, Hanyu N, Tashiro F, Nishimura S. Proc Natl Acad Sci USA. 1985;82:4758–4762. doi: 10.1073/pnas.82.14.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanyu N, Kuchino Y, Nishimura S. EMBO J. 1986;5:1307–1311. doi: 10.1002/j.1460-2075.1986.tb04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kervestin S, Frolova L, Kisselev L, Jean-Jean O. EMBO Reports. 2001;2:680–684. doi: 10.1093/embo-reports/kve156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karamyshev A, Ito K, Nakamura Y. FEBS Lett. 1999;457:483–488. doi: 10.1016/s0014-5793(99)01089-3. [DOI] [PubMed] [Google Scholar]
- 19.Liang A, Brunen-Nieweler C, Muramatsu T, Kuchino Y, Beier H, Heckmann K. Gene. 2001;262:161–168. doi: 10.1016/s0378-1119(00)00538-2. [DOI] [PubMed] [Google Scholar]
- 20.Inagaki Y, Doolittle W F. Nucl Acids Res. 2001;29:921–927. doi: 10.1093/nar/29.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone C A, Knight R D, Landweber L F. Curr Biol. 2001;11:65–74. doi: 10.1016/s0960-9822(01)00028-8. [DOI] [PubMed] [Google Scholar]
- 22.Knight R D, Landweber L F. Cell. 2000;101:569–572. doi: 10.1016/s0092-8674(00)80866-1. [DOI] [PubMed] [Google Scholar]
- 23.Muramatsu T, Heckmann K, Kitanaka C, Kuchino Y. FEBS Lett. 2001;488:105–109. doi: 10.1016/s0014-5793(00)02391-7. [DOI] [PubMed] [Google Scholar]
- 24.Frolova L, Seit-Nebi A, Kisselev L. RNA (New York, NY) 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuite M, Akhmaloka, Firoozan M, Duarte J A B, Grant C M. In: Post-Transcriptional Control of Gene Expression. McCarthy J, Tuite M, editors. Berlin: Springer; 1991. pp. 611–622. [Google Scholar]
- 26.Stansfield I, Jones K M, Kushnirov V V, Dagkesamanskay A R, Poznyakovski A I, Paushkin S V, Nierras C R, Cox B S, Ter-Avanesyan M D, Tuite M F. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman F. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Karamyshev A, Karamycheva Z, Ito K, Matsufuji S, Nakamura Y. Biochemistry (Moscow) 1999;64:1648–1658. [PubMed] [Google Scholar]
- 30.Cox R A, Hirst W. Biochem J. 1976;160:505–519. doi: 10.1042/bj1600505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frolova L, Le Goff X, Rasmussen H H, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni A-L, Celis J E, Philippe M, et al. Nature (London) 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 32.Asai D J, Forney J D. Tetrahymena thermophila. London: Academic; 2000. [Google Scholar]
- 33.Czaplinski K, Majlesi N, Banerjee T, Peltz S W. RNA (New York, NY) 2000;6:730–743. doi: 10.1017/s1355838200992392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tate W P, Mannering S A. Mol Microbiol. 1996;21:213–219. doi: 10.1046/j.1365-2958.1996.6391352.x. [DOI] [PubMed] [Google Scholar]
- 35.Ito K, Uno M, Nakamura Y. Proc Natl Acad Sci USA. 1998;95:8165–8169. doi: 10.1073/pnas.95.14.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uno M, Ito K, Nakamura Y. Proc Natl Acad Sci USA. 2002;99:1819–1824. doi: 10.1073/pnas.032457599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inagaki Y, Blouin C, Doolittle W F, Roger A J. Nucleic Acids Res. 2002;30:532–544. doi: 10.1093/nar/30.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]